Abstract
The capsular polysaccharides of Cryptococcus neoformans have historically been divided into three components namely, glucuronoxylomannan (GXM), galactoxylomannan (GalXM) and mannoprotein (MP) but their relative spatial-geographical relationship in the capsule is unknown. To explore this problem would require the capacity for visualizing these components in the capsule. Prior studies have reported serological reagents to GXM and GalXM but no antibodies are available against MPs. Consequently, we immunized Balb/c mice with C. neoformans recombinant mannoprotein 98 and recovered twelve monoclonal antibodies (mAbs) of which one, an IgG2a designated 18F2, bound to intact cells by immunofluorescence. mAb 18F2 bound to the cell wall surface in acapsular and encapsulated cells. Using mAb 18F2 and previously generated antibodies to GXM and GalXM we have established the localization of three capsular components GXM, GalXM and one type of mannoprotein, MP98 on the C. neoformans cell. The results show that MP98, like GalXM, is found near the cell wall and this information allows us to begin to discern the geography of the cryptococcal capsule.
Key words: C. neoformans, MP98, chitin deacetylase, monoclonal antibodies, localization
Introduction
Cryptococcus neoformans is a fungal pathogen that is a relatively frequent cause of disease in persons with impaired cell-mediated immunity.1,2 In the 1980s, C. neoformans emerged as a major pathogen for patients with AIDS and more recently is increasingly associated with disease in patients with organ transplants.3–5
C. neoformans is unusual among the pathogenic fungi in that it has a polysaccharide capsule that contributes to virulence by being antiphagocytic, serving as an antioxidant and interfering with immunity.1,6 Historically, the C. neoformans capsular polysaccharide was believed to have three components known as glucuronoxylomannan (GXM), galactoxylomannan (GalXM) and mannoproteins (MP).5,7,8 A recent study proposed that GalXM be renamed to ‘glucuronoxylomannogalactan’ due to the presence of glucuronic acid.9 Although we recognize that the term GalXM is inadequate for this polysaccharide we continue to use GalXM for continuity in the literature, and due to concern that until the structure is fully solved additional renaming may be necessary.9,10
C. neoformans capsular composition has been inferred based largely on analysis of shed exopolysaccharides that accumulate in culture supernatants.11 Currently, there is no direct evidence for a structural role of GalXM and MP in capsule assembly or architecture. Earlier studies using acapsular strain cap67 suggested that GalXM and MPs are not covalently bound to the cell wall.12 MPs are thought to be produced intracellularly and then released into the periplasmic space between the cell membrane and cell wall, where they diffuse slowly through the wall to such extracellular environments as the cell wall and capsule.12,13 However, other studies have shown putative GPI-anchored MPs that are potentially cross-linked to β1,6 glucan in the cell wall.8
Historically, mannoproteins were considered a minor component of the capsule but there is little direct experimental evidence to support this belief. MPs were identified in culture filtrates by 13C-NMR analysis of the GXM-free portion of the GalXM enriched fraction using concanavalin A affinity chromatography.14,15 Recent studies to elucidate the structural features of two mannoproteins, MP88 and MP98 revealed conserved motifs such as a signal sequence, a functional domain, a serine/threonine-rich region and a site for attachment of a glycosylphosphatidylinositol (GPI) anchor. These mannoproteins contain extensive O-mannosylation within the serine/threonine region.8,16
C. neoformans MPs are highly immunogenic.8,17,18 MP antigens have been implicated in the induction of pro-inflammatory responses against C. neoformans by their association with delayed type hypersensitivity (DTH).19 These proteins can regulate the expression of cytokines such as IL-12, IL-6, IL-10, IFNγ, IL-8 and TNFα, each of which is associated with effective responses to cryptococcal infection.14,20–25 MPs elicit a protective cell mediated immune response against C. neoformans and other medically important fungi such as Candida albicans by promoting the maturation and activation of dendritic cells which in turn activate CD4+ and CD8+ T-lymphocytes.21,25 Mannosylation is required for optimal T-cell stimulation,17 which is consistent with the finding that MP effects are mediated through host mannose-binding lectin18 or mannose receptors.8,26,27
Several C. neoformans MPs have been isolated in culture filtrates. Levitz et al. identified a 98 kDa mannoprotein (MP98) that was reactive with a part of murine T-cell hybridomas.28 The encoding gene (chitin deacetylase 2, CDA2) was cloned and MP98 was found to contain the same conserved motifs as mentioned above. Additionally, the protein also has a domain with similarity to chitin deacetylases from other fungal species, including Saccharomyces cerevisiae, Mucor rouxii and Phycomyces blakesleeanus.28 Chitin deacetylases function to convert chitin to chitosan, the deacetylated form of chitin. A recent study found that C. neoformans encodes three chitin deacetylases and a polysaccharide deacetylase. As part of that study chitin deacetylase deletion strains cda1Δ, cda2Δ, cda3Δ and a polysaccharide deacetylase fpd1Δ were generated.29 The results revealed that deletion of a single chitin deacetylase does not adversely affect the ability of C. neoformans to produce chitosan, whereas the triple deletion cda1Δcda2Δcda3Δ and quadruple deletion cda1Δcda2Δcda3Δfpd1Δ abolished chitosan production. Strains with the triple and quadruple deletions were shown to have incomplete mother daughter cell separation, were sensitive to cell wall inhibitors, had increased capsule size and were unable to retain melanin.29
In this study, we have generated monoclonal antibodies (mAb) to MP98 and used them to study the cellular location of MP98 relative to other capsule components. The results provide the first simultaneous localization of the GXM, GalXM and MP in the C. neoformans capsule and demonstrate that they occupy spatially separate and discrete regions.
Results
Strains deleted of MP98 (CDA2) and CDA1.
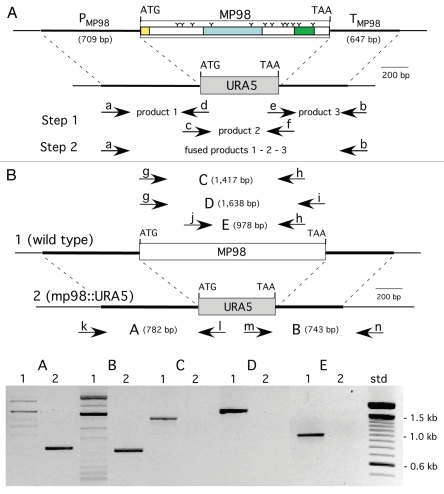
Deletion cassettes were constructed by PCR fusion, substituting the ORF of a CDA with that of URA5, as shown in (Fig. 1A) for MP98(CDA2). Requisite for gene replacement were the 600–700 bp of promoter and terminator sequences of each CDA gene, which also enabled transcription of URA5. Ura+ transformed cells of strain JEC34 were screened by PCR to determine if gene replacement had occurred. Primers specific to CDA and URA5 DNA sequence, when used in combination yielded novel PCR products as predicted (Fig. 1B) for the deletion of the MP98(CDA2) gene and its replacement with the URA5 gene (data not shown for the CDA1 deletion/replacement). A second assay was done to confirm that the MP98 protein was also missing from the JEC34 mp98 (cda2)::URA5 strain, as shown in (Fig. 2). This bioassay incorporated the specificity of the T-cell hybridoma P1D6 in recognizing an epitope on the MP98(CDA2) protein; recognition activates IL-2 secretion.28 No MP98(CDA2) protein was detected in cell homogenates of the JEC34 mp98::URA5 strain, while clearly evident in the untransformed JEC34 samples. A similar bioassay was not available for the CDA1 deleted strain.
Figure 1.
Construction of MP98(CDA2) deletion in strain JEC 34 (serotype D). PC R was used to fuse the promoter region of MP98 (PMP98) and its terminator (TMP98) with the ORF of URA5 to generate a cassette for gene replacement, as shown in (A). Primers are designated in lower case (a–h) and their sequences are given in Table 1. Domains of the MP98(CDA2) protein (signal peptide, catalytic and Serine/Threonine-rich) are boxed with “Y” used to identify putative N-glycosylation sites.28 As shown in (B), the strategy to confirm gene replacement for Ura+ transformants was by PC R. Genomic DNAs were used as templates with five sets of primers (g–n, Table 1) to generate products (A–E). Shown below the maps is an electrophoretogram comparing PC R products from wild-type control DNA (1) with a JEC34 Ura+ transformant (2). Each of the five PC R products of the transformant were indicative of gene replacement and a new genotype (mp98::URA5).
Figure 2.
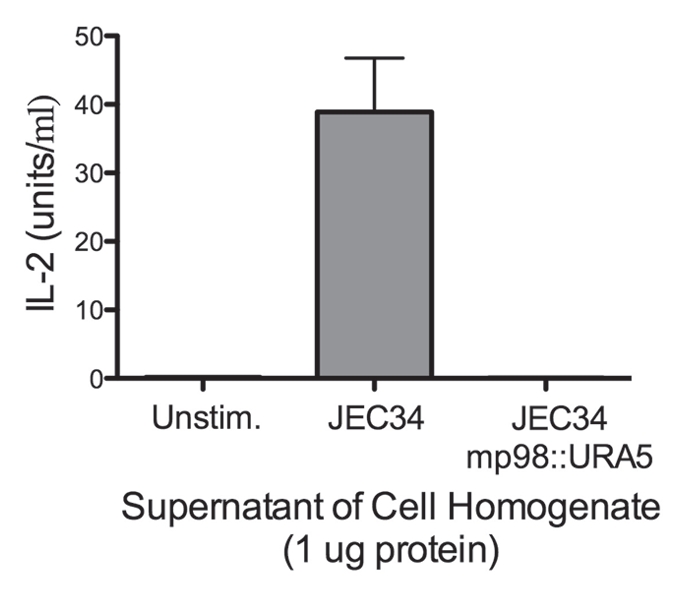
Strain deleted of MP98(CDA2) lacks MP98 protein. Supernatant of a cell homogenate of untransformed strain JEC34 was compared to that of the Ura+ transformant with genotype mp98(cda2)::URA5 ascribed by PCR analysis in Figure 1. Unstimulated sample had no protein added. Values are the average of four measurements. This bioassay incorporated the specificity of the T-cell hybridoma P1D6 in recognizing an epitope on the MP98(CDA2) protein; recognition activates IL-2 secretion.
Generation and characterization of mAbs to MP98.
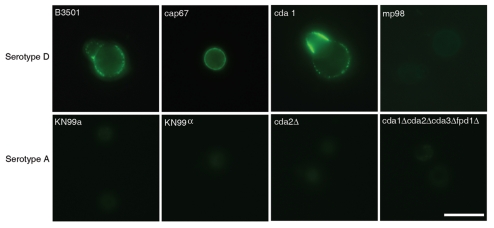
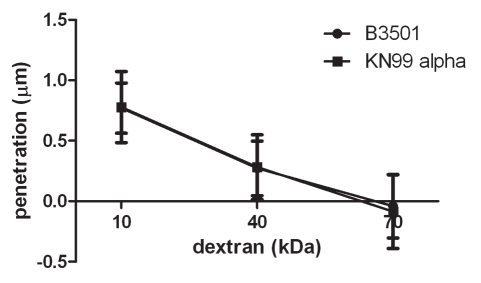
We screened hybridoma supernatants by ELISA and recovered twelve hybridoma cell lines producing mAbs of the IgG1, IgG2a and IgG2b isotypes that reacted specifically with MP98. Three of these mAbs recognized a recombinant mannosylated MP98 produced from P. pastoris, while nine of them recognized only the unglycosylated protein produced in E. coli by ELISA. However, when these antibodies were tested for reactivity to proteins in C. neoformans lysates and recombinant glycosylated and unglycosylated MP98 by immunoblot, no signal was detected (data not shown). Immunofluorescence was used on B3501 and cap67 C. neoformans cells as a secondary measure of binding. We found that only mAb, 18F2 (IgG2a), gave a positive fluorescence signal; this antibody bound to the unglycosylated form of MP98 produced in E. coli by ELISA. The mAb 18F2 binding pattern was punctate and the most intense fluorescence was present during budding. To determine if the mAb 18F2 was binding the MP98 protein, we first tested several chitin deacetylase (CDA) mutants of serotype A including cda2Δ, the quadruple deletion strain cda1Δcda2Δcda3Δfpd1Δ and the parental strains KN99a and KN99α. The mAb 18F2 did not bind to the parental or the mutant strains. We then tested a second set of mutant strains, cda1::URA5 and cda2::URA5 constructed in the serotype D strain JEC34. The results revealed that mAb 18F2 bound to the cda1::URA5 mutant strain which contains the CDA2 gene that encodes for the MP98 protein. However, the antibody did not bind to the mp98(cda2)::URA5 mutant strain demonstrating that mAb 18F2 binds to MP98 and that this antibody is specific for MP98 encoded by the CDA2 allele of the JEC34 (serotype D) background (Fig. 3). To rule out a false negative result based on differences on capsule penetration we measured dextran penetration for each strain. The capsule thicknesses of B3501 and KN99α was first measured by India ink staining, the sizes were 1.389 ± 0.266 µm and 0.894 ± 0.243 µm respectively (n = 70). After normalization of the capsule thickness, dextran penetration for B3501 and KN99α was shown to be essentially identical (Fig. 4). This suggests that antibody reactivities for MP98 in C. neoformans strains were not due to the hindrance of mAb penetration. We also tested whether the mAb 18F2 bound to strains of other serotypes. We tested strains H99 (serotype A), strain 123-97 (serotype B), strain 106-97 (serotype C), acapsular strain cap67 (derived from serotype D), strain B3501 (serotype D) and strain 92-203 (serotype AD). No reactivity was observed with any of the encapsulated strains of the other serotypes (data not shown). mAb 18F2 was found at or near the cell bodies for all serotype D and strains derived from serotype D.
Figure 3.
mAb 18F2 is limited to strain JEC21. Immunofluorescence using anti-MP98 MAb 18F2 with serotype D and serotype A, C. neoformans strains. MP98 is located in a punctate pattern in B3501 and near the cell wall in cap67 (top parts). The cda1::URA5 mutant shows an accumulation of MP98 protein in the emerging bud. The negative control, mp98(cda2)::URA5 mutant shows only background fluorescence. Parental strains KN99a and KN99α, mutant cda2Δ and the cda1Δcda2Δcda3Δfpd1Δ quadruple mutant each display only background fluorescence (lower parts). Scale bar is 5 µm.
Figure 4.
Capsules from B3501 and KN99α had similar permeability to dextran. A dextran penetration assay was performed using tetramethylrhodamine-labeled dextran of molecular weights 10, 40 and 70 kDa. The penetration of dextrans into the capsule was similar for both strains.
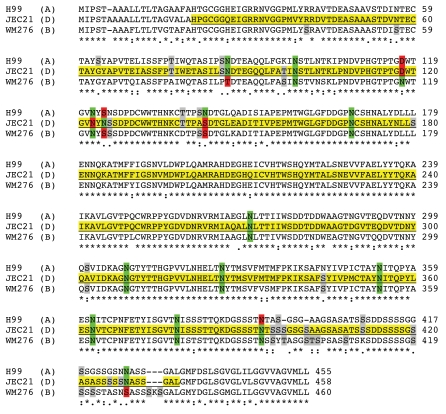
A comparison of the predicted MP98 protein sequences from JEC21 (serotype D), H99 (serotype A) and WM276 (serotype B) strain revealed 42 amino acid differences in the relevant region of MP98 that was cloned in E. coli and used for immunizing mice. These differences suggest a possible explanation for the lack of cross-serotype reactivity. A few differences affect potential N- and O-glycosylation sites, which could dramatically alter antibody specificity or block antibody binding if the epitope is masked by glycosylation (Fig. 5).
Figure 5.
Alignment of MP98 protein sequences encoded by H99, JEC 21 and WM 276 (Serotype B) C. neoformans strains. Sequence highlighted in yellow is for the MP98 protein encoded by the JEC21 allele that was expressed in E. coli and used for generating mAbs. The green areas are putative N-linked glycosylation sites. The red areas are not N-glycosylated in protein encoded by other alleles. The gray areas are possible sites of O-glycosylation differences among protein sequences.
Localization of MP98 on cells.
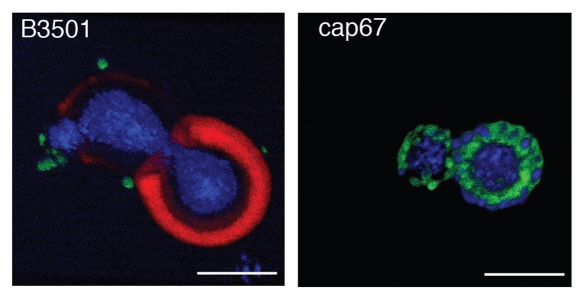
Three-dimensional confocal microscopy reconstructions revealed that for B3501 and cap67 cells, mAb 18F2 for MP98 was located at or near the cell wall. Immunofluorescence studies of B3501 using mAb 18B7 for GXM localized this polysaccharide in the capsule.30 The polyclonal antibody for GalXM localized this polysaccharide in discrete pockets on the outer edge of the capsule as previously described.31 In the acapsular strain cap67, MP98 and GalXM were in close proximity to each other near the cell wall (Fig. 6).
Figure 6.
Reconstruction of capsular components. 3D confocal reconstructions from Z-series stacks of B3501 and cap67 cells. Green: GalXM, red: GXM, blue: MP98. Scale bar is 5 µm.
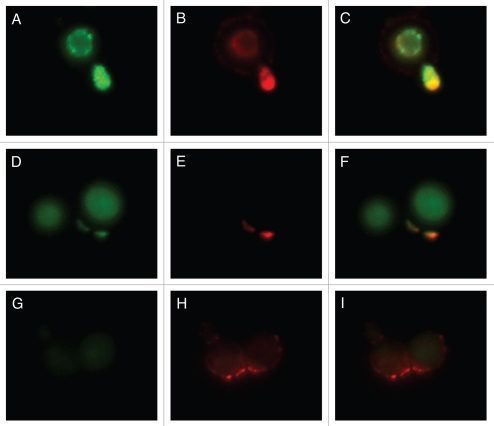
Whereas mAb 18F2 is specific for MP98, Con A should bind to all MPs. Therefore, we investigated whether binding of mAb 18F2 localized with Con A colocalized by incubating B3501 and the MP98 mutant, cda2::URA5 with Con A and the mAb 18F2. The results reveal that in strain B3501, mAb 18F2 colocalizes with Con A and that MP98 is especially present in regions of budding. Con A is also present on the capsular edge but no colocalization with mAb 18F2 is seen in this area (Fig. 7A–C). We also found colocalization of mAb 18F2 and Con A near the cell wall with a concentration near bud scars (Fig. 7D–F). As expected, mAb 18F2 did not specifically bind to the MP98 deletion strain; however, Con A binding was seen reflecting reactivity with MPs other than MP98 (Fig. 7G–I).
Figure 7.
Colocalization studies of MP98 and other mannoproteins using mAb 18F2 and Con A lectin respectively. C. neoformans strain B3501 parts (A–F) and MP98 mutant strain parts (G–I). Left parts are C. neoformans treated with mAb 18F2 (green), middle parts Con A (red) and right parts are the merged images. In B3501 (parts A–F) Con A stains the mannoproteins of cell wall and mannoproteins found on the capsular edge, the mAb 18F2 specifically recognizes and binds to MP98. Parts (A–C) also show that MP98 is mostly localized next to the cell wall during budding. Parts (D–F) shows that 18F2 localizes to bud scars. Parts (G–I) shows that 18F2 is specific for MP98 but that other MPs are available to bind to Con A. Scale bar is 5 µm.
Discussion
C. neoformans mannoproteins have an important role in inducing T-cell mediated immune responses that contribute to host protection against this pathogen.32 MPs bind to the mannose receptor (MR) and elicit the release of inflammatory cytokines.18,26,27,29 It was suggested that MPs could enhance HIV replication in peripheral blood mononuclear cells (PBMCs).32 MPs isolated from C. neoformans capsule supernatants manifest immunomodulatory properties.14,27 MPs have also been used as potential vaccine components, which have proven to be beneficial in elucidating some of their functions.14
Mannosylation enhances the immunogenicity of MP by facilitating recognition by MRs on dendritic cells (DCs), resulting in efficient uptake, processing and presentation of antigen to T cells.17 Here we used unglycosylated recombinant MP98 produced in E. coli to immunize mice and generate monoclonal antibodies. Despite the recovery of numerous mAbs reactive with MP98 by ELISA, only one mAb, 18F2, bound to cells by immunofluorescence microscopy and was useful for our studies since it yielded strong signals. The lack of reactivity of these mAbs by immunoblot or immunofluorescence (except for mAb 18F2) suggests that the ELISA screen selected primarily for mAbs that recognize epitopes only when the protein is immobilized to polystyrene. This could reflect exposure of hidden epitopes when the protein undergoes conformation changes upon adsorption to the plastic support that are not present or not accessible when the protein is in solution or on immunoblot membranes.
mAb 18F2 bound in a punctate pattern to encapsulated strains. The cell wall surface binding of mAb 18F2 on strain cap67 and B3501 was consistent with and confirmatory of prior studies that placed MP in the cell wall of acapsular strains.7,33 Additionally, mAb 18F2 appeared to be serotype D strain specific since it did not bind to MP98 of serotype A background, namely strains KN99a or KN99α. To rule out the possibility that differences in capsule size or polysaccharide structure from different serotypes affected the ability of mAb 18F2 to penetrate, we investigated capsule permeability using a dextran penetration assay. Dextran penetration for the serotype D and A strains tested was essentially the same, suggesting that the inability of mAb 18F2 to bind to serotype A was not a false negative caused by permeability differences. The most reasonable explanation for our findings is that mannoproteins are antigenically diverse among different serotypes of Cryptococcus. A sequence alignment comparing sequences from JEC21 (serotype D), H99 (serotype A) and WM276 (serotype B) strain revealed 42 amino acid differences in the relevant region of MP98 that was cloned in E. coli and used to vaccinate mice. The sequence polymorphisms include potential N-glycosylation sites that could dramatically alter antibody specificity. We also explored the specificity of mAb 18F2 by carrying out co-localization experiments with Con A, a mannose-binding lectin which is known to recognize glycosylated MP.27 mAb 18F2 and ConA colocalized on B3501 cells especially during budding and along bud scars and not for the MP98 mutant. We also detected that Con A was able to bind to other types of mannoproteins along the capsular edge and no colocalization was detected with mAb 18F2 in this area. The absence of competition between mAb 18F2 and ConA implies that they bind to separate epitopes.
To understand the spatial relationship of GXM, GalXM and MP on the C. neoformans capsule, we attempted to localize each using specific antibodies to each of the three components and observing their binding by immuofluorescence. The results provide the first geographical distribution of these three components in the capsule, with Abs to both GalXM and MP showing a punctuate-like appearance near the cell wall consistent with material that is being transported to the extracellular space. These results provide strong support for the view that capsule-associated GalXM and MP are probably materials in the process of transport to the extracellular space while only GXM is an integral capsular component.
Materials and Methods
Animals and hybridomas.
Balb/c, 6–8 week old, female mice were obtained from the National Cancer Institute (NCI). Mice were injected with 5 µg of recombinant MP98 expressed in either Escherichia coli or Pichia pastoris, systems that yield non-glycosylated and glycosylated proteins, respectively.17 Primary immunizations were done by mixing the proteins with complete Freund's adjuvant (CFA). Serum titers were measured by ELISA and mice were subsequently challenged with 5 µg of antigen in incomplete Freund's adjuvant (IFA) at day 14. Mice were rested for a month to allow titers to MP to decrease. The spleen used for fusion came from a mouse injected with the non-glycosylated MP98 from E. coli. Twenty four hours prior to fusion, mice were injected with 5 µg MP98 in IFA. Mice were sacrificed by asphyxiation with CO2, the spleen was harvested and splenocytes were fused with the Ag8.8 myeloma cell line. All animal experiments were done according to institutional guidelines.
ELISA for MP.
Ab binding to MP98 expressed in E. coli and P. pastoris was measured by ELISA. Briefly, Costar® polystyrene plates were coated with 5 µg/ml of MP98 proteins in phosphate-buffered saline (PBS) and blocked with 200 µl of 1% bovine serum albumin in PBS. Hybridomas making mAb to MP98 were identified by ELISA. Binding of the Abs was detected by isotype-specific alkaline-phosphatase-conjugated goat anti-mouse IgM, IgG (H+L) and IgA reagents. For all steps, incubations were done at 37°C for 1 h and absorbances were measured with a microtiter plate reader at 405 nm (Labsystems Multiskan, Franklin, MA).
Strains.
C. neoformans strains cap67 (acapsular), B3501 (serotype D) and 24067 (serotype D) were obtained from the American Type Tissue Collection (Manassas, VA). Strains 123-97 (serotype B), 106-97 (serotype C), were obtained from Dr. Thomas Mitchell at Duke University and strain 92–203 (serotype AD) was obtained from Dr. Mary Brandt at CDC. C. neoformans strains KN99α, KN99a, cda2Δ, cda1Δcda2Δcda3Δfpd1Δ derived from the KN99 background29 were obtained from Dr. Jennifer Lodge. Strains cda1::URA5 and cda2::URA5 derived from the JEC34 background was generated as described below.
Bioinformatics.
DNA and protein sequences for CDA2 and MP98, respectively were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/) for strain JEC21 (serotype D). Protein sequences were from the Broad Institute (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/MultiHome.html) for strain H99 (serotype A) and from Dr. James Kronstad for WM276 (serotype B). Alignments were made using CLUSTAL 2.0.1 multiple sequence alignment with default settings (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
Construction of strains deleted of the MP98(CDA2) and CDA1 genes.
PCR was used to generate and step-wise fuse into one fragment DNAs flanking the MP98(CDA2) or CDA1 ORF and the URA5 ORF of Cryptococcus, as depicted in Figure 1A. Primer sequences that were used are provided in Table 1. The template for the URA5 gene was linearized pPM8 plasmid DNA36 and for the promoter and terminator regions, flanking the MP98 and CDA1 genes was genomic DNA of Cryptococcus strain B3501. DNA was amplified using BIO-X-ACT polymerase (Bioline). Each cycle consisted of 45 s denaturation at 95°C, 45 s annealing at 55°C and 1–2.5 min extension at 72°C. Before the first cycle, denaturation was at 95°C for 5 min and after the last cycle, extension time was increased to 7 min. PCR products were separated by electrophoresis in agarose gels, stained, excised and purified using a gel extraction kit (QIAGEN). The final PCR product for each deletion cassette was transformed into C. neoformans strain JEC34(MATa, ura5)37 by electroporation in 0.4 cm cuvettes (BioRad) following the protocol of.38 Selection of Ura+ transformants was on YNB (6.7 g yeast nitrogen base without amino acids, 2% glucose) medium plus agar with incubation at 30°C for 3 d. Further culturing was done in liquid YNB medium at 30°C with shaking. Genomic DNAs of the Ura+ transformants and JEC34 were purified using a MasterPure™ Yeast DNA Kit (Epicentre Biotechnologies) according to the manufacturer's instructions. Replacement of the MP98 gene with the URA5 gene was confirmed by PCR using primers (g–n) listed in Table 1. Sizing of the products was by electrophoresis in agarose gels. The strain displaying the expected PCR fragments shown in Figure 1B was named JEC34 mp98::URA5. Similarly, the strain confirmed by PCR (data not shown) to be deleted of the CDA1 gene was named JEC34 cda1::URA5.
Table 1.
Primers to make MP98(CDA2) deletion cassette and confirm gene replacement
| Primer | Sequence1 | |
| a2 | NFJEC21 | tga tgc cga gga ggt tat |
| b | MP98R | cat tcg taa tcc cgt gcc |
| c | MP98URA5-F | cgc ctt tta atc aca cag gca ctt gat cAT GