Abstract
Our limited ability to improve the survival of patients with heart failure is due, in part, to the inability of the mammalian heart to meaningfully regenerate itself. The recent identification of distinct families of multipotent cardiovascular progenitor cells from endogenous as well as exogenous sources, such as embryonic and induced pluripotent stem cells, has raised much hope that therapeutic manipulation of these cells may lead to regression of many forms of cardiovascular disease. While the exact source and cell type remains to be clarified, our greater understanding of the scientific underpinning behind developmental cardiovascular progenitor cell biology has helped to clarify the origin and properties of diverse cells with putative cardiogenic potential. In this review, we highlight recent advances in the understanding of cardiovascular progenitor cell biology from embryogenesis to adulthood and their implications for therapeutic cardiac regeneration. We believe that a detailed understanding of cardiogenesis will inform future applications of cardiovascular progenitor cells in heart failure therapy and regenerative medicine.
Keywords: cardiovascular disease, stem cells, regeneration, cardiac development, cardiomyocytes
Introduction
“Education consists mainly of what we have unlearned.”
–Mark Twain (1835-1910)
The view of the mammalian heart as a post mitotic organ, incapable of generating new heart muscle, has dominated cardiovascular science and medicine for over a century. To those who care for patients with cardiovascular diseases, their observations would give no reason to challenge this view. For instance, following the proximal occlusion of a major coronary artery, a billion or more cardiomyocytes are typically lost and replaced by noncontractile scar tissue.1 The injured heart attempts, ineffectively, to compensate for the loss of functioning myocardium, resulting in a downward spiral of adverse cardiac remodeling, neurohormonal activation, and ultimately, congestive heart failure.2 However, as Mark Twain aptly alluded to, sometimes even our most basic assertions are found to be imprecise, askew, or just plainly wrong. Several practices in the recent history of cardiovascular medicine serve as prime examples: avoidance of beta blockers in heart failure, use of antiarrhythmics for suppression of ventricular ectopy following myocardial infarction, and estrogen replacement therapy for cardiovascular disease prevention are among the doctrines that have been “unlearned” in recent years.
Likewise, rapid advances in stem cell and regenerative biology have prompted the scientific community to reconsider the assumption that the infarcted myocardium is incapable of self-repair. In this regard, the concept that new myocytes might be generated in the diseased myocardium by administering exogenous stem cells elicited such overwhelming excitement by the medical community that treatment of cardiac patients with autologous stem cells were expedited into clinical trials.3 While the exact benefit of such treatment remains to be defined, it is clear that an effective, safe, and durable therapy for cardiomyocyte replacement will require a detailed understanding of the fundamental biology of cardiac progenitor cells. In this review, we examine the current knowledge of adult mammalian heart regeneration, provide an overview of cardiovascular progenitor cell biology, and highlight important questions that remain unanswered. A comprehensive understanding of developmental cardiovascular progenitor cells will help to inform our future regenerative strategies regardless of the source or type of cell employed.
Mammalian Cardiac Regeneration
The animal kingdom is abundant with examples of spontaneous organ regeneration following injury.4 With respect to cardiac regeneration, the zebrafish has captured the most attention from the scientific community due to its remarkable capacity to replace substantial portions of its heart following ventricular amputation.5 By contrast, the regenerative capacity of the mammalian heart is inadequate to compensate for the loss of cardiomyocytes following myocardial infarction. While many new vascular and connective tissue cells continue to populate the heart during postnatal development and into adult life, cardiomyocyte proliferation slows dramatically after birth.6 Following injury, cardiomyocytes bordering the infarct zone rarely divide, although transgenic manipulation of specific genes and a small number of extrinsic factors have been shown to increase cardiomyocyte division in mice.7-9 For this and other reasons, the notion that mammalian cardiac tissue may harbor regenerative potential has been vigorously debated.
To explore this question in humans, Beltrami and colleagues examined myocardial cells from the hearts of 13 patients who died shortly after a myocardial infarction.10 They found co-localization of metaphase chromosomes or Ki-67 labeling (a nuclear protein associated with cell proliferation) in cells that stained positively with cardiac sarcomeric protein and concluded that cardiomyocyte division had occurred in a human post-injured heart. In their samples, Ki-67 was expressed in 4% of myocytes in the zone that bordered the infarct, and in 1% of myocytes in regions distant from the scar. Compared to an age-matched control group, Ki-67 expression in infarcted hearts was 84 and 28 times higher in these regions, respectively, suggesting that significant myocyte proliferation may occur during normal aging and becomes further amplified in the context of a myocardial infarction. Based on these findings, the authors estimated that the entire population of human myocytes may turnover every 4.5 years.10,11
The results from Beltrami and colleagues, however, need to be examined in the context of normal postnatal cardiomyocyte maturation. Unlike many other differentiated cell types, cardiomyocytes often undergo a final round of nuclear division without cytokinesis during the first decade of life, resulting in ~25% of human cardiomyocytes becoming binucleated.12 Cardiomyocytes also retain the ability to undergo DNA synthesis without nuclear division (i.e. endoreduplication). As a result, many cardiomyocyte nuclei are polyploid, containing twice or even higher multiples of the normal content of DNA.13 These confounding issues regarding cardiomyocyte DNA synthesis and nuclear mitosis raise concern that the findings observed by Beltrami and colleagues may reflect, in part, the occurrence of endoreduplication or acytokinesis in the injured heart.14,15
These issues were addressed recently by Bergmann and colleagues using a novel method of radiocarbon dating of DNA.16 As a result of aboveground nuclear testing during the Cold War, atmospheric levels of carbon-14 (14C) rose dramatically, ultimately becoming incorporated into the food chain and eventually into the DNA of all plant and animal cells. Following the signing of the Limited Nuclear Test Ban Treaty in 1963, levels of atmospheric 14C dropped precipitously, back to pre-test levels, but residual 14C from the initial testing still persisted in the DNA of cells “born” during that era. By exploiting this unusual period in human history, these investigators have effectively determined the lifespan of a variety of cell types within the human body,16-20 and in the process, provide the most definitive evidence to date that human cardiomyocytes are renewed during postnatal life.16
Applying the 14C approach to examine human heart specimens, they showed that the chronological age of their subjects was slightly older than the mean age of their cardiomyocyte DNA. In patients born before the Cold War, the concentration of 14C in their cardiomyocyte DNA exceeded the atmospheric concentration of 14C at the time of their birth, whereas patients born during or after the nuclear testing had a lower concentration of 14C, firmly establishing that human cardiomyocytes synthesize new DNA after birth. By meticulously sorting cardiomyocyte nuclei and excluding cells with more than a single set of chromosomes, the investigators were able to account for the effects of cardiomyocyte binucleation and polyploidy. Taking these factors into consideration, they concluded that cardiomyocyte renewal does occur in humans,16 albeit much more slowly than what was suggested by Beltrami and colleagues.10 Mathematical modeling suggested that cardiomyocyte renewal is an age-dependent process, with rates of ~1% turnover per year at age 20, declining to ~0.4% of total cardiomyocytes per year by the age of 75. Over an average human lifespan, this translates to about half of an individual's cardiomyocytes being replaced.
If cardiomyocyte renewal does occur in mammals, whether these cells originate from a stem/progenitor cell pool or are derived from already differentiated cardiomyocytes remains an open question. To investigate whether resident cardiac stem cells or precursor cells can participate in regeneration, Hsieh and colleagues studied cardiomyocyte turnover in mice using a tamoxifen driven Cre-lox system that induced expression of an enhanced green fluorescent protein (eGFP) in differentiated cardiomyocytes only.21 An early “pulse” of tamoxifen induced labeling of cardiomyocytes, detectable by eGFP fluorescence. This was followed by a “chase” period (e.g. aging over one year, myocardial infarction, and pressure overload) during which unlabeled progenitor cells could differentiate into cardiomyocytes. If new cardiomyocytes are formed from progenitors, there should be a decrease in the percentage of eGFP-expressing cardiomyocytes over time as new unlabeled cardiomyocytes replace them. When the mice were analyzed one year after tamoxifen treatment but without injury, the investigators found no detectable replacement of cardiomyocytes by progenitor cells during normal aging. The authors then looked for repair by stem/progenitor cells after cardiac stress, which was induced either through myocardial infarction or through pressure overload by aortic banding. After either form of cardiac stress, the percentage of eGFP+ cardiomyocytes decreased, suggesting that damaged cells were replaced with new cardiomyocytes that had differentiated from unrecombined progenitors. It was estimated that ~15% of the cardiomyocytes in the peri-infarct areas and ~5% of the cardiomyocytes in infarct-remote areas were derived from unrecombined progenitors. In hearts that were challenged with pressure-overloading, ~5% of cardiomyocytes were found to arise from unlabeled progenitors. These results suggest that progenitor cells are recruited to become cardiomyocytes after injury, but not during normal aging. In addition, the authors were able to detect small numbers of cells in the infarcted hearts staining positive for stem cell markers such as c-Kit and Nkx2.5, further adding plausibility to the notion that the new cardiomyocytes may have been formed from a cardiac stem/progenitor cell pool. Although the regenerative potential of the mammalian heart appears to be quite limited, and clearly is not sufficient to cope with the widespread loss of cardiomyocytes seen in a myocardial infarction, these studies point toward an unexpected source of cells in the adult heart with cardiogenic potential.
