Abstract
Despite considerable progress over recent years, the prognosis of invasive aspergillosis (IA) remains unfavorable, reflecting an incomplete understanding of Aspergillus pathogenesis and suboptimal antifungal efficacy in vivo. Mammalian host systems including rodents and rabbits are important tools in elucidating antifungal drug activity and the immunopathogenesis of IA. Nonetheless, they are hampered by limitations that impose a “bottleneck” in mass screening of novel antifungal compounds and putative Aspergillus virulence factors including their cost, labor intensity and ethical constraints. Drosophila melanogaster is an invertebrate host with a long track record of genetic studies and a simple yet highly conserved innate immune system. Herein, we describe our experience using this fly model as a facile, non-laborious, inexpensive pathosystem for high-throughput screening of novel antifungal compounds and putative Aspergillus mutants, and studying antifungal innate immunity. We present three infection protocols (i.e., injection, rolling, ingestion) that introduce Aspergillus either directly into the hemolymph or at different epithelial surfaces of Toll-deficient Drosophila flies. As a proof of principle, we demonstrate attenuated virulence of known hypovirulent Aspergillus strains and protection of Aspergillus-infected flies given oral Aspergillus-active agents such as voriconazole. These protocols can be adapted for similar studies of other fungal pathogens. Crossing and generation of Toll-deficient Drosophila flies takes three weeks; Aspergillus conidial preparation takes three days; fly inoculation depending on the infection assay takes one to 6–8 hours; and assessment of fly survival, Aspergillus strain virulence, Drosophila innate host parameters and/or drug activity takes 4–8 days.
Key words: Drosophila, Aspergillus, mini-host model, virulence, antifungal efficacy
Introduction
Since the 1990s, invasive aspergillosis (IA) has emerged as the leading cause of infection-related death in recipients of hematopoietic stem cell and solid organ transplants.1,2 Despite significant advances in diagnosis and antifungal therapy over recent years, the prognosis of patients who develop IA remains poor, reflecting its incompletely understood pathogenesis, suboptimal diagnosis and the mediocre efficacy of modern antifungal drugs in vivo. Traditionally, small mammals such as mice, rats, guinea pigs and rabbits have served as invaluable research tools in modeling human IA and studying Aspergillus virulence and antifungal drug activity against IA.3–6 Because of their immunological and anatomical similarities to humans and the ability to precisely control the host and its environment, these biosystems are considered the gold standard for pathogenetic and pharmacological studies of IA.
Nevertheless, use of small mammals as host models of human IA is restricted by several factors. With regard to pathogenetic studies, although they are amenable to reverse genetics via generation of knockout mutants,7 use of large-scale forward genetic strategies with these models is difficult for logistical, economical and ethical reasons. Not surprisingly, only one Aspergillus mutant is typically tested at a time using these models via comparison of its virulence with that of the isogenic wild-type (WT) parental Aspergillus strain in a small number of animals. In this era in which sequencing of the genome of Aspergillus and other medically important fungi (e.g., Candida, Cryptococcus) has been completed, and tools that facilitate generation of a larger number of mutants are becoming increasingly sophisticated,8–10 alternative methods of large-scale screening of fungal virulence traits and assessment of their contribution to the pathogenesis of IA are required. Successful use of large-scale screens may uncover potential novel targets for diagnosis and treatment of IA. However, the same logistical difficulties in investigating the immunopathogenesis of IA also impede assessment of the activity of antifungals or combination of them in mammalian models of IA. Not unexpectedly, use of these animal models is typically limited to testing a limited number of drugs or drug combinations and only one Aspergillus isolate at a time in small numbers of animals.
Over the past 10 years, pioneering studies have demonstrated that various fungi including Aspergillus fumigatus can cause fatal infections in an array of simpler invertebrate hosts, such as the fruit fly Drosophila melanogaster,11–15 the greater wax moth Galleria mellonella,16 the roundworm Caenorhabditis elegans,17 and the free-living soil amoeba Dictyostelium discoideum.18 Elegant studies using these invertebrate pathosystems have shown that important aspects of innate immunity are evolutionarily conserved in mammalian hosts.19–21 For example, 50–60% of protein homologs involved in pathogen recognition, signal transduction and innate immunity in humans also exist in D. melanogaster and C. elegans. For these reasons, as well as because the genomes of D. melanogaster and C. elegans have been sequenced and research tools such as full-genome microarrays and RNA interference libraries can be used with these hosts,22 these invertebrates have gained significant favor in studying microbial pathogenesis and host defense.
D. melanogaster does not have an adaptive immune system but its innate immune pathways are evolutionary conserved through mammals.11,12,19,20,23,24 The imd (immune deficiency) and Toll cascades, which are important for host defense against Gram-negative bacteria and fungi/Gram-positive bacteria respectively are considered the fly counterparts to the human tumor necrosis factor receptor signaling and Toll-like receptor pathways respectively.19 Following fungal challenge in Drosophila flies, peptidoglycan recognition protein SA and Gramnegative binding protein 1 activate the serpin Persephone, which cleaves the serpin Spatzle and then activates the transmembrane receptor Toll. Upon activation, Toll recruits the adaptor proteins MyD88 and Tube and the threonine/serine kinase Pelle.19 The resultant intracellular proteolytic cascade results in degradation of the Ik-B-like protein Cactus and nuclear translocation of the NFκB-like transcriptional factors Dorsal and Dif, which induce expression of antifungal peptide-related genes such as drosomycin and metchnikowin.19
Because the Toll pathway is significantly conserved between Drosophila and humans and because flies can grow, and be manipulated and analyzed in larger numbers in a more time-efficient manner and with significantly less labor and cost than conventional mammalian models can, Drosophila is a useful model for high-throughput screening of Aspergillus virulence factors and of compounds for activity against IA.25 For this purpose, three infection assays described herein deliver Aspergillus conidia (1) directly into the fly hemolymph (needle pricking [injection] assay), (2) on the fly exterior surface (rolling assay) or (3) in the fly gastrointestinal epithelium (ingestion assay). All three assays have high inter-operator and intra-operator reproducibility, are easy to perform, and result in higher mortality rates in Toll-deficient flies than in WT flies, which are resistant to IA.26 Aspergillus strains shown to be hypovirulent in mammalian hosts also exhibit attenuated virulence in Toll-deficient Drosophila flies.26 In fact, a recent comparative analysis of hypovirulent Aspergillus strains in mice and flies revealed a high rate of concordance in Aspergillus virulence between the two species.27 In addition, administration of voriconazole, the preferred drug for treatment of IA in humans,28 significantly protects flies against IA, as demonstrated by decreased mortality rates and tissue fungal burdens.26
In addition to A. fumigatus, the protocols described below can be adapted with modifications to the study of virulence, host defense against and antifungal activity against other molds (e.g., Aspergillus terreus,29 Zygomycetes species,30,31 Fusarium species,32 Scedosporium species32) and yeasts (e.g., Candida albicans,15,33 Cryptococcus neoformans13) although some of these pathogens have key differences. For example, C. albicans does not infect Toll-deficient flies following ingestion; it only does so following injection.33 Also, infection of OregonR WT flies with Zygomycetes species leads to higher mortality rates in WT Drosophila flies than does infection with other molds and yeasts.30 These fungus-specific differences in Drosophila susceptibility to fungal infection provide ample opportunities for investigating fungus-specific host-pathogen interaction questions using this fruit fly mini-host model.
Despite having the benefit of facilitation of testing large numbers of animals quickly and inexpensively, the Drosophila model has its limitations. For instance, precisely quantifying the number of conidia that infect each fly as well as the amount of antifungal drug ingested by individual flies is difficult. Furthermore, studying the pharmacokinetic and pharmacodynamic properties of tested compounds is not feasible in Drosophila flies; instead, this requires a larger mammalian host. Moreover, to extend testing of antifungal activity to compounds that are not orally absorbed, injection of such agents into Drosophila flies using micro-pipettes is currently under development. Finally, because thermotolerance is a universal virulence trait across pathogenic fungi,34 infection of Drosophila flies and maintenance of them at 29°C may limit the study of certain fungal virulence traits. For example, studies showed that an Aspergillus strain lacking the gene that regulates the expression of the nucleolar protein CgrA, a key regulator of Aspergillus thermotolerance, was hypovirulent in mice infected with it35 but fully virulent in Toll-deficient Drosophila flies infected and maintained at 29°C suggesting that certain aspects of fungal virulence in mammals may not be accurately modeled in mini-hosts. Thus, the lack of a virulence of an Aspergillus mutant at 29°C (fly model) does not mean that it is not virulent when tested at 37°C (mouse model) and vice versa.
Experimental Design
Because 10- to 15-day-old Drosophila flies have significantly higher mortality rates after Aspergillus infection than do 2- to 4-day-old flies,36 consistent use of flies in the latter age range in all experiments is critical. In addition, female flies are typically used because they are larger than male flies and relatively more resistant to injection injury. To minimize potential sex-dependent effects on infection susceptibility, we use female flies in all experiments. Finally, to decrease the influence of circadian rhythm on innate immune responses in Drosophila flies, we perform all experiments with this model in the morning.
Fungal strain selection.
We use the Aspergillus fumigatus clinical isolate AF293 WT strain for the infection assays and drug protection experiments because it was the strain used in the Aspergillus genome sequencing project.8 For Aspergillus virulence studies, we have used (1) the gliotoxin-deleted A. fumigatus strain ΔgliP and its isogenic WT Aspergillus strain AF293,37 and (2) the alb1-deleted A. fumigatus strain B-5233/RGD12-8 and its isogenic WT Aspergillus strain, B-5233.38
Fly selection.
We use WT OregonR flies, which have a functional Toll cascade and thus are inherently resistant to Aspergillus challenge, as controls. Also, we generate Tlr632/TlI-RXA flies by crossing Tlr632/TM6B and TlI-RXA/TM6B Toll-deficient flies as described below (Steps 1–8). Because Tlr632 is a thermosensitive loss-of-function allele with a strong phenotype at 29°C, flies should be maintained at that temperature following Aspergillus infection.
Materials
Reagents.
-
Fungal strains: Aspergillus fumigatus clinical isolate AF293 wild-type (WT) strain and gliP-deleted Aspergillus fumigatus strain derived from AF293; alb1-deleted Aspergillus fumigatus strain B-5233/RGD12-8 and its isogenic WT Aspergillus strain, B-5233; Candida kefyr American Type Culture Collection 66028.
NOTE: Aspergillus and Candida agar plates and conidial suspensions should be disposed of as biohazardous material.
-
Adult fly lines: OregonR WT flies; Tlr632/TM6B and TlI-RXA/TM6B Toll-deficient flies.
NOTE: Infected flies are killed by freezing at −20°C and disposed of as biohazardous material.
Yeast extract (BD Biosciences, cat. no. 211931)
Dextrose (Sigma, cat. no. D9434)
Bacto Agar (BD Biosciences, cat. no. 214030)
NaCl (AMRESCO, cat. no. 0241)
MgSO4 (AMRESCO, cat. no. 0662)
Phosphate-buffered saline (Sigma, cat. no. P5368)
Hank's balanced salt solution (Sigma, cat. no. H8264)
Hexamethyldisilazane (HMDS; Sigma, cat. no. H4875)
Autoclaved water
Ethanol (Sigma, cat. no. 2777649)
Acetone (Sigma, cat. no. 650501)
Glycerol (Sigma, cat. no. G9012)
Formalin solution, neutral buffered, 10% (Sigma, cat. no. HT501320)
TRIzol reagent (Invitrogen, cat. no. 15596-026)
Voriconazole (Sigma, cat. no. PZ0005)
Terbinafine (Sigma, cat. no. T8826)
Fly food reagents: agar, sucrose, yeast and cornmeal (Genesee Scientific)
Vials (Genesee Scientific, cat. no. 32-116)
Inactive dry yeast particles (Genesee Scientific, cat. no. 62-106) Equipment.
Paintbrush (size 0)
Tungsten stainless steel needle (tip diameter, 0.01 mm), held in a pin vise (Ernest F. Fullam Inc., cat. no. 54270)
Stereoscopic microscope (Stemi 2000, Carl Zeiss Inc.) equipped with a controllable CO2 flow pad
Spatula (Sigma, cat. no. Z283282)
Sterile disposable Petri dishes 100 × 15 mm (BD Biosciences, cat. no. 351029)
Bunsen burner (Sigma, cat. no. Z270296)
29°C and 37°C incubators (Precision Scientific)
Fly incubators with high humidity capacity (60–75%), adjustable temperature and a 12 hour light/12 hour dark cycle
Bead-beater homogenizer (Biospec Products, cat. no. 1107900)
DNeasy Kit (Qiagen, cat. no. 69504)
Glass spreaders (Sigma, cat. no. S4522)
Hemocytometer
Reagent setup.
YAG agar plates. Add 5 g of yeast extract (wt/vol), 10 g of dextrose (wt/vol), 15 g of agar (wt/vol), 10 ml of 1M MgSO4 (vol/vol), 2 ml of vitamin mix (vol/vol) and 1 ml of trace elements (vol/vol) in 1 l of distilled water and autoclave. Pour the autoclaved medium on 100 × 15 mm sterile disposable Petri dishes (∼20 ml/dish) or in empty fly vials (∼15 ml/vial) and let overnight at room temperature to solidify. Store at 4°C for up to 2–3 months until use.
Fly food. Prepare conventional fly food with 1% agar (wt/vol), 3% yeast (wt/vol), 0.6% sucrose (wt/vol), 4.4% cornmeal (wt/vol), 0.11% methylparaben (tegosept; wt/vol) and 0.36% propionic acid (vol/vol).
Antibiotics stock. Prepare stock solutions of 40 mg/ml voriconazole and terbinafine diluted in distilled water.
Procedure
Fly preparation: crossing of Tlr632/TM6B and TlI-RXA/TM6B Toll-deficient Drosophila alleles for generation of Tlr632/TlI-RXA mutants (timing 21–28 d).
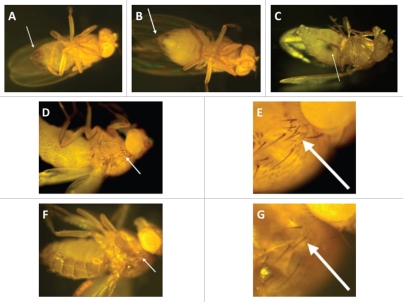
(1) Distinguish male (Fig. 1A) and female (Fig. 1B) flies based on their genitalia.
Figure 1.
Toll-deficient Drosophila flies. Image of a (A) male and a (B) female D. melanogaster. The arrows point to their genitalia. (C) A female virgin D. melanogaster. The arrow points to the embryonic residue that is present in virgin female flies within the first 8–12 h after eclosion. (D and E) The “multiple-hair type” of bristle seen in flies with the TM6B balancer, such as TlI-RXA/TM6B and Tlr632/TM6B flies (E is an image of the bristle in D at a higher magnification). (F and G) The “double-hair type” of bristle seen in flies without the TM6B balancer, such as Tlr632/TlI-RXA flies (G is an image of the bristle in F at a higher magnification).
(2) Identify virgin female flies according to the dark mark on the ventral abdomen (Fig. 1C), which is an embryonic residue that is excreted from their gastrointestinal tract upon maturation. Typically, female flies are considered virgins during the first 8–12 h after eclosion. Afterwards, they mature and become reproductively active. Hence, another way to collect virgin females besides looking at this abdominal mark, is to make sure that you completely remove every single fly from the vials in the morning (e.g., 9:00 a.m.) and then come back to collect them before the completion of the 8 hour critical post-eclosion period (e.g., 4:00 p.m.).
(3) Drosophila genotypes typically include a so-called “balancer” that is used to provide flies with unique phenotypic characteristics for distinguishing different fly crossings phenotypically. For example, a balancer may provide a specific eye color or wing or bristle pattern. In the Tlr632/TM6B and TlI-RXA/TM6B flies the balancer is called TM6B. A fly with this balancer has a “multiple hair-type” bristle in its upper lateral thorax/torso (Fig. 1D and E), whereas flies without TM6B have a “double hair-type” bristle (Fig. 1F and G).
(4) Maintain vials containing the stock Tlr632/TM6B flies in which the TlI-RXA/TM6B × TlI-RXA/TM6B cross could lead to the following three genotypic combinations: Tlr632/TM6B, Tlr632/Tlr632 and TM6B/TM6B. TM6B/TM6B flies do not have a viable phenotype so they never appear in the vials; Tlr632/TM6B flies are similar to their ancestors and capable of reproduction; Tlr632/Tlr632 flies, although having a viable phenotype (despite their developmental defects), are sterile. Differentiate between Tlr632/TM6B and Tlr632/Tlr632 flies by identifying the balancer TM6B (Step 3). Hence, Tlr632/TM6B flies have the “multiple hair-type” bristle whereas Tlr632/Tlr632 flies have the “double hair-type” bristle.
(5) Maintain vials containing the stock TlI-RXA/TM6B flies in which the TlI-RXA/TM6B × TlI-RXA/TM6B cross could lead to the following three genotypic combinations: TlI-RXA/TM6B, TlI-RXA/TlI-RXA and TM6B/TM6B. TM6B/TM6B and TlI-RXA/TlI-RXA flies do not have viable phenotypes so they never appear in the vials; TlI-RXA/TM6B flies are similar to their ancestors and capable of reproduction.
NOTE: TlI-RXA is a null allele of Toll, whereas Tlr632 is a strong loss-of-function allele accounting for why homozygous TlI-RXA mutants are not viable but homozygous Tlr632 mutants are.
(6) Obtain TlI-RXA/TlI-RXA Drosophila mutants for use in the infection experiments by either crossing female virgin Tlr632/TM6B flies with male TlI-RXA/TM6B flies (virgin ♀ Tlr632/TM6B × ♂ TlI-RXA/TM6B) or crossing female virgin TlI-RXA/TM6B flies with male Tlr632/TM6B flies (virgin ♀ TlI-RXA/TM6B × ♂ Tlr632/TM6B).
(7) Four genotypic combinations may result from the Tlr632/TM6B × TlI-RXA/TM6B cross: (1) TM6B/TM6B flies, which are not viable; (2) TlI-RXA/TM6B flies, which are dark gray and have the “multiple hair-type” bristle; (3) Tlr632/TM6B flies, which are light brown and have the “multiple hair-type” bristle; (4) Tlr632/TlI-RXA flies, which are light brown and have the “double hair-type” bristle (Figs. 1F and G).
(8) Keep transferring the Tlr632/TM6B × TlI-RXA/TM6B cross to new vials every 3–4 days so that the female flies lay their eggs in multiple vials. This will increase the yield of Tlr632/TlI-RXA flies. Hydrate vials that appear to be dry by adding distilled water to them.
Fungal inoculum preparation (timing 3 d).
(9) Streak frozen glycerol stocks of A. fumigatus AF293 (or the hypovirulent Aspergillus strain of interest) onto YAG agar plates and incubate at 37°C for 24 h.
(10) Inoculate single colonies from the fresh plates onto new YAG agar plates and incubate at 37°C for 72 h until a uniform lawn of Aspergillus conidia forms onto the agar surface.
(11) Collect Aspergillus conidia from the surface of the agar plate by adding 0.5 ml of autoclaved water and using a glass spreader. Count the conidia using a hemocytometer. Prepare working solutions of Aspergillus conidia at various concentrations depending on the experimental design (range: 107–1010 conidia/ml).
Fly infection assays (timing 1 to 6–8 h).
Depending on the question to be answered, choose one of three infection assays: injection, rolling and ingestion.
(12) Needle pricking (injection) assay (timing 1 h).
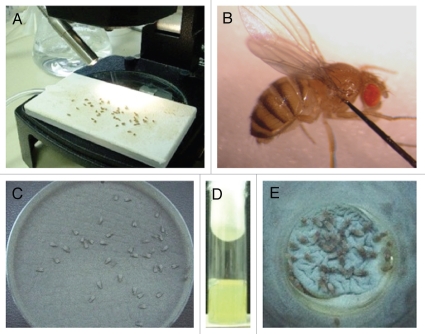
(A) Anesthetize flies by placing them on a CO2 fly pad (Fig. 2A). Handle the flies with a paintbrush to avoid injury.
Figure 2.
Infection assays of aspergillosis in Toll-deficient D. melanogaster. (A) Anesthetized flies on a CO2-flow fly pad. (B) Injection assay. A CO2-anesthetized fly was pricked at its dorsolateral thorax with a 0.1 mm diameter needle previously dipped in a concentrated Aspergillus conidial solution. (C) Rolling assay. Anesthetized flies were rolled on a Petri dish covered by a fresh layer of Aspergillus conidia for 2 min. At the end of rolling, Aspergillus uniformly covered the fly surface. (D) Fifteen milliliters of a sterile YAG medium that was allowed to solidify in an empty vial. (E) Ingestion assay. Flies feeding on the surface of a fresh lawn of Aspergillus conidia pre-grown in a YAG-containing vial.
(B) Sterilize a tungsten needle (tip diameter, 0.01 mm) with a flame, and wait for it to cool off before dipping the tip of the needle into the Aspergillus conidial suspension. (PROBLEM)
(C) Prick the dorsolateral thorax of each fly by inserting the needle midway into the thorax along the anteroposterior axis (Fig. 2B). This assay takes 5–10 min per 10 injected flies. Inject 30–50 female flies per group of interest.
(D) Return the injected flies to the vials containing fly food. To prevent flies from sticking in the food and dying as a result, place the vials on their sides until the flies recover from anesthesia (this usually takes a few minutes). For control purposes, inject a different group of 30–50 female flies with a sterilized needle that has not been dipped in an Aspergillus solution (septic injury control).
(E) Observe the injected flies over the next 3 h. Flies that die during this 3-hour period (typically <5%) are considered to have died of injection injury (not Aspergillus infection) and should be excluded from the analysis.
NOTE: Dead flies are typically found on the surface of the fly food. At death, they appear smaller than normal and dehydrated.
(F) Maintain the flies at 29°C, the temperature at which their susceptibility to microbial challenge is maximal.
NOTE: Tlr632 is a thermosensitive loss-of-function allele with a strong phenotype at this temperature.
(G) Proceed to Step 15 for assessment of virulence and pathogenicity or to Steps 21–25 for assessment of antifungal drug activity.
(13) Rolling assay (timing 1–2 h).
(A) Anesthetize flies by placing them on a CO2 fly pad for 3–4 min. (PROBLEM) Handle flies with a paintbrush to avoid injury.
(B) Transfer 30–50 anesthetized female flies onto the surface of a YAG plate with a fresh layer of Aspergillus conidia pregrown for 3 d as described in Step 10.
(C) Roll the anesthetized flies on the YAG agar plate surface for 2 min to uniformly cover them with Aspergillus conidia (Fig. 2C).
NOTE: Cover the Petri dish with its lid to avoid dispersion of conidia in the air.
(D) A key difference between the rolling and injection assays is that, as shown in Figure 2C, flies are covered with a high Aspergillus inoculum during rolling. To prevent continuous conidial exposure and rapid death of a significant number of flies within 24 h after rolling, place them in “temporary” vials for 1–2 h to allow for a substantial number of conidia to fall off their surfaces and wings into these vials. Again, to prevent flies from sticking in the food in these vials, place the vials on their sides until the flies recover from anesthesia.
(E) After this 1- to 2-hour period, transfer the flies to new vials and maintain them at 29°C. If flies are transferred directly to vials without the intermediate 1–2 hour step, 24 hours later, the fly-food surface will be covered with Aspergillus conidia that fall from the flies after they recover from anesthesia and move around in the vial, and a substantial number of flies will die because of this 24-hour continuous exposure to Aspergillus conidia.
(F) For control purposes, roll a different group of 30–50 female flies on empty sterile disposable Petri dishes (rolling-associated injury control).
(G) Flies that die within 3 h after rolling (typically <1%) are considered to have died of the rolling procedure and should not be included in subsequent analyses.
(H) Proceed to Step 15 for assessment of virulence and pathogenicity or to Steps 21–25 for assessment of antifungal drug activity.
(14) Ingestion assay (timing 6–8 h).
(A) Prepare special fly vials without fly food by adding YAG medium to empty vials (Fig. 2D).
(B) Modify step 10 by allowing Aspergillus to grow in these YAG-containing fly vials. Specifically, add 100–200 µl of a 108 conidia/ml solution to the surface of the YAG medium. A fresh conidial layer forms after a 72 h incubation period at 37°C.
(C) Place 30–50 female flies into the vials and let them feed on Aspergillus conidia for 6–8 h (Fig. 2E). (PRO BLEM)
NOTE: The ingestion assay does not require fly anesthetization.
(D) Because a few flies are exposed to Aspergillus conidia on their surface after this 6–8 h period (that is, a slight rolling exposure may occur in some flies) perform steps 13D and 13E to remove Aspergillus conidia from the flies.
(E) For control purposes, place a different group of 30–50 female flies into vials containing YAG medium without Aspergillus conidia for 6–8 h (starvation control).
(F) Flies that die within 3 h after completion of the ingestion assay (typically <1%) are considered to have died because of procedure and should be excluded from subsequent analyses.
(G) Proceed to Step 15 for assessment of virulence and pathogenicity or to Steps 21–25 for assessment of antifungal drug activity.
Virulence/pathogenicity assessment (timing 4–8 d).
(15) Assess virulence/pathogenicity using any of the three infection assays described above by determining the: (A) fly survival rate, (B) tissue fungal burden using real-time quantitative polymerase chain reaction (qPCR), histopathological analysis and scanning electron microscopy and/or (C) fly host immune responses.
(A) Fly survival (timing 4–8 d). (i) After infection incubate flies at 29°C and transfer them to new fly-food vials every 2 d.
(ii) Count live flies at intervals of 3–6 hours after inoculation. Flies begin to die about 48 h after infection in all assays but the progression of mortality thereafter is faster for the injection assay, followed by the ingestion and rolling assays.
(iii) Exclude from analysis flies that have died within 3 h of any of the infection assays as death in such cases is not caused by Aspergillus infection but is likely the result of excessive injury and/or stress produced by the procedure. Of note is that the percentage of excluded flies should not exceed 5% with the injection assay or 1% with the rolling and ingestion assays.
(iv) Perform at least three independent experiments using 30–50 flies per group and statistical analysis of differences in fly survival rate using the Kaplan-Meier survival method.
(B) Fungal burden using qPCR, histopathological analysis and scanning electron microscopy (timing 4–8 d). (i) For qPCR analysis, store groups of 20 flies of interest at −80°C until proceeding with DNA extraction. Collect flies at various time-points after Aspergillus inoculation for comparative analysis of tissue fungal burden.
(ii) When ready to proceed, wash flies twice with 0.85% NaCl to remove conidia from their exterior (which could skew the qPCR results) and homogenize them.
(iii) Extract DNA using the DNeasy tissue kit.
(iv) Analyze the DNA samples using primers and dual-labeled fluorescent hybridization probes specific for the A. fumigatus 18S rRNA gene (GenBank accession number. AB008401): (i) forward, 5′-GGC CCT TAA ATA GCC CGG T-3′; (ii) reverse, 5′-TGA GCC GAT AGT CCC CCT AA-3′; (iii) probe, 5′-FAM-AGC CAG CGG CCC GCA AAT G-TAMRA-3′. The threshold cycle (CT) for each sample is then interpolated from a standard seven-point curve of CT values prepared by spiking naïve, uninfected flies with 1 × 101 to 1 × 107 AF293 conidia.
(v) Report the qPCR results as conidial equivalents of A. fumigatus DNA.39 Perform all experiments in triplicate and analyze each DNA sample in duplicate.
(vi) For histopathological analysis, fix flies using 10% buffered formalin, and embed them in paraffin wax. Stain the tissue sections with Grocott-Gomori methenamine-silver nitrate or hematoxylin-eosin and examine them for visible hyphal burden.
(vii) For scanning electron microscopy, place flies of interest in 70% acetone for 4 h and then transfer them to 100% acetone for a 4 h or overnight incubation. Next, transfer flies to a new 100% acetone solution and incubate them for at least 4 h and then transfer them into a 1:1 100% acetone:HMDS solution for a 4 h or overnight incubation. After this incubation, transfer flies to a 100% HMDS solution for at least 4 h and then to a new 100% HMDS solution for a 4 h or overnight incubation. Let flies air-dry on paper tissue and place them on mounting pads in the desired orientation. Handle flies carefully as they become very brittle after these incubations.
NOTE: All incubations should take place at room temperature.
(C) Fly host responses (timing 4–8 d). (i) Collect Aspergillus-infected and uninfected control flies at various time points after inoculation following induction of anesthesia using CO2.
(ii) For subsequent RNA extraction, grind and homogenize 20 flies of interest in 1 ml of Trizol reagent and store the homogenates at −80°C until proceeding with qPCR or microarray analysis.
(iii) For subsequent protein analysis, grind and homogenize 20 flies of interest in 0.5 ml of phosphate-buffered saline or Hank's balanced salt solution for western blot analysis or enzyme-linked immunosorbent assay.
Preparation of antifungal drug containing fly-food vials (timing 1–2 d).
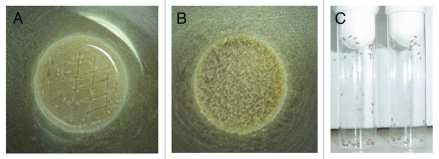
(16) Sterilize a spatula with a flame and make horizontal and vertical abrasions on the surface of the fly food (Fig. 3A).
Figure 3.
Preparation of antifungal-drug containing food vials and fly fasting. (A) A spatula is used to create 2- to 3-mm-deep abrasions on the surface of the fly food. Afterward, 200 µl of the antifungal drug of choice is added to the surface. (B) Dry yeast particles are then added to the surface of the drug-containing vial until they are entirely soaked by the antifungal agent. (C) Before exposure to the drug-containing food vials, flies are placed in empty vials for 6–8 hours so that they can starve.
NOTE: The abrasions should be superficial, not exceeding 2–3 mm in depth.
(17) Calculate the concentration of the antifungal drug and add the drug to the surface of the fly food (Fig. 3A).
NOTE: An optimal drug volume is 200 µl. If a higher volume is added, it will not be absorbed by the fly food and the yeast particles, and the flies will become stuck in the food and die. Prepare a highconcentration stock solution of the drug (e.g., 40 mg/ml). In doing so, even if a very high concentration of the drug or a drug combination (e.g., voriconazole plus terbinafine) is added to the fly food adding a volume higher than 200 µl will not be required.
(18) After addition of the 200 µl drug volume, fill a 1 ml pipette tip with dry inactive yeast particles and slowly drop them onto the surface of the vial. Do not add all the yeast particles at once; add a small number, let them soak into the drug and then add more. Continue until all yeast particles are soaked into the drug volume (Fig. 3B). (PROBLEM)
NOTE: Addition of yeast particles is critical for ingestion of the drug. If the antifungal drug is added directly to the fly food without any yeast particles, two problems may arise. First, absorption of the drug will be suboptimal and erratic, as flies will not eat much of the drug-containing fly food without yeast particles. Second, the flies will be much more likely to become stuck in the food because the food will not fully absorb the drug and it will be sticky.
(19) After preparation of the vials, let them sit for 24–48 h at room temperature to dry completely before use; otherwise the flies will become stuck in the food and die. (PROBLEM)
(20) After the vials dry transfer them at 4°C to (1) maintain them for long periods (i.e., because they not dry quickly at 4°C) and (2) protect the drug or drugs against degradation. After preparing the vials and letting them dry for 24–48 h (Step 19), they are ready for use in antifungal protection experiments.
Antifungal drug activity assessment (timing 4–8 d).
(21) Place female flies in empty vials for 6–8 hours to starve them (Fig. 3C). This will facilitate improved ingestion of the antifungal drug-containing food (PROBLEM).
(22) After this 6- to 8-hour fasting period, transfer flies into a drug-containing vial and let them feed on the drug-containing food for 24 h before infecting them with Aspergillus. This will result in detectable drug levels in the flies prior to infection, increasing the likelihood of demonstration of drug efficacy. Alternatively, in addition to prophylaxis with the antifungal drug for 24 h prior to infection, the activity of the drug can be assessed in treatment by initiating exposure of the flies to the immediately after infection. When transferring anesthetized flies after infection, place them at the side of the vial to keep them from sticking in the fly food until they recover from anesthesia.
(23) Continue transferral of infected flies to new drug-containing vials every 24 h.
(24) For control purposes, infect a different group of 30–50 female flies with Aspergillus and place them in vials with fly food that does not contain any antifungal drugs.
(25) Count the live flies every 3–6 h after infection with Aspergillus and compare the survival rates in drug-treated and untreated control flies. Carry out at least three independent experiments using 30–50 flies per group and perform statistical analysis of the differences in fly survival using the Kaplan-Meier method.
(26) Assess the tissue fungal burden using qPCR, histopathological analysis and scanning electron microscopy in groups of drug-treated and untreated control flies as described above (Step 15B).
Antifungal drug bioassay (timing 1–2 d).
(27) Place groups of 20 drug-exposed flies at −20°C until use.
(28) Grind and homogenize flies in 0.85% NaCl with a bead-beater homogenizer.
(29) Prepare YAG agar plates and add 5 ml of a 104 C. kefyr ATCC 66028 sterile saline solution to each plate. Allow the inoculum to coat the entire plate prior to aspirating off the fluid with a sterile pipette.
(30) Allow the plates to dry at ambient temperatures for 1 h, and drill a well on the surface of YAG agar plates.
(31) Instill 200 µl of the fly homogenate into drilled wells on the surface of the YAG agar plate that has been previously inoculated with C. kefyr ATCC 66028.
(32) Measure the zone of growth inhibition in millimeters after 24 h of incubation at 37°C and compare it with the zone of growth inhibition caused by known drug concentrations.40
Timing
Steps 1–8: fly preparation, including collection of newly eclosed flies, crossing of Toll-deficient alleles and allowing flies to age to 2–4 days old, 3–4 wk.
Steps 9–11: fungal inoculum preparation, 3 d.
Step 12: needle pricking (injection) assay, 1 h.
Step 13: rolling assay, 1–2 h.
Step 14: ingestion assay, 6–8 h.
Step 15: virulence/pathogenicity assessment including monitoring of fly survival and tissue fungal burden using qPCR/histopathological analysis/scanning electron microscopy, 4–8 d.
Steps 16–20: preparation of antifungal drug-containing fly food vials, 1–2 d.
Steps 21–26: antifungal drug activity assessment including monitoring of fly survival and tissue fungal burden using qPCR/histopathological analysis/scanning electron microscopy, 4–8 d.
Steps 27–32: antifungal drug bioassay, 1–2 d
Problem Handling
Steps 4–8.
Transfer flies to new vials every 3–4 d to renew the stocks continuously.
Among the critical components of fly hatching are optimal humidity and temperature. For optimal humidity, every 3–5 d (depending on how dry the vials appear to be), supplement vials with distilled water to the point of covering the vial surface. For optimal temperature, maintain flies at 25°C. This increases the yield of emerging adult flies. Extreme temperatures inhibit fly hatching and the emergence of adult flies; for example, male flies may become sterile at temperatures above 29°C, whereas temperatures below 20°C will slow hatching and emergence of adult flies. At optimal temperature and humidity, the time span from when female flies lay their eggs in fly food to when adult flies emerge is about 10 d.
20–30 flies is an optimal number for placement in each vial; flies do not produce many eggs if they are overcrowded in vials (>50/vial) so avoiding overcrowding is advisable.
Crossing male flies with virgin female flies at a ratio of 2:3 is optimal. For example, place six male flies with nine virgin female flies in each vial. The reason for this is that placement of more male flies will disturb the female flies and prevent them from producing as many eggs as they would at the optimal male:female ratio. Hence, do not place more than 10 male flies and 15 virgin female flies in a vial.
Step 12B.
To ensure uniform inoculation of all 30–50 flies per group, regularly vortex the Aspergillus suspension between fly inoculations.
Step 13A.
The reason for anesthetizing flies 3–4 min prior to rolling (instead of the few seconds otherwise required to anesthetize them) is to keep the flies from waking up during the 2 min rolling procedure. This facilitates uniform exposure to Aspergillus conidia. In contrast, if flies recover from anesthesia during rolling, exposure to Aspergillus is not uniform because the flies move around in the Petri dish.
Step 14C.
Longer feeding times result in fly death because of dehydration and starvation, as conidia do not constitute an optimal nutritional medium for flies. For example, a 24 h feeding period using these vials results in fly mortality rate of about 50%.
Step 18.
Add the number of yeast particles necessary to saturate them with the drug (Fig. 3B) but not more or fewer. If more than the required number of yeast particles is added, the flies will eat the surface yeast particles that are not soaked with the drug, as the drug will only soak the yeast particles at the bottom. This will lead to suboptimal exposure of the flies to the drug. On the other hand, if fewer than the required number of yeast particles is added, the flies will become stuck in the food because the number of yeast particles will not be sufficient to soak the added drug volume.
Step 19.
Prepare vials in sets of 5–10 at a time depending on how many will be required for the experiments of each week. If drug-containing vials are prepared but not used within 5–10 days, they will dry excessively and not be suitable for use.
Step 21.
Do not let flies starve for more than 6–8 hours because the majority of them will die of starvation. For instance, a 24 hour fasting period will result in death of 50–75% of flies.
Results
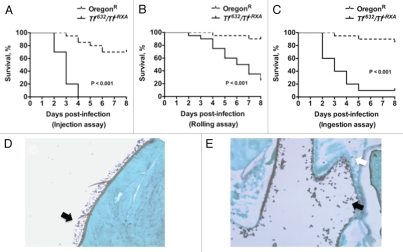
Aspergillus challenge in any of the three infection assays described above results in reproducibly higher mortality rates in Toll-deficient than in WT Drosophila flies Fig. 4A–C).26 Because it delivers Aspergillus conidia directly into the fly hemolymph, the injection assay is the most acute of these three infection models. Mortality after injection of Aspergillus is inoculum-dependent; hence, injection with a needle dipped in a 1 × 107 conidia/ml solution delivers about 700–800 conidia per fly, leading to survival rates of about 60% and 30% at days 3 and 6 post-infection, respectively. In contrast, injection with a needle dipped in a 1 × 1010 conidia/ml solution delivers about 20,000 conidia per fly, resulting in a mortality rate of 100% by day 6 post-infection (Fig. 4A).26 In comparison, the rolling and ingestion assays deliver Aspergillus conidia to epithelial surfaces, specifically, the skin (Fig. 4D) and gastrointestinal tract (Fig. 4E), respectively. Thus, because of the requirement for Aspergillus invasion through mucosal surfaces, the tempo of experimental infection in these two infection assays is more protracted than that in the injection assay (Fig. 4B and C), leading to less acute and lower mortality rates.26 This characteristic of the rolling and ingestion assays may be beneficial in effectively identifying virulence attributes of fungal strains with attenuated virulence using these assays.
Figure 4.
Toll-deficient Drosophila flies are susceptible to Aspergillus challenge. Shown are Kaplan-Meier survival curves for OregonR WT and Tlr632/TlI-RXA flies infected with AF293 using the (A) injection, (B) rolling and (C) ingestion assays. (D) Aspergillus conidia covering a fly's exterior surface (arrow) following the rolling assay. (E) Aspergillus conidia in the lumen of a fly's gastrointestinal tract (black arrow) following the ingestion assay. Later on, conidia invade through the gastrointestinal tract (white arrow) and cause disseminated infection.
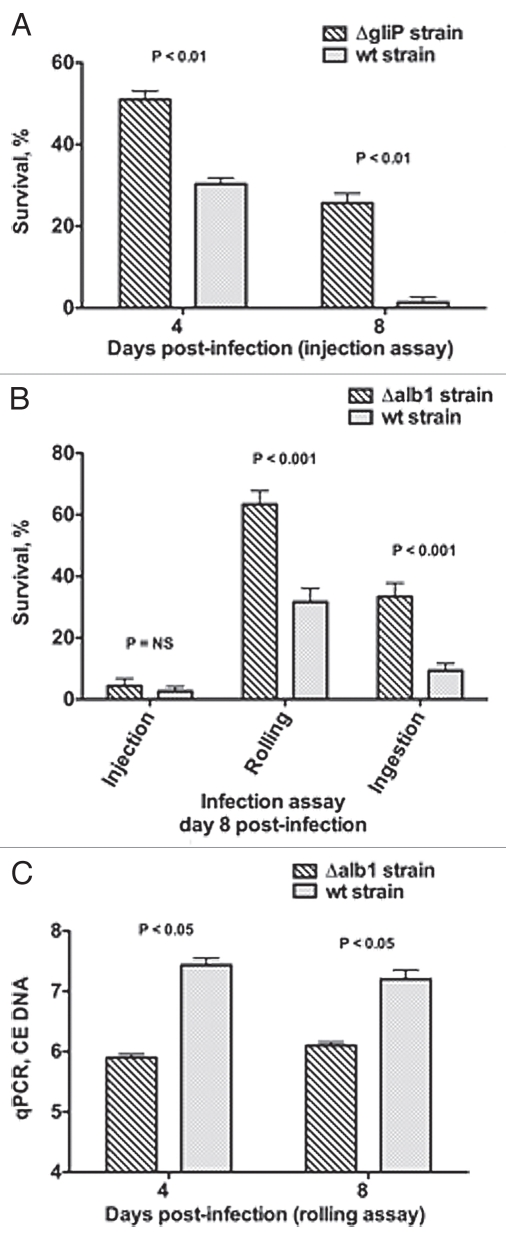
Infection of Toll-deficient Drosophila flies with Aspergillus strains shown to be hypovirulent in mammalian models of IA results in improved survival rates and less acute infection progression than does infection with WT Aspergillus strains. Two examples of such hypovirulent mutants are worth mentioning: (1) the ΔgliP Aspergillus strain that lacks gliotoxin, a virulence factor by induction of host-cell apoptosis and by impairment of phagocyte effector functions,41,42 and (2) the albino Δalb1 Aspergillus strain that lacks melanin production, another virulence factor by quenching free oxygen radicals and inhibiting conidial phagocytosis by neutrophils.38 Similar to that shown in rodent models of IA,37,38 injection of Toll-deficient Drosophila flies with ΔgliP Aspergillus results in better survival rates than does infection with its isogenic WT strain (Fig. 5A).37 In addition, Toll-deficient Drosophila flies infected by rolling in or ingestion of (but not injection of) Δalb1 Aspergillus have lower mortality rates (Fig. 5B) and tissue fungal burdens (Fig. 5C) than do flies infected with its isogenic WT strain.26 This differing behavior of (1) Δalb1 as a function of the mode of introduction of infection and (2) Δalb1 and ΔgliP when injected emphasizes the effect that the site of conidial inoculation and relative virulence potential of various strains have on the acuity of Aspergillus infection in Drosophila flies. It also offers an opportunity for studying differential induction of host immune responses against Aspergillus conidia when infection is introduced into Drosophila flies via various epithelial surfaces.43
Figure 5.
Evaluation of Aspergillus virulence in Toll-deficient Drosophila flies. (A) Survival rates in Tlr632/TlI-RXA flies following injection of the hypovirulent ΔgliP Aspergillus strain or its isogenic WT strain AF293. (B) Survival rates in Tlr632/TlI-RXA flies eight days after infection with the hypovirulent Δalb1 Aspergillus strain compared with infection with its isogenic WT strain B-5233 in the three infection assays. (C) qPCR analysis of the tissue fungal burden in Tlr632/TlI-RXA flies infected with Δalb1 Aspergillus or its isogenic WT strain in the rolling assay. CE, conidial equivalent of Aspergillus fumigatus DNA.
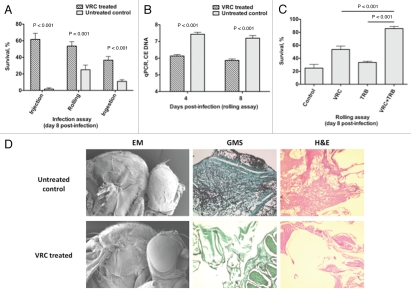
Voriconazole treatment in Aspergillus-infected Toll-deficient Drosophila flies results in reproducible protection against mortality (Fig. 6A) and decreases the tissue fungal burden as determined using qPCR (Fig. 6B), and as observed in histopathological analysis and scanning electron microscopy (Fig. 6D).26 Levels of voriconazole in flies can be detected using a simple antifungal drug bioassay as described above (Steps 27–32). Furthermore, Aspergillus-infected flies given a combination of orally absorbable antifungals known to act synergistically against Aspergillus spp. in vitro (i.e., voriconazole and terbinafine)44 have better survival rates than do flies given single drugs alone (Fig. 6C),26 supporting the role of the Drosophila mini-host model in performing in vivo testing of various antifungal combinations. However, in terms of studying antifungal drug pharmacology, the significant differences in metabolism (e.g., transport, oxidation, pharmacokinetics) among all mini-host models and mammals preclude the use of Drosophila in dose-response and, potentially, toxicity assessment. For example, if DNA methylation is the potential mode of action of an antifungal, flies, in view of their “methylase-deficient” background, may not be suitable for assessing drug efficacy.45
Figure 6.
Voriconazole protects Toll-deficient Drosophila flies against Aspergillus infection. (A) Survival rates in untreated control and voriconazole-treated Tlr632/TlI-RXA flies 8 days after infection with AF293 in the three infection assays. (B) qPCR analysis of the tissue fungal burden in untreated control and voriconazole-treated Tlr632/TlI-RXA flies infected in the rolling assay. (C) Survival rates in untreated control and Tlr632/TlI-RXA flies given voriconazole, terbinafine or a combination of the two drugs 8 days after infection with AF293 in the rolling assay. (D) Histopathological and scanning electron microscopic analysis of the difference in tissue fungal burden in untreated control and voriconazole-treated Tlr632/TlI-RXA flies. VRC, voriconazole; TRB, terbinafine; EM, electron microscopy; GMS, Grocott-Gomori methenamine-silver nitrate stain; H&E, hematoxylin-eosin stain; CE, conidial equivalent of Aspergillus fumigatus DNA.
In summary, despite its shortcomings, the Toll-deficient Drosophila fly model is an inexpensive, easy-to-use heterologous host suitable for quickly studying Aspergillus virulence, antifungal innate immune responses and the efficacy of orally absorbed antifungals against Aspergillus spp. and other fungal pathogens. The level of conservation of key cellular, immune and developmental processes from Drosophila to mammals and the fact that Drosophila has been behind many fundamental modern biological discoveries makes this mini-host well suited for further advances in the study of important areas in experimental mycology.
Acknowledgements
We thank Dr. Tony Ip for providing us with the wild-type and Toll-deficient Drosophila melanogaster and Dr. Georg Halder for helping us introduce this model into the Kontoyiannis laboratory. We also thank Drs. Russell Lewis, Georgios Chamilos, Ronen Ben-Ami, Greg Lamaris and Nathaniel Albert for their outstanding contributions in establishing Drosophila models of infection by several fungal pathogens in addition to Aspergillus species and for substantially expanding the applicability of this fly model. This research is supported in part by the National Institutes of Health through MD Anderson's Cancer Center Support Grant CA016672.
Abbreviations
- IA
invasive aspergillosis
- WT
wild-type
- HMDS
hexamethyldisilazane
- qPCR
real-time quantitative polymerase chain reaction
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/13311
References
- 1.Kontoyiannis DP, Bodey GP. Invasive aspergillosis in 2002: an update. Eur J Clin Microbiol Infect Dis. 2002;21:161–172. doi: 10.1007/s10096-002-0699-z. [DOI] [PubMed] [Google Scholar]
- 2.Kontoyiannis DP, Marr KA, Park BJ, Alexander BD, Anaissie EJ, Walsh TJ, et al. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients 2001–2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clin Infect Dis. 2010;50:1091–1100. doi: 10.1086/651263. [DOI] [PubMed] [Google Scholar]
- 3.Lewis RE, Prince RA, Chi J, Kontoyiannis DP. Itraconazole preexposure attenuates the efficacy of subsequent amphotericin B therapy in a murine model of acute invasive pulmonary aspergillosis. Antimicrob Agents Chemother. 2002;46:3208–3214. doi: 10.1128/AAC.46.10.3208-3214.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petraitis V, Petraitiene R, Lin P, Calis K, Kelaher AM, Muray HA, et al. Efficacy and safety of generic amphotericin B in experimental pulmonary aspergillosis. Antimicrob Agents Chemother. 2005;49:1642–1645. doi: 10.1128/AAC.49.4.1642-1645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vallor AC, Kirkpatrick WR, Najvar LK, Bocanegra R, Kinney MC, Fothergill AW, et al. Assessment of Aspergillus fumigatus burden in pulmonary tissue of guinea pigs by quantitative PCR, galactomannan enzyme immunoassay and quantitative culture. Antimicrob Agents Chemother. 2008;52:2593–2598. doi: 10.1128/AAC.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van de Sande WW, Mathot RA, ten Kate MT, van Vianen W, Tavakol M, Rijnders BJ, et al. Combination therapy of advanced invasive pulmonary aspergillosis in transiently neutropenic rats using human pharmacokinetic equivalent doses of voriconazole and anidulafungin. Antimicrob Agents Chemother. 2009;53:2005–2013. doi: 10.1128/AAC.01556-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pradel E, Ewbank JJ. Genetic models of pathogenesis. Annu Rev Genet. 2004;38:347–363. doi: 10.1146/annurev.genet.38.072902.092528. [DOI] [PubMed] [Google Scholar]
- 8.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 9.Jones T, Federspiel NA, Chibana H, Dungan J, Kalman S, Magee BB, et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci USA. 2004;101:7329–7334. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 12.Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: differential expression of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apidianakis Y, Rahme LG, Heitman J, Ausubel FM, Calderwood SB, Mylonakis E. Challenge of Drosophila melanogaster with Cryptococcus neoformans and role of the innate immune response. Eukaryot Cell. 2004;3:413–419. doi: 10.1128/EC.3.2.413-419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mylonakis E, Casadevall A, Ausubel FM. Exploiting amoeboid and non-vertebrate animal model systems to study the virulence of human pathogenic fungi. PLoS Pathog. 2007;3:101. doi: 10.1371/journal.ppat.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alarco AM, Marcil A, Chen J, Suter B, Thomas D, Whiteway M. Immune-deficient Drosophila melanogaster: a model for the innate immune response to human fungal pathogens. J Immunol. 2004;172:5622–5628. doi: 10.4049/jimmunol.172.9.5622. [DOI] [PubMed] [Google Scholar]
- 16.Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun. 2005;73:3842–3850. doi: 10.1128/IAI.73.7.3842-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mylonakis E, Ausubel FM, Perfect JR, Heitman J, Calderwood SB. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc Natl Acad Sci USA. 2002;99:15675–15680. doi: 10.1073/pnas.232568599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steenbergen JN, Nosanchuk JD, Maliaris SD, Casadevall A. Cryptococcus neoformans virulence is enhanced after growth in the genetically malleable host Dictyostelium discoideum. Infect Immun. 2003;71:4862–4872. doi: 10.1128/IAI.71.9.4862-4872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennan CA, Anderson KV. Drosophila: the genetics of innate immune recognition and response. Annu Rev Immunol. 2004;22:457–483. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nat Immunol. 2002;3:121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- 21.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 22.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci USA. 2001;98:12590–12599. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in drosophila. EMBO J. 2002;21:2568–2579. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leclerc V, Reichhart JM. The immune response of Drosophila melanogaster. Immunol Rev. 2004;198:59–71. doi: 10.1111/j.0105-2896.2004.0130.x. [DOI] [PubMed] [Google Scholar]
- 25.Lionakis MS, Kontoyiannis DP. Fruit flies as a minihost model for studying drug activity and virulence in Aspergillus. Med Mycol. 2005;43:111–114. doi: 10.1080/13693780400020030. [DOI] [PubMed] [Google Scholar]
- 26.Lionakis MS, Lewis RE, May GS, Wiederhold NP, Albert ND, Halder G, et al. Toll-deficient Drosophila flies as a fast, high-throughput model for the study of antifungal drug efficacy against invasive aspergillosis and Aspergillus virulence. J Infect Dis. 2005;191:1188–1195. doi: 10.1086/428587. [DOI] [PubMed] [Google Scholar]
- 27.Chamilos G, Bignell EM, Schrettl M, Lewis RE, Leventakos K, May GS, et al. Exploring the concordance of Aspergillus fumigatus pathogenicity in mice and Toll-deficient flies. Med Mycol. 2010;48:506–510. doi: 10.3109/13693780903225813. [DOI] [PubMed] [Google Scholar]
- 28.Herbrecht R, Denning DW, Patterson TF, Bennett JE, Greene RE, Oestmann JW, et al. Voriconazole versus amphotericin B for primary therapy of invasive aspergillosis. N Engl J Med. 2002;347:408–415. doi: 10.1056/NEJMoa020191. [DOI] [PubMed] [Google Scholar]
- 29.Ben-Ami R, Lamaris GA, Lewis RE, Kontoyiannis DP. Interstrain variability in the virulence of Aspergillus fumigatus and Aspergillus terreus in a Toll-deficient Drosophila fly model of invasive aspergillosis. Med Mycol. 2010;48:310–317. doi: 10.1080/13693780903148346. [DOI] [PubMed] [Google Scholar]
- 30.Chamilos G, Lewis RE, Hu J, Xiao L, Zal T, Gilliet M, et al. Drosophila melanogaster as a model host to dissect the immunopathogenesis of zygomycosis. Proc Natl Acad Sci USA. 2008;105:9367–9372. doi: 10.1073/pnas.0709578105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pongas GN, Ben-Ami R, Lewis RE, Walsh TJ, Kontoyiannis DP. Culture medium composition affects the lethality of Cunninghamella bertholletiae in a fly model of mucormycosis. Antimicrob Agents Chemother. 2009;53:4569. doi: 10.1128/AAC.00994-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamaris GA, Chamilos G, Lewis RE, Kontoyiannis DP. Virulence studies of Scedosporium and Fusarium species in Drosophila melanogaster. J Infect Dis. 2007;196:1860–1864. doi: 10.1086/523765. [DOI] [PubMed] [Google Scholar]
- 33.Chamilos G, Lionakis MS, Lewis RE, Lopez-Ribot JL, Saville SP, Albert ND, et al. Drosophila melanogaster as a facile model for large-scale studies of virulence mechanisms and antifungal drug efficacy in Candida species. J Infect Dis. 2006;193:1014–1022. doi: 10.1086/500950. [DOI] [PubMed] [Google Scholar]
- 34.Tekaia F, Latge JP. Aspergillus fumigatus: saprophyte or pathogen? Curr Opin Microbiol. 2005;8:385–392. doi: 10.1016/j.mib.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 35.Bhabhra R, Miley MD, Mylonakis E, Boettner D, Fortwendel J, Panepinto JC, et al. Disruption of the Aspergillus fumigatus gene encoding nucleolar protein CgrA impairs thermotolerant growth and reduces virulence. Infect Immun. 2004;72:4731–4740. doi: 10.1128/IAI.72.8.4731-4740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kontoyiannis DP, Lionakis MS, Halder G. 14th Focus on Fungal Infections. New Orleans LA, USA: 2004. Toll pathway in Drosophila melanogaster: A possible role to study the impact of immune senescence in poor responses against Aspergillus fumigatus. Abstract # 31. [Google Scholar]
- 37.Spikes S, Xu R, Nguyen CK, Chamilos G, Kontoyiannis DP, Jacobson RH, et al. Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J Infect Dis. 2008;197:479–486. doi: 10.1086/525044. [DOI] [PubMed] [Google Scholar]
- 38.Tsai HF, Chang YC, Washburn RG, Wheeler MH, Kwon-Chung KJ. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J Bacteriol. 1998;180:3031–3038. doi: 10.1128/jb.180.12.3031-3038.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowman JC, Abruzzo GK, Anderson JW, Flattery AM, Gill CJ, Pikounis VB, et al. Quantitative PCR assay to measure Aspergillus fumigatus burden in a murine model of disseminated aspergillosis: demonstration of efficacy of caspofungin acetate. Antimicrob Agents Chemother. 2001;45:3474–3481. doi: 10.1128/AAC.45.12.3474-3481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Odds FC, Dupont B, Rinaldi MG, Stevens DA, Warnock DW, Woestenborghs R. Bioassays for itraconazole blood levels: an interlaboratory collaborative study. J Antimicrob Chemother. 1999;43:723–727. doi: 10.1093/jac/43.5.723. [DOI] [PubMed] [Google Scholar]
- 41.Stanzani M, Orciuolo E, Lewis R, Kontoyiannis DP, Martins SL, St. John LS, et al. Aspergillus fumigatus suppresses the human cellular immune response via gliotoxin-mediated apoptosis of monocytes. Blood. 2005;105:2258–2265. doi: 10.1182/blood-2004-09-3421. [DOI] [PubMed] [Google Scholar]
- 42.Coméra C, André K, Laffitte J, Collet X, Galtier P, Maridonneau-Parini I. Gliotoxin from Aspergillus fumigatus affects phagocytosis and the organization of the actin cytoskeleton by distinct signalling pathways in human neutrophils. Microbes Infect. 2007;9:47–54. doi: 10.1016/j.micinf.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart JM, Lemaitre B, et al. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity. 2000;13:737–748. doi: 10.1016/s1074-7613(00)00072-8. [DOI] [PubMed] [Google Scholar]
- 44.Ryder NS, Leitner I. Synergistic interaction of terbinafine with triazoles or amphotericin B against Aspergillus species. Med Mycol. 2001;39:91–95. doi: 10.1080/mmy.39.1.91.95. [DOI] [PubMed] [Google Scholar]
- 45.Jowett T, Wajidi MF, Oxtoby E, Wolf CR. Mammalian genes expressed in Drosophila: a transgenic model for the study of mechanisms of chemical mutagenesis and metabolism. EMBO J. 1991;10:1075–1081. doi: 10.1002/j.1460-2075.1991.tb08047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]