Cardiac Progenitors in the Rodent Adult Heart
The persistence of cardiac progenitors in the adult heart would provide an avenue to direct regeneration of cardiomyocytes that are lost due to cardiac injury and an opportunity to bypass the need for cell transplantation, one of the major challenges in regenerative medicine. These potential advantages led investigators to search for novel heart muscle forming cells resident in the postnatal and adult myocardium (Table 1).
Table 1.
Characteristics of Resident Populations of Cardiac Progenitors and Stem Cells
| Cell Type (References) | Frequency | Self-Renewal | Clonogenic | Multi-Potent | Markers | Differentiation Protocol | Functional Characterization | Effect After Transplantation |
|---|---|---|---|---|---|---|---|---|
| Lin- c-Kit+ Stem Cells (22, 23, 45) | ~1 per 1 × 104 myocytes | Yes | Yes | Yes | + c-Kit ± Nkx2.5, Gata4, Mef2 ± Sarcomeric proteins –Lin, CD20, CD34, CD45 –SM Markers |
Differentiation medium (6 days) plus intracardiac injection | + Endothelial proteins + SM proteins + Sarcomeric proteins + Action potentials –Organized sarcomeres –Striations –Spontaneous contraction |
Bands of regenerating myocardium and partial functional improvement following myocardial infarction |
| Sca-1+ Stem Cells (27-29, 53-55) | ~3 per 1 × 103 myocytes | Unknown | Unknown | Unknown | + Sca-1, Gata4, Mef2c, CD31 –c-Kit, Lin, Nkx2.5, CD34, CD45 –Sarcomeric proteins |
5-azacytidine or oxytocin (4 weeks) | + Nkx2.5 ± Sarcomeric proteins ± Organized sarcomeres ± Striations ± Ca2+ transients ± Spontaneous contraction |
Engraftment of Sca-1+ cells in the infarcted myocardium |
| Cardiac Side Population Cells (32-35) | ~1 per 3 × 104 cardiac cells | Unknown | Unknown | Unknown | + Abcg2/Mdr1, Sca-1, Mef2a/c ± c-Kit, Nkx2.5, Gata4 CD34, CD45 –CD31 –Sarcomeric proteins |
Co-culture with adult cardiomyocytes (2-3 weeks) | + Sarcomeric proteins + Organized sarcomeres + Ca2+ transients + E-M coupling + Spontaneous contraction |
Unknown |
| Cardiospheres (46-50) | Unknown | Yes | Yes | Yes | + c-Kit, Sca-1, Flk1, vWF, CD31, CD34, CD90, CD105 + Sarcomeric proteins – Mdr1, CD45, CD133 |
Intracardiac injection | + Endothelial proteins + SM proteins + Sarcomeric proteins + Organized Sarcomeres + Ca2+ transients* + Action potentials* + Spontaneous contraction* |
Bands of regenerating myocardium and partial functional improvement following myocardial infarction |
| Isl1+ Progenitors (92-93) | 500-600 per rat heart | Yes | Yes† | Yes† | + Isl1, Nkx2.5, Gata4 –c-Kit, Sca-1, CD31, Tbx5 –Sarcomeric proteins –SM markers |
Co-culture with neonatal cardiomyocytes (3-5 days) | + Sarcomeric proteins + Organized sarcomeres + Ca2+ transients + Action potentials + E-M coupling + Spontaneous contraction |
Unknown |
This table summarizes the results of several studies that examined the phenotypes and functional traits of putative endogenous cardiac stem and progenitor cell populations.
these features were observed in human cardiospheres upon co-culture with adult rat cardiomyocytes.
clonality and multipotency determined for embryonic Isl1+ progenitors.
Abbreviations: E-M, electro-mechanical; SM, smooth muscle; vWF, von Willebrand factor.
To identify a progenitor cell population in the adult heart, a number of investigators have utilized a combination of cell surface markers traditionally used to identify stem cell populations in other tissues. Beltrami and colleagues reported isolating clonogenic, self-renewing cells that are capable of differentiating into cardiomyocytes, vascular smooth muscle cells, and endothelial cells.22 These cells are negative for many blood lineage markers (Lin-), and positive for c-Kit, the receptor for stem cell factor. In the adult rat myocardium, c-Kit+ cells are rare (1 per 10,000 myocytes), and heterogeneous, with a minority (7%-10%) expressing early cardiac transcription factors such as GATA4, Mef2, and Nkx2.5. In vitro, c-Kit+ clones differentiate into cells that biochemically resemble mature cardiomyocytes, although functionally, organized sarcomeres and spontaneous contractile activity are not observed.
Although results from transplantation studies using adult c-Kit+ cells in animal models have been mixed,23 Beltrami and colleagues observed a band of regenerating myocardium and a contribution of labeled cells to blood vessels when c-Kit+ cells were injected into the infarct border zone of hearts from syngeneic rats after myocardial infarction.22 The tagged cells expressing sarcomeric proteins were small relative to cardiomyocytes, but they exhibited visible striations and expressed N-cadherin and connexin 43, components of intercalated discs. What is the developmental origin of these putative cardiac progenitor cells? They may represent a developmental remnant from a multipotent mesodermal cell population that has persisted in the heart throughout embryonic and postnatal development or, alternatively, itinerant bone marrow-derived cells that are mobilized in response to myocardial injury.24 In support of the latter hypothesis, transplantation of GFP-labeled, c-Kit+ bone marrow-derived mononuclear cells into sublethally irradiated wild-type adult mice revealed that c-Kit+ cells in the adult injured heart are derived mostly from the transplanted marrow cells.25,26
Concomitant with the Beltrami study, Oh and colleagues isolated another resident population of putative adult cardiac progenitor cells characterized by the expression of stem cell antigen 1 (Sca-1).27,28 Sca-1+ cells express several early cardiac transcription factors including GATA4 and Mef2, as well as telomerase reverse transcriptase, which has been associated with the potential for self-renewal. However, they do not express Nkx2.5 or genes encoding cardiac sarcomeric proteins. Although these cells do not spontaneously differentiate in vitro into cardiomyocytes, treatment with the genome-wide demethylating agent, 5-azacytidine,27,29 or oxytocin,29 for 4 weeks generated a small subpopulation of cells (<5%) that expressed the cardiac transcription factor Nkx2.5 and cardiac contractile proteins. Phenotypically, sarcomeric organization was seen and the cells began to beat spontaneously. A chronotropic response to a pharmacological agent (i.e. isoproterenol) was observed as well.29 It remains unclear whether the cells that adopt some characteristics of cardiac differentiation represent a homogenous or, more likely, heterogeneous population of cells.
When administered intravenously to mice following ischemia-reperfusion injury, cardiac Sca-1+ cells home to the heart and can be identified in the infarct border zone two weeks after injury.27 These cells express contractile proteins and connexin 43, suggesting that they undergo differentiation into cardiomyocytes. However, evidence of fusion between Sca-1+ cells and host cardiomyocytes has been observed in up to 50% of the cells,27,28 a finding that has not been widely observed with c-Kit+ stem cells.23,30 Additional studies are needed to evaluate the clonogenic potential and capacity for self-renewal of Sca1+ cardiac cells, and to determine whether a subpopulation exists with restricted developmental potential to differentiate into cardiac progenitors or cardiomyocytes.
Finally, a population of cells with stem cell-like properties has been identified in bone marrow, muscle, and skin by their ability to exclude Hoechst dye and certain anticancer drugs, resulting in a characteristic appearance on density dot plots generated during fluorescence-activated cell sorting (FACS) that led to the name “side population” or SP to describe this pool of cells.31 Multiple groups have identified a subset of Sca-1+, SP cells in adult mouse hearts marked by the expression of Abcg2 and Mdr1, two genes belonging to the ATP-binding cassette (ABC) transporter superfamily that constitute the molecular basis for the dye efflux.32-35 While their clonogenic potential, capacity for self-renewal, and developmental origin remain to be determined, upon coculture with adult rat ventricular cardiomyocytes, these cells demonstrate not only biochemical differentiation, as evidenced by the expression of cardiac transcription factors and contractile proteins, but also functional cardiomyogenic differentiation, as determined by sarcomeric organization, intracellular calcium transients, and cellular contraction.34
Depending on the study examined, these three populations of adult cardiac progenitor cells (c-Kit+, Sca-1+, SP) represent 0.005-2% of the total cellular content in the heart, enter the cell cycle when growth of the heart is attenuated, proliferate in culture, and form cells expressing cardiomyogenic markers.11 They appear phenotypically distinct from one another and show differential expression of surface markers.36-38 In some instances, long-term culture of these cells is required to generate sufficient cell numbers for experimentation raising concern that phenotypic drift could arise as an artifact of in vitro cell culture. Furthermore, differences in the experimental approaches and readouts employed in these various studies have contributed to discrepancies in defining the relative cardiomyogenic potential of resident cardiac progenitor cells. To date, the exact lineage relationships between these adult cardiac progenitor cell populations and embryonic cardiac progenitor cells remain unknown. Despite these questions, injection of adult cardiac stem cells directly into infarcted mouse or rat myocardium has been reported to provide short-term improvement in heart function. It is possible that the reported functional improvement is due to a prosurvival paracrine effect or enhanced angiogenesis.39,40 Indeed, clinical trials using bone marrow-derived stem cell infusions into patients with myocardial infarction have suggested that paracrine factors may be responsible for the transient improvement in cardiac function in humans.41
Cardiac Progenitors in the Human Adult Heart
The presence of endogenous cardiac progenitors in the rodent adult heart has prompted studies into whether similar populations exist in the human adult heart. As was demonstrated in rodents, a heterogeneous population of cardiac cells defined by the expression of the primitive stem cell markers c-Kit, MDR1, or a Sca-1-like epitope could also be found in human cardiac specimens from patients with aortic stenosis,42 myocardial infarction,43 and in the post-mortem hearts of patients who had undergone cardiac transplantation.44 These cells lacked early indicators of bone marrow cell differentiation and did not express markers of differentiated cardiomyocytes, smooth muscle, fibroblasts, or endothelial cells. When isolated by FACS, the human cardiac c-Kit+ subset was reported to give rise to cardiomyocytes, vascular smooth muscle cells, and endothelial cells in vitro, and following transplantation into immunodefficient mice.45
Concurrently, Messina and colleagues described the isolation of a heterogeneous population of cells from human atrial and ventricular surgical biopsy specimens that form clonal, multicellular spherical clusters in suspension culture termed “cardiospheres” (CSps).46 Cardiospheres contain cells positive for c-Kit, Sca-1, and the kinase insert domain receptor (KDR), are capable of long-term self-renewal, and appear to give rise to both endothelial and smooth muscle cell types.46,47 When cocultured with neonatal rat cardiomyocytes, they form beating clusters. Cardiosphere-derived cells have more recently been isolated with improved efficiency from endomyocardial right ventricular biopsies from adult patients, approaching cell numbers that would be required for transplantation.48 Thus far, transplantation studies in post-infarct pigs,49 and SCID mice,50 show short term improvements in cardiac function, likely mediated through paracrine effects of the injected cells,50 as few surviving cells can be identified beyond three weeks of transplantation.50,51
The lack of known cell surface markers specific to cardiac progenitors has raised questions regarding the cardiac specificity of the currently described adult cardiac progenitor cells. Pouly and colleagues reported that at least some of the c-Kit+ cells found in human adult heart specimens may in fact be mast cells, based on immunohistochemical staining of endomyocardial biopsy and right atrial appendage samples.52 Colocalization experiments demonstrated that all the c-Kit+ cells they isolated from heart biopsies stained positive for leukocyte common antigen (CD45), suggesting hematopoietic origin. Their perivascular location and expression of c-Kit suggested that these cells could be mast cells. This phenotype was subsequently confirmed by positive staining for tryptase, an enzyme specific to mast cells.
Additionally, multiple groups have identified cells with the capacity to proliferate and form cardiomyocytes in adherent cell culture on the basis of their ability to bind an anti-mouse Sca-1 antibody.27-29,53-55 Sca-1, however, is not a known determinant on human cells, a detail that has sparked debate as to their legitimacy as cardiac progenitors. To date, there is no consensus on the best marker for identification of adult cardiac stem cells and the molecular mechanisms promoting their self-renewal and differentiation into the various lineages of the heart are largely unknown. Ultimately, in vivo lineage tracing studies in animal models will be required to validate any putative cardiac stem/progenitor cell population both during normal aging and in the setting of cardiac injury.
Origins of Cardiac Progenitors in the Developing Heart
Recent advances in conditional gene targeting techniques and the availability of tissue specific enhancers and promoters have allowed us to track the stepwise commitment of cardiac progenitor cells and their intermediates before their terminal differentiation. Such in vivo genetic fate mapping has increasingly been used to identify novel stem cells and related progenitors, and has contributed significantly to our understanding of cell lineage diversification within the heart.56
Until recently, the formation of cardiac, smooth muscle and endothelial cell lineages in the heart had largely been ascribed to distinct populations of embryonic precursors.57 Early lineage-tracing studies in avian model systems suggested that a common muscle cell precursor exists for both the working myocardium and the conduction system.58,59 Likewise, another common progenitor, the hemangioblast, was found to give rise to endothelial and blood cell lineages, a finding which indicated that a common precursor might also exist for endothelial cells in the heart.60-62 Recently, a growing body of evidence from multiple independent laboratories instead suggests the existence of a common multipotent precursor for the diverse muscle and non-muscle lineages of the heart.63-65 This clonal model of heart lineage diversification implies a stem cell paradigm for the generation of the diverse cardiovascular cell types, similar to that of hematopoiesis where a single stem cell is able to reconstitute the entire hematopoietic system through extensive self-renewal and subsequent multilineage differentiation.66,67
It is generally accepted that all vertebrates follow a similar developmental paradigm in which pluripotent cells undergo germ layer commitment to become further differentiated into organ fields as development proceeds. The earliest precursors for heart-forming cells form in the vertebrate mesoderm68 and, upon entering the precardiac mesoderm stage of development, transition from expressing the T-box transcription factor brachyury T (Bry) to expressing mesoderm posterior 1 (Mesp1).69,70 As these early cardiac mesodermal cells contribute to the developing heart, their transcriptional program determines the lineage specification that will follow. Mesp1+ cells encompass all cardiac progenitor cells, but have not yet committed to the cardiac fate, as some also give rise to derivatives of the paraxial mesoderm and skeletal muscle of the head and neck.71 During their migration, Mesp1+ cardiac precursor cells expand rapidly, ultimately segregating into two spatially and temporally distinct cardiogenic heart fields exhibiting unique time courses of differentiation and distinct regional contributions to the embryonic heart. It is at this stage that heart precursor cells commit irreversibly to the cardiac lineage and become cardiac progenitor cells expressing key cardiac transcription factors such as GATA4, Nkx2.5, and Islet-1 (Isl1).
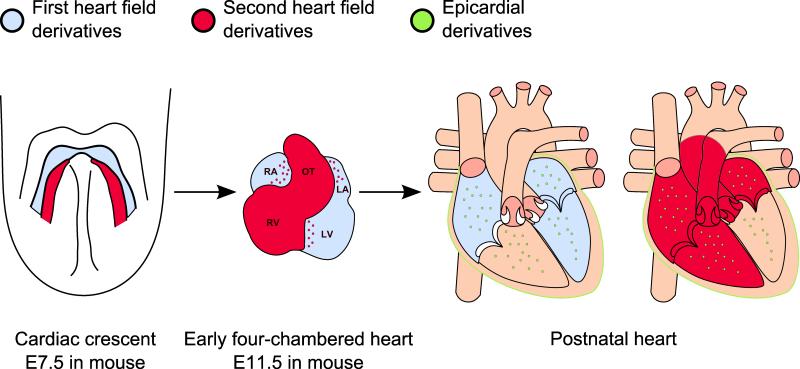
The first population of cells to migrate to the heart-forming region, referred to as the first, or primary, heart field originates bilaterally in the anterior splanchnic mesoderm and gives rise to a group of cardiovascular precursors that form the cardiac crescent (Figure 1). The cardiac crescent goes on to fuse in the midline, forming the linear heart tube, and ultimately gives rise to the majority of the left ventricle, the atrioventricular canal, and parts of the atria.72 The second, or anterior, heart field appears in the extra-crescent tissue and is largely derived from a population of cells located dorsal and anterior to the heart tube in the pharyngeal and splanchnic mesoderm. As the linear heart tube elongates and undergoes rightward looping, cardiovascular progenitors from the second heart field migrate into the heart tube, contributing cells that will form the main parts of the atria, the right ventricle, and the outflow tract myocardium.73-76
Figure 1. Embryologic contributions to mammalian heart development.
The heart primordium is first recognizable as the cardiac crescent (left panel), a structure derived from first heart field (FHF, blue) cardiogenic precursors. The cells of the cardiac crescent join in the midline to form the linear heart tube which undergoes rightward looping to form the primitive chambers of the mammalian heart (middle panel). By this time, precursor cells that form in the second heart field (SHF, red) have migrated into the rostral and caudal portions of the developing heart. In the postnatal heart (right panel), progenitors from the FHF contribute primarily to the atria (LA, RA) and the left ventricle (LV). SHF derivatives contribute mainly to the atria, outflow tract (OT), and right ventricle (RV). Epicardial progenitors (green) also contribute to a minor portion of cardiomyocytes in all four heart chambers.
The heart subsequently receives important contributions from two additional sources, the cardiac neural crest and the proepicardium. Cardiac neural crest cells contribute to normal development of the outflow tract and great vessels, as well as to essential components of the cardiac autonomic nervous system.77,78 Proepicardial cells migrate onto the surface of the heart, giving rise to the epithelial sheet of cells known as the epicardium. Some epicardial cells undergo an epithelial-to-mesenchymal transformation and migrate into the subjacent myocardium, contributing to the development of coronary smooth muscle and to the surrounding cardiac fibroblasts of the interstitium and adventitia.79-83 Recently, the epicardium has also been shown to be a source of cardiomyocytes.84,85 In addition to contributing to the cellular makeup of the heart, the epicardium and myocardium mutually engage in both paracrine and direct cellular interactions that are required for the growth and development of each compartment.86-89
Heart Field Progenitors in Cardiac Development
Lineage tracing experiments in mice have demonstrated that most of the early second heart field myocardial, smooth muscle, and endothelial cells can be traced to multipotent heart progenitors that express the LIM-homeodomain transcription factor Isl1 (Figure 2).64,76,90 Isl1 is transiently expressed in cardiac mesoderm, and while it is not strictly cardiac-specific, its expression has been used to identify cardiac progenitor cells because its downregulation coincides with the expression of terminal cardiac differentiation markers.76 Using genetically labeled embryonic stem (ES) cells for the Isl1 lineage, Moretti and colleagues showed that ES cell-derived Isl1+ progenitors can differentiate into the above three cardiac lineages after a brief period of in vitro expansion on a mesenchymal feeder layer.64 Evidence supporting the possibility that human ISL-1+ cardiac progenitors are multipotent has also been provided by their isolation from human ES cells followed by in vitro differentiation.91 Additional studies in the mouse embryo have confirmed that Isl1+ cardiovascular progenitors contribute to all cells in the right ventricle, to portions of the atria, ventricular septum and conduction system, and to a minor proportion of the left ventricular free wall which is derived largely from first heart field progenitors.64,76,92 Notably, a subset of Isl1+ undifferentiated progenitors remains embedded in the embryonic heart after its formation and a few cells are still detectable shortly after birth in the compartments that arise from Isl1+ second lineage precursors during cardiac development.92,93 Unlike the other putative resident cardiac progenitor populations discussed previously, these cells do not express c-Kit or Sca-1, although they do express Nkx2.5 upon differentiation. Their ability to self-renew in vitro on a cardiac mesenchymal feeder layer and to be stimulated to differentiate into fully mature functional cardiomyocytes, with electrophysiologic characteristics of fully differentiated cardiomyocytes including responsiveness to β-adrenergic agonists, indicates that these cells may represent remnant cardiac progenitors from their embryonic Isl1+ precursors.
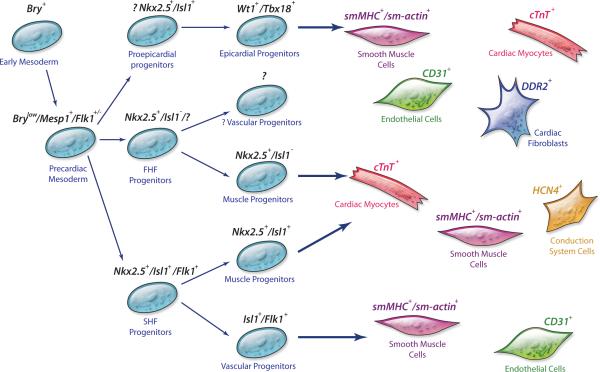
Figure 2. Proposed cellular hierarchy of cardiac progenitor cells and their lineage diversification.
Precursors for heart-forming cells in the vertebrate mesoderm transition from expressing brachyury T to Mesp1 when they enter the precardiac mesoderm stage of development. As these early cardiac mesodermal cells contribute to the developing heart, their transcriptional program determines their further lineage specification. Within the second heart field, Isl1, together with Nkx2.5 and Flk1, defines multipotent Isl1+ cardiovascular progenitor cells that can give rise to myocardial, conduction system, smooth muscle, and endothelial lineages. A subset of precursors derived from Isl1+ progenitors may function as more restricted bipotent progenitors, displaying myocardial and smooth muscle potential or endothelial and smooth muscle potential. The developmental potential of the first heart field progenitors is largely uncharacterized. Epicardial progenitor cells are marked by Wt1 and/or Tbx18. These cells have been shown to give rise to cardiomyocytes, smooth muscle, endothelial cells, and fibroblasts in the heart. Abbreviations: Bry, brachyury T; CD31 (PECAM 1), platelet/endothelial cell adhesion molecule; cTnT, cardiac troponin T; DDR2, discoidin domain receptor 2; FHF, first heart field; Flk1, fetal liver kinase 1 (vascular endothelial growth factor receptor 2); HCN4, potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4; Isl1, Islet-1 transcription factor, LIM/homeodomain; Mesp1, mesoderm posterior 1; Nkx 2.5, NK2 transcription factor related, locus 5; SHF, second heart field; sm-actin, smooth muscle actin; smMHC, smooth muscle myosin heavy chain; Tbx18, T-box transcription factor 18; Wt1, Wilms’ tumor protein. (Illustration Credit: Cosmocyte/Cameron Slayden)
Concurrent with these Isl1 studies, we reported the isolation of a bipotent cardiac progenitor cell population from mouse embryos and differentiated ES cells using a cardiac-specific enhancer element of the homeobox transcription factor gene Nkx2.5, a marker expressed in both the first and second heart fields.63 Nkx2.5+ cells obtained from ES cultures display high proliferative capacity and express modest levels of the stem cell markers c-Kit and Sca-1, but do not express endothelial markers, suggesting that the endothelial and myogenic lineages are largely segregated by the time Nkx2.5 is expressed. However, other lineage tracing studies in mice have shown that some, if not the majority, of the endocardial cells are descendants of embryonic Nkx2.5 progenitor cells.94 Furthermore, genome-wide transcriptional analysis demonstrated that embryo-derived Nkx2.5 progenitor cells express vascular-endothelial markers, a finding that further supports residual capacity for endothelial differentiation in Nkx2.5+ cells.95 Whether the Nkx2.5+ progenitors,63 and the previously mentioned Isl1+ progenitors,64 represent completely distinct populations or descendants from a common progenitor remains to be fully determined. Further lineage tracing experiments using cells doubly marked for Isl1 and Nkx2.5 will help to rigorously define the relationship between these populations.
To date, it has not been possible to isolate and characterize the developmental potential of purified populations of primary heart field progenitors because of an absence of molecular markers unique to that field. The identification of specific sets of markers for the first and second heart field lineages would be extremely valuable, as it is unclear whether cardiac regeneration of the left ventricle, a first heart field-derived structure, will require purified populations of first heart field progenitors. Furthermore, while it has been postulated that a common primordial cardiovascular progenitor that gives rise to progenitors for both the first and second heart fields exists, the identity of such common progenitor remains elusive.
Evidence for the existence of such a common progenitor comes from studies using retrospective clonal analysis,96,97 as well as genetic fate mapping experiments.65 Studying the in vitro differentiation of mouse ES cells, Kattman and colleagues labeled cells positive for Bry as well as fetal liver kinase 1 (Flk1), the cell surface receptor that encodes the vascular endothelial growth factor receptor 2 (VEGFR2), to identify a heterogeneous population of cells that exhibited cardiomyogenic potential.65 Upon ES cell differentiation, two distinct waves of Bry+Flk1+ cells were identified within the developing embryoid body. The first wave of Bry+Flk1+ cells contributed to the hemangioblast, a population of mesodermally derived cells that contribute precursors to the hematopoietic and vascular compartments.98 The second wave of Bry+Flk1+ cells contained clones with cardiomyocyte, vascular smooth muscle, and endothelial potential. Some colonies became positive for the second heart field marker Isl1, while others were negative for Isl1, but expressed Tbx5, a T-box transcription factor associated with derivatives of the first heart field.99 Similar findings have since been replicated using human ES cells in a parallel set of experiments, where a population of progenitor cells positive for KDR, the human ortholog of Flk1, have been shown to display cardiac, endothelial, and vascular smooth muscle potential in vitro and, following transplantation, in vivo.100 Taken together, these experiments point toward a common early progenitor that gives rise to the first and second heart field lineages.
Epicardial Progenitors in Cardiac Development
Along with the two heart fields, the importance of the proepicardial organ and epicardium to cardiovascular development and cell speciation has become increasingly appreciated in recent years. Under defined conditions, adult epicardial cells derived from rodents and humans can undergo epithelial-to-mesenchymal transformation, migrating into the subjacent myocardium to differentiate into smooth muscle and endothelial cells.80-83 Interestingly, two independent studies have recently drawn attention to a previously unrecognized role of epicardial derivatives in cardiogenesis, providing evidence for the existence of an epicardium-derived cardiac progenitor cell population (Figure 1, 2). These progenitors are marked by the transcription factors Wilms’ tumor protein (Wt1)84 and/or Tbx18,85 although the use of Tbx18 to uniquely label epicardial progenitor cells has recently been challenged because concomitant myocardial Tbx18 expression was found to be present.101 These cells not only contribute to cardiomyocytes in all four chambers of the adult mouse heart, but may also have multipotent potential, as some Tbx18 clones from mouse embryos are able to give rise to both cardiomyocytes and smooth muscle cells under defined culture conditions.85 What is the relationship between epicardium-derived cardiac progenitors and other embryonic progenitors? Fate mapping experiments performed by Zhou and colleagues found evidence for a robust contribution from Nkx2.5+ and Isl1+ precursors to Wt1+ proepicardial cells, positioning the Wt1+ proepicardial lineage as an early branch from the multipotent Nkx2.5+/Isl1+ progenitor lineages.84,102 However, proepicardial cells do not actively express either Nkx2.5 or Isl1, suggesting that these markers and Wt1 are either transiently coexpressed or sequentially expressed earlier in development. Supporting this possibility, in an ES model system, Zhou and colleagues were able to document transient coexpression of Wt1 with Nkx2.5 in ES derived cardiac progenitors.84 In contrast to the Wt1+ progenitors, Cai and colleagues found little overlap between Tbx18 and Isl1 lineages in the embryo, suggesting that Tbx18 and Isl1 are, at least in part, distinct cardiac progenitor populations and contribute in a complementary fashion to heart formation.85
While epicardial progenitors theoretically provide an attractive population of cells to be used in cardiac regeneration or repair, their ability to produce functional cardiomyocytes in an infarcted heart remains to be demonstrated. In zebrafish, the epicardium promotes cardiac regeneration by invading the wound site and creating a dense vascular network.103 Therefore, it is possible that the epicardium's greatest contribution to cardiac regeneration lies not in its ability to deliver new cardiomyocytes to the injured myocardium, but in its ability to promote endogenous cardiac progenitor cell expansion and neovascularization by paracrine signaling to surrounding cells. Future research on the regenerative role of epicardium-derived progenitors should include an assessment of both the epicardium's ability to directly contribute cardiomyocytes and its role in mediating paracrine effects.
Applications of Cardiac Progenitors
For cardiac progenitor cells to play a significant role in the field of cardiac regenerative medicine, they will need to be recruited or transplanted to the site of injury in sufficient numbers and directed to differentiate into fully mature and functional cardiomyocytes (Figure 3). One strategy to approach the challenge of obtaining sufficient quantities of cells needed for transplantation is to derive cardiac progenitors from pluripotent ES cells which represent an expandable and renewable source of multiple progenitors. ES cells can be directed by a variety of methods to undergo stepwise differentiation to mesoderm and then to cardiac progenitors, and in this regard have been instrumental in characterizing cardiac progenitor populations during the early stages of development and lineage commitment, which are difficult to study in the embryo. Importantly, ES-derived cardiomyocytes not only share molecular markers with native cardiomyocytes, but ultrastructural, electrophysiological, and mechanical studies of ES cell progeny indicate that they also exhibit all the hallmarks of cardiomyocytes.104 Nonetheless, ES-derived cardiomyocytes exhibit less organized sarcomeric structures than adult cardiomyocytes, a phenotype reminiscent of immature cardiomyocytes. The issue of cellular maturation will need to be addressed if ES-derived cardiomyocytes are to be employed in therapeutic applications to promote contractility in the failing heart.105,106
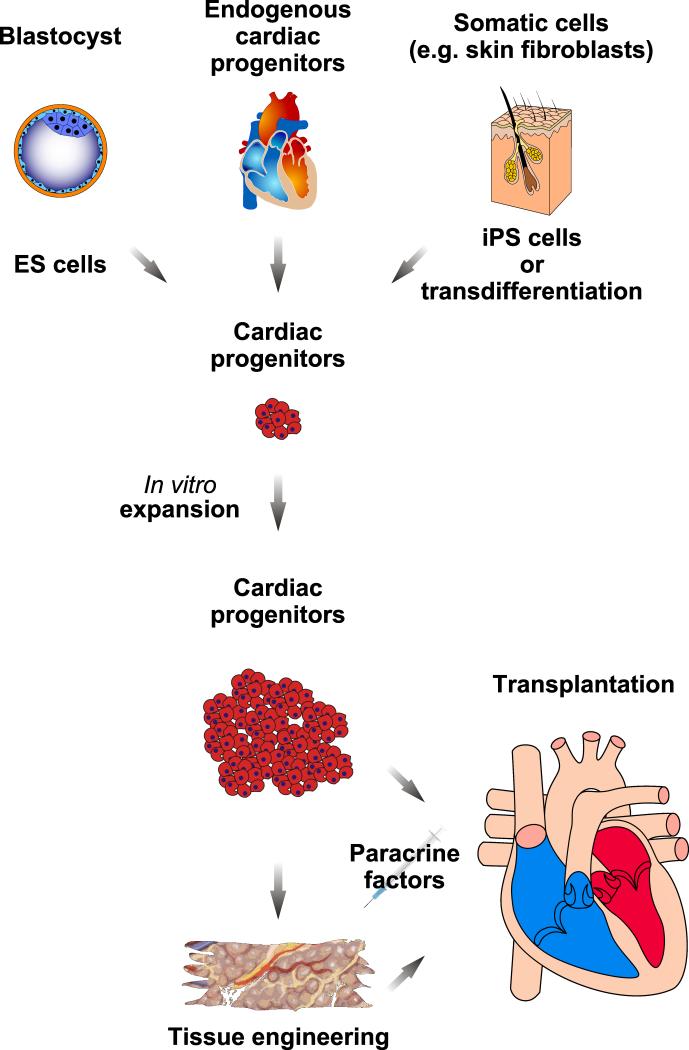
Figure 3. Strategies for delivering cardiac cell therapy.
Conceptually, cardiovascular progenitor cells could be derived from human ES cells or isolated from cardiac biopsy specimens. Alternatively, they could be generated by reprogramming a patient's own somatic cells either directly (i.e. transdifferentiation) into cardiac progenitors/cardiomyocytes, or by generating iPS cells followed by differentiation into cardiac progenitors. Following expansion, they could be directly implanted into the heart or used for the generation of an engineered tissue graft. The addition of extracellular factors, either in vitro or in vivo, may enhance cardiomyocyte survival, trigger cardiac lineage-specific differentiation of endogenous or exogenous cardiac progenitor cells, or promote cardiac progenitor and/or cardiomyocyte proliferation.
Recently, much excitement has been generated by the discovery that human somatic cells, through the ectopic expression of three or four defined transcription factors, can be reprogrammed into induced pluripotent stem cells (iPS), exhibiting all of the cardinal features of pluripotent ES cells.107,108 Apart from their potential application in regenerative medicine, patient-derived iPS cells may be particularly useful for bedside-to-bench research by allowing for the creation of disease-specific cell lines for which animal model systems are either lacking or inadequate.109 The use of autologous, patient-derived iPS cells for regenerative purposes would circumvent issues related to immunocompatibility and bypass many of the ethical considerations associated with generating ES cell lines. The original practice of virally inserting genes into human somatic cells has also been recently overcome, eliminating concerns regarding the potential hazards of viral integration into the human genome.110-114 Despite these potential advantages, patient-specific iPS cell lines require weeks if not months to generate and also differ epigenetically from ES cells, a distinction which may interfere with their ability to form mature, functional cardiomyocytes.115,116 The use of pluripotent stem cells, such as ES and iPS cells, also poses a significant risk of teratoma formation unless the progenitor cells can be isolated with an exceptional degree of purity, a task that has not been satisfactorily achieved with any pluripotent stem cell lineage thus far.
While the identification of human cardiac progenitor cell-specific surface markers would help to overcome this issue, to date no single surface marker has been identified which readily distinguishes cardiac progenitors from other differentiated progenies or their pluripotent precursors. One solution to overcome this challenge is to directly reprogram exogenous cells such as fibroblasts to a relatively restricted mesodermal or cardiac-restricted progenitor state through induced expression of a defined set of cardiogenic transcription factors. This strategy has already been used successfully to generate pancreatic cells,117 functional neurons,118 and cardiomyocytes.119 Induced expression of two cardiac transcription factors, Gata4 and Tbx5, has been used to direct mesoderm to cardiomyocytes,120 and the combination of Gata4, Tbx5 and Mef2c can direct reprogramming of cardiac or dermal fibroblasts into cells that very closely resemble adult cardiomyocytes.119 Importantly, the reprogramming of fibroblasts directly to cardiomyocytes does not appear to proceed via dedifferentiation to a progenitor cell state, perhaps explaining why transcription factors associated with cardiac progenitors such as Isl1 and Nkx2.5 are not required for induction of this process. Further validation of this reprogramming approach to generate autologous cardiomyocytes should help to simplify purification procedures required for cell transplantation since potentially fewer harmful contaminating cells would be involved.
Other unique challenges in cardiac regenerative medicine include the assembly of differentiated cardiac cells into the specific three-dimensional structures of the mature heart. In the ventricle, a complex alignment of cardiac myocytes exists designed to create the high degree of force generation needed to propel blood rapidly out of the heart during systole. Cardiomyocytes must also be coupled to each other by intercalated disks whose gap junction proteins facilitate spreading of the electrical impulse from one fiber to another. To improve cardiac function and prevent arrhythmogenesis, transplanted cells will have to stably engraft, align, and couple with the myocardium of the host in a coordinated fashion. Given the architectural complexity required, a bioengineered tissue graft may be the most ideal way to introduce regenerating cardiomyocytes to the injured heart.
Perspectives
While achieving meaningful cardiac regeneration in patients may not yet be feasible, incremental advances in our understanding of developmental and stem cell biology continue to move medical science closer toward this noble goal. Along the way, some of our most basic premises underlying cardiac development and cardiomyocyte renewal are being redefined. Dissecting the biologically complex roles of cardiac progenitor cell populations, both in cardiogenesis and in regeneration, will require a complete understanding of the developmental logic that accounts for the establishment of the diverse cell lineages within the heart. In this respect, developing a detailed human cardiovascular cell lineage map, with the molecular markers needed to identify and purify distinct cell populations with cardiogenic potential, is an important step in the right direction. Meanwhile, we anticipate that many of our current paradigms and dogmas will be unlearned before cardiac regeneration becomes a clinical reality.
Acknowledgments
We would like to thank Dipayan Chaudhuri, R. Sharon Chinthrajah, Serge Gregoire, Markus Krane, Marcello Panagia, Vivek Puppala, Alex Yi, and members of the Wu laboratory for valuable feedback on the manuscript.
Sources of Funding A.C.S. is supported by NIH Training Grant T32 HL07208-33. S.M.W. is supported by grants from the NIH/NHLBI (HL081086, HL100408), NIH Director's New Innovator Award (OD004411), and Harvard Stem Cell Institute.
Non-standard Abbreviations and Acronyms
- 14C
carbon-14
- Abcg2
ATP-binding cassette sub-family G member 2
- Bry
brachyury T
- c-Kit
stem cell factor receptor
- CSp
cardiosphere
- cTnT
cardiac troponin T
- DDR2
discoidin domain receptor 2
- eGFP
enhanced green fluorescent protein
- ES
embryonic stem cell
- FACS
fluorescence-activated cell sorting
- FHF
first heart field
- Flk1
fetal liver kinase 1
- GATA4
GATA binding factor 4
- HCN4
potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4
- iPS
induced pluripotent stem cell
- Isl1
Islet-1 transcription factor, LIM/homeodomain
- KDR
kinase insert domain receptor
- Mdr1
multi-drug resistance 1
- Mef2
myocyte enhancer factor 2
- Mesp1
mesoderm posterior 1
- Nkx 2.5
NK2 transcription factor related, locus 5
- Sca-1
stem cell antigen 1
- SHF
second heart field
- sm-actin
smooth muscle actin
- smMHC
smooth muscle myosin heavy chain
- SP
side population
- Tbx5
T-box transcription factor 5
- Tbx18
T-box transcription factor 18
- VEGFR2
vascular endothelial growth factor receptor 2
- vWF
von Willebrand factor
- Wt1
Wilms’ tumor protein
Footnotes
Subject codes [6] Cardiac development, [7] Chronic ischemic heart disease, [130] Animal models of human disease, [139] Developmental biology, [154] Myogenesis
Disclosures None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Robey TE, Saiget MK, Reinecke H, Murry CE. Systems approaches to preventing transplanted cell death in cardiac repair. J Mol Cell Cardiol. 2008;45:567–581. doi: 10.1016/j.yjmcc.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sigurdsson A, Swedberg K. The role of neurohormonal activation in chronic heart failure and postmyocardial infarction. Am Heart J. 1996;132:229–234. [PubMed] [Google Scholar]
- 3.Hansson EM, Lindsay ME, Chien KR. Regeneration next: toward heart stem cell therapeutics. Cell Stem Cell. 2009;5:364–377. doi: 10.1016/j.stem.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Odelberg SJ. Unraveling the molecular basis for regenerative cellular plasticity. PLoS Biol. 2004;2:E232. doi: 10.1371/journal.pbio.0020232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 6.Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev. 2007;87:521–544. doi: 10.1152/physrev.00032.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engel FB, Schebesta M, Duong MT, Lu G, Ren S, Madwed JB, Jiang H, Wang Y, Keating MT. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005;19:1175–1187. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn B, del Monte F, Hajjar RJ, Chang YS, Lebeche D, Arab S, Keating MT. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med. 2007;13:962–969. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 9.Bersell K, Arab S, Haring B, Kühn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 10.Beltrami AP, Urbanek K, Kajstura J, Yan SM, Finato N, Bussani R, Nadal-Ginard B, Silvestri F, Leri A, Beltrami CA, Anversa P. Evidence that human cardiac myocytes divide after myocardial infarction. N Engl J Med. 2001;344:1750–1757. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]
- 11.Anversa P, Kajstura J, Leri A, Bolli R. Life and death of cardiac stem cells: a paradigm shift in cardiac biology. Circulation. 2006;113:1451–1463. doi: 10.1161/CIRCULATIONAHA.105.595181. [DOI] [PubMed] [Google Scholar]
- 12.Olivetti G, Cigola E, Maestri R, Corradi D, Lagrasta C, Gambert SR, Anversa P. Aging, cardiac hypertrophy and ischemic cardiomyopathy do not affect the proportion of mononucleated and multinucleated myocytes in the human heart. J Mol Cell Cardiol. 1996;28:1463–1477. doi: 10.1006/jmcc.1996.0137. [DOI] [PubMed] [Google Scholar]
- 13.Adler CP, Friedburg H. Myocardial DNA content, ploidy level and cell number in geriatric hearts: post-mortem examinations of human myocardium in old age. J Mol Cell Cardiol. 1986;18:39–53. doi: 10.1016/s0022-2828(86)80981-6. [DOI] [PubMed] [Google Scholar]
- 14.Meckert PC, Rivello HG, Vigliano C, González P, Favaloro R, Laguens R. Endomitosis and polyploidization of myocardial cells in the periphery of human acute myocardial infarction. Cardiovasc Res. 2005;67:116–123. doi: 10.1016/j.cardiores.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 15.Herget GW, Neuburger M, Plagwitz R, Adler CP. DNA content, ploidy level and number of nuclei in the human heart after myocardial infarction. Cardiovasc Res. 1997;36:45–51. doi: 10.1016/s0008-6363(97)00140-5. [DOI] [PubMed] [Google Scholar]
- 16.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisén J. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spalding KL, Buchholz BA, Bergman LE, Druid H, Frisén J. Forensics: age written in teeth by nuclear tests. Nature. 2005;437:333–334. doi: 10.1038/437333a. [DOI] [PubMed] [Google Scholar]
- 18.Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisén J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 19.Spalding KL, Arner E, Westermark PO, Bernard S, Buchholz BA, Bergmann O, Blomqvist L, Hoffstedt J, Naslund E, Britton T, Concha H, Hassan M, Ryden M, Frisén J, Arner P. Dynamics of fat cell turnover in humans. Nature. 2008;453:783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- 20.Bhardwaj RD, Curtis MA, Spalding KL, Buchholz BA, Fink D, Björk-Eriksson T, Nordborg C, Gage FH, Druid H, Eriksson PS, Frisén J. Neocortical neurogenesis in humans is restricted to development. Proc Natl Acad Sci U S A. 2006;103:12564–12568. doi: 10.1073/pnas.0605177103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh PCH, Segers VFM, Davis ME, MacGillivray C, Gannon J, Molkentin JD, Robbins J, Lee RT. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med. 2007;13:970–974. doi: 10.1038/nm1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, Leri A, Kajstura J, Nadal-Ginard B, Anversa P. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 23.Zaruba MM, Soonpaa M, Reuter S, Field LJ. Cardiomyogenic potential of C-kit(+)-expressing cells derived from neonatal and adult mouse hearts. Circulation. 2010;121:1992–2000. doi: 10.1161/CIRCULATIONAHA.109.909093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massberg S, Schaerli P, Knezevic-Maramica I, Köllnberger M, Tubo N, Moseman EA, Huff IV, Junt T, Wagers AJ, Mazo IB, von Andrian UH. Immunosurveillance by hematopoietic progenitor cells trafficking through blood, lymph, and peripheral tissues. Cell. 2007;131:994–1008. doi: 10.1016/j.cell.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fazel S, Cimini M, Chen L, Li S, Angoulvant D, Fedak P, Verma S, Weisel RD, Keating A, Li RK. Cardioprotective c-kit+ cells are from the bone marrow and regulate the myocardial balance of angiogenic cytokines. J Clin Invest. 2006;116:1865–1877. doi: 10.1172/JCI27019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fazel SS, Chen L, Angoulvant D, Li SH, Weisel RD, Keating A, Li RK. Activation of c-kit is necessary for mobilization of reparative bone marrow progenitor cells in response to cardiac injury. FASEB J. 2008;22:930–940. doi: 10.1096/fj.07-8636com. [DOI] [PubMed] [Google Scholar]
- 27.Oh H, Bradfute SB, Gallardo TD, Nakamura T, Gaussin V, Mishina Y, Pocius J, Michael LH, Behringer RR, Garry DJ, Entman ML, Schneider MD. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc Natl Acad Sci U S A. 2003;100:12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh H, Chi X, Bradfute SB, Mishina Y, Pocius J, Michael LH, Behringer RR, Schwartz RJ, Entman ML, Schneider MD. Cardiac muscle plasticity in adult and embryo by heart-derived progenitor cells. Ann N Y Acad Sci. 2004;1015:182–189. doi: 10.1196/annals.1302.015. [DOI] [PubMed] [Google Scholar]
- 29.Matsuura K, Nagai T, Nishigaki N, Oyama T, Nishi J, Wada H, Sano M, Toko H, Akazawa H, Sato T, Nakaya H, Kasanuki H, Komuro I. Adult cardiac Sca-1-positive cells differentiate into beating cardiomyocytes. J Biol Chem. 2004;279:11384–11391. doi: 10.1074/jbc.M310822200. [DOI] [PubMed] [Google Scholar]
- 30.Dawn B, Stein AB, Urbanek K, Rota M, Whang B, Rastaldo R, Torella D, Tang XL, Rezazadeh A, Kajstura J, Leri A, Hunt G, Varma J, Prabhu SD, Anversa P, Bolli R. Cardiac stem cells delivered intravascularly traverse the vessel barrier, regenerate infarcted myocardium, and improve cardiac function. Proc Natl Acad Sci U S A. 2005;102:3766–3771. doi: 10.1073/pnas.0405957102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hierlihy AM, Seale P, Lobe CG, Rudnicki MA, Megeney LA. The post-nfatal heart contains a myocardial stem cell population. FEBS Lett. 2002;530:239–243. doi: 10.1016/s0014-5793(02)03477-4. [DOI] [PubMed] [Google Scholar]
- 33.Martin CM, Meeson AP, Robertson SM, Hawke TJ, Richardson JA, Bates S, Goetsch SC, Gallardo TD, Garry DJ. Persistent expression of the ATP-binding cassette transporter, Abcg2, identifies cardiac SP cells in the developing and adult heart. Dev Biol. 2004;265:262–275. doi: 10.1016/j.ydbio.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 34.Pfister O, Mouquet F, Jain M, Summer R, Helmes M, Fine A, Colucci WS, Liao R. CD31- but Not CD31+ cardiac side population cells exhibit functional cardiomyogenic differentiation. Circ Res. 2005;97:52–61. doi: 10.1161/01.RES.0000173297.53793.fa. [DOI] [PubMed] [Google Scholar]
- 35.Pfister O, Oikonomopoulos A, Sereti KI, Sohn RL, Cullen D, Fine GC, Mouquet F, Westerman K, Liao R. Role of the ATP-binding cassette transporter Abcg2 in the phenotype and function of cardiac side population cells. Circ Res. 2008;103:825–835. doi: 10.1161/CIRCRESAHA.108.174615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murry CE, Reinecke H, Pabon LM. Regeneration gaps: observations on stem cells and cardiac repair. J Am Coll Cardiol. 2006;47:1777–1785. doi: 10.1016/j.jacc.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Evans SM, Mummery C, Doevendans PA. Progenitor cells for cardiac repair. Semin Cell Dev Biol. 2007;18:153–160. doi: 10.1016/j.semcdb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 38.Parmacek MS, Epstein JA. Pursuing cardiac progenitors: regeneration redux. Cell. 2005;120:295–298. doi: 10.1016/j.cell.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 39.Yoon CH, Koyanagi M, Iekushi K, Seeger F, Urbich C, Zeiher AM, Dimmeler S. Mechanism of improved cardiac function after bone marrow mononuclear cell therapy: role of cardiovascular lineage commitment. Circulation. 2010;121:2001–2011. doi: 10.1161/CIRCULATIONAHA.109.909291. [DOI] [PubMed] [Google Scholar]
- 40.Gnecchi M, Zhang Z, Ni A, Dzau VJ. Paracrine mechanisms in adult stem cell signaling and therapy. Circ Res. 2008;103:1204–1219. doi: 10.1161/CIRCRESAHA.108.176826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer GP, Wollert KC, Lotz J, Steffens J, Lippolt P, Fichtner S, Hecker H, Schaefer A, Arseniev L, Hertenstein B, Ganser A, Drexler H. Intracoronary bone marrow cell transfer after myocardial infarction: eighteen months’ follow-up data from the randomized, controlled BOOST (BOne marrOw transfer to enhance ST-elevation infarct regeneration) trial. Circulation. 2006;113:1287–1294. doi: 10.1161/CIRCULATIONAHA.105.575118. [DOI] [PubMed] [Google Scholar]
- 42.Urbanek K, Quaini F, Tasca G, Torella D, Castaldo C, Nadal-Ginard B, Leri A, Kajstura J, Quaini E, Anversa P. Intense myocyte formation from cardiac stem cells in human cardiac hypertrophy. Proc Natl Acad Sci U S A. 2003;100:10440–10445. doi: 10.1073/pnas.1832855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urbanek K, Torella D, Sheikh F, De Angelis A, Nurzynska D, Silvestri F, Beltrami CA, Bussani R, Beltrami AP, Quaini F, Bolli R, Leri A, Kajstura J, Anversa P. Myocardial regeneration by activation of multipotent cardiac stem cells in ischemic heart failure. Proc Natl Acad Sci U S A. 2005;102:8692–8697. doi: 10.1073/pnas.0500169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quaini F, Urbanek K, Beltrami AP, Finato N, Beltrami CA, Nadal-Ginard B, Kajstura J, Leri A, Anversa P. Chimerism of the transplanted heart. N Engl J Med. 2002;346:5–15. doi: 10.1056/NEJMoa012081. [DOI] [PubMed] [Google Scholar]
- 45.Bearzi C, Rota M, Hosoda T, Tillmanns J, Nascimbene A, De Angelis A, Yasuzawa-Amano S, Trofimova I, Siggins RW, LeCapitaine N, Cascapera S, Beltrami AP, D'Alessandro DA, Zias E, Quaini F, Urbanek K, Michler RE, Bolli R, Kajstura J, Leri A, Anversa P. Human cardiac stem cells. Proc Natl Acad Sci U S A. 2007;104:14068–14073. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Messina E, De Angelis L, Frati G, Morrone S, Chimenti S, Fiordaliso F, Salio M, Battaglia M, Latronico MVG, Coletta M, Vivarelli E, Frati L, Cossu G, Giacomello A. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ Res. 2004;95:911–921. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- 47.Davis DR, Zhang Y, Smith RR, Cheng K, Terrovitis J, Malliaras K, Li TS, White A, Makkar R, Marbán E. Validation of the cardiosphere method to culture cardiac progenitor cells from myocardial tissue. PLoS One. 2009;4:e7195. doi: 10.1371/journal.pone.0007195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith RR, Barile L, Cho HC, Leppo MK, Hare JM, Messina E, Giacomello A, Abraham MR, Marban E. Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation. 2007;115:896–908. doi: 10.1161/CIRCULATIONAHA.106.655209. [DOI] [PubMed] [Google Scholar]
- 49.Johnston PV, Sasano T, Mills K, Evers R, Lee ST, Smith RR, Lardo AC, Lai S, Steenbergen C, Gerstenblith G, Lange R, Marban E. Engraftment, differentiation, and functional benefits of autologous cardiosphere-derived cells in porcine ischemic cardiomyopathy. Circulation. 2009;120:1075–1083. doi: 10.1161/CIRCULATIONAHA.108.816058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chimenti I, Smith RR, Li TS, Gerstenblith G, Messina E, Giacomello A, Marban E. Relative roles of direct regeneration versus paracrine effects of human cardiosphere-derived cells transplanted into infarcted mice. Circ Res. 2010;106:971–980. doi: 10.1161/CIRCRESAHA.109.210682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terrovitis J, Lautamaki R, Bonios M, Fox J, Engles JM, Yu J, Leppo MK, Pomper MG, Wahl RL, Seidel J, Tsui BM, Bengel FM, Abraham MR, Marban E. Noninvasive quantification and optimization of acute cell retention by in vivo positron emission tomography after intramyocardial cardiac-derived stem cell delivery. J Am Coll Cardiol. 2009;54:1619–1626. doi: 10.1016/j.jacc.2009.04.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pouly J, Bruneval P, Mandet C, Proksch S, Peyrard S, Amrein C, Bousseaux V, Guillemain R, Deloche A, Fabiani JN, Menasché P. Cardiac stem cells in the real world. J Thorac Cardiovasc Surg. 2008;135:673–678. doi: 10.1016/j.jtcvs.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 53.Tang YL, Shen L, Qian K, Phillips MI. A novel two-step procedure to expand cardiac Sca-1+ cells clonally. Biochem Biophys Res Commun. 2007;359:877–883. doi: 10.1016/j.bbrc.2007.05.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Vliet P, Roccio M, Smits AM, van Oorschot AA, Metz CH, van Veen TA, Sluijter JP, Doevendans PA, Goumans MJ. Progenitor cells isolated from the human heart: a potential cell source for regenerative therapy. Neth Heart J. 2008;16:163–169. doi: 10.1007/BF03086138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smits AM, van Vliet P, Metz CH, Korfage T, Sluijter JP, Doevendans PA, Goumans MJ. Human cardiomyocyte progenitor cells differentiate into functional mature cardiomyocytes: an in vitro model for studying human cardiac physiology and pathophysiology. Nat Protoc. 2009;4:232–243. doi: 10.1038/nprot.2008.229. [DOI] [PubMed] [Google Scholar]
- 56.Martin-Puig S, Wang Z, Chien KR. Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell. 2008;2:320–331. doi: 10.1016/j.stem.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 57.Laugwitz KL, Moretti A, Caron L, Nakano A, Chien KR. Islet1 cardiovascular progenitors: a single source for heart lineages? Development. 2008;135:193–205. doi: 10.1242/dev.001883. [DOI] [PubMed] [Google Scholar]
- 58.Mikawa T. Cardiac lineages. In: Harvey RP, Rosenthal N, editors. Heart Development. Academic Press; San Diego, Calif: 1999. pp. 19–33. [Google Scholar]
- 59.Gourdie RG, Wei Y, Kim D, Klatt SC, Mikawa T. Endothelin-induced conversion of embryonic heart muscle cells into impulse-conducting Purkinje fibers. Proc Natl Acad Sci U S A. 1998;95:6815–6818. doi: 10.1073/pnas.95.12.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi K, Kennedy M, Kazarov A, Papadimitriou JC, Keller G. A common precursor for hematopoietic and endothelial cells. Development. 1998;125:725–732. doi: 10.1242/dev.125.4.725. [DOI] [PubMed] [Google Scholar]
- 61.Fehling HJ, Lacaud G, Kubo A, Kennedy M, Robertson S, Keller G, Kouskoff V. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development. 2003;130:4217–4227. doi: 10.1242/dev.00589. [DOI] [PubMed] [Google Scholar]
- 62.Kouskoff V, Lacaud G, Schwantz S, Fehling HJ, Keller G. Sequential development of hematopoietic and cardiac mesoderm during embryonic stem cell differentiation. Proc Natl Acad Sci U S A. 2005;102:13170–13175. doi: 10.1073/pnas.0501672102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien Cl, Schultheiss TM, Orkin SH. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 64.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 65.Kattman SJ, Huber TL, Keller Gordon M. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 66.Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 67.Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 68.Rawles ME. The heart-forming areas of the early chick blastoderm. Physiol Zool. 1943;16:22–42. [Google Scholar]
- 69.Kitajima S, Takagi A, Inoue T, Saga Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development. 2000;127:3215–3226. doi: 10.1242/dev.127.15.3215. [DOI] [PubMed] [Google Scholar]
- 70.Saga Y, Kitajima S, Miyagawa-Tomita S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc Med. 2000;10:345–352. doi: 10.1016/s1050-1738(01)00069-x. [DOI] [PubMed] [Google Scholar]
- 71.Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki Ji, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 72.Dehaan R. Migration patterns of the precardiac mesoderm in the early chick embrvo. Exp Cell Res. 1963;29:544–560. doi: 10.1016/s0014-4827(63)80016-6. [DOI] [PubMed] [Google Scholar]
- 73.Abu-Issa R, Waldo K, Kirby ML. Heart fields: one, two or more? Dev Biol. 2004;272:281–285. doi: 10.1016/j.ydbio.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 74.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–837. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 75.Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 76.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Epstein JA, Buck CA. Transcriptional regulation of cardiac development: implications for congenital heart disease and DiGeorge syndrome. Pediatr Res. 2000;48:717–724. doi: 10.1203/00006450-200012000-00003. [DOI] [PubMed] [Google Scholar]
- 78.Hutson MR, Kirby ML. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin Cell Dev Biol. 2007;18:101–110. doi: 10.1016/j.semcdb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poelmann R, Lie-Venema H, Gittenberger-de Groot A. The role of the epicardium and neural crest as extracardiac contributors to coronary vascular development. Tex Heart Inst J. 2002;29:255–261. [PMC free article] [PubMed] [Google Scholar]
- 80.Wada AM, Smith TK, Osler ME, Reese DE, Bader DM. Epicardial/Mesothelial cell line retains vasculogenic potential of embryonic epicardium. Circ Res. 2003;92:525–531. doi: 10.1161/01.RES.0000060484.11032.0B. [DOI] [PubMed] [Google Scholar]
- 81.Smart N, Risebro CA, Melville AAD, Moses K, Schwartz RJ, Chien KR, Riley PR. Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature. 2007;445:177–182. doi: 10.1038/nature05383. [DOI] [PubMed] [Google Scholar]
- 82.van Tuyn J, Atsma D, Winter E, van der Velde-van Dijke I, Pijnappels D, Bax N, Knaän-Shanzer S, Gittenberger-de Groot A, Poelmann R, van der Laarse A, van der Wall E, Schalij M, de Vries A. Epicardial cells of human adults can undergo an epithelial-to-mesenchymal transition and obtain characteristics of smooth muscle cells in vitro. Stem Cells. 2007;25:271–278. doi: 10.1634/stemcells.2006-0366. [DOI] [PubMed] [Google Scholar]
- 83.Gittenberger-de Groot AC, Vrancken Peeters M, Mentink M, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 1998;82:1043–1052. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- 84.Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen TH, Chang TC, Kang JO, Choudhary B, Makita T, Tran CM, Burch JB, Eid H, Sucov HM. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Dev Biol. 2002;250:198–207. doi: 10.1006/dbio.2002.0796. [DOI] [PubMed] [Google Scholar]
- 87.Stuckmann I, Evans S, Lassar AB. Erythropoietin and retinoic acid, secreted from the epicardium, are required for cardiac myocyte proliferation. Dev Biol. 2003;255:334–349. doi: 10.1016/s0012-1606(02)00078-7. [DOI] [PubMed] [Google Scholar]
- 88.Winter E, Gittenberger-de Groot A. Epicardium-derived cells in cardiogenesis and cardiac regeneration. Cell Mol Life Sci. 2007;64:692–703. doi: 10.1007/s00018-007-6522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 90.Qyang Y, Martin-Puig S, Chiravuri M, Chen S, Xu H, Bu L, Jiang X, Lin L, Granger A, Moretti A, Caron L, Wu X, Clarke J, Taketo MM, Laugwitz KL, Moon RT, Gruber P, Evans SM, Ding S, Chien Kenneth R. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 91.Bu L, Jiang X, Martin-Puig S, Caron L, Zhu S, Shao Y, Roberts DJ, Huang PL, Domian IJ, Chien KR. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- 92.Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, Chen J, Evans SM. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304:286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, Platoshyn O, Yuan JX, Evans S, Chien KR. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stanley EG, Biben C, Elefanty A, Barnett L, Koentgen F, Robb L, Harvey RP. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3'UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int J Dev Biol. 2002;46:431–439. [PubMed] [Google Scholar]
- 95.Masino AM, Gallardo TD, Wilcox CA, Olson EN, Williams RS, Garry DJ. Transcriptional regulation of cardiac progenitor cell populations. Circ Res. 2004;95:389–397. doi: 10.1161/01.RES.0000138302.02691.be. [DOI] [PubMed] [Google Scholar]
- 96.Meilhac SM, Esner M, Kelly RG, Nicolas JF, Buckingham ME. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev Cell. 2004;6:685–698. doi: 10.1016/s1534-5807(04)00133-9. [DOI] [PubMed] [Google Scholar]
- 97.Meilhac SM, Kelly RG, Rocancourt D, Eloy-Trinquet S, Nicolas JF, Buckingham ME. A retrospective clonal analysis of the myocardium reveals two phases of clonal growth in the developing mouse heart. Development. 2003;130:3877–3889. doi: 10.1242/dev.00580. [DOI] [PubMed] [Google Scholar]
- 98.Huber TL, Kouskoff V, Joerg Fehling H, Palis J, Keller G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature. 2004;432:625–630. doi: 10.1038/nature03122. [DOI] [PubMed] [Google Scholar]
- 99.Takeuchi JK, Ohgi M, Koshiba-Takeuchi K, Shiratori H, Sakaki I, Ogura K, Saijoh Y, Ogura T. Tbx5 specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development. 2003;130:5953–5964. doi: 10.1242/dev.00797. [DOI] [PubMed] [Google Scholar]
- 100.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 101.Christoffels VM, Grieskamp T, Norden J, Mommersteeg MTM, Rudat C, Kispert A. Tbx18 and the fate of epicardial progenitors. Nature. 2009;458:E8–E9. doi: 10.1038/nature07916. [DOI] [PubMed] [Google Scholar]
- 102.Zhou B, Gise Av, Ma Q, Rivera-Feliciano J, Pu WT. Nkx2-5- and Isl1-expressing cardiac progenitors contribute to proepicardium. Biochem Biophys Res Commun. 2008;375:450–453. doi: 10.1016/j.bbrc.2008.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 104.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 105.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, Livne E, Binah O, Itskovitz-Eldor J, Gepstein L. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mummery C, Ward-van Oostwaard D, Doevendans P, Spijker R, van den Brink S, Hassink R, van der Heyden M, Opthof T, Pera M, de la Riviere AB, Passier R, Tertoolen L. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–2740. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 107.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 108.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 109.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, Dorn T, Goedel A, Höhnke C, Hofmann F, Seyfarth M, Sinnecker D, Schömig A, Laugwitz KL. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 110.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010 doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ieda M, Fu J-D, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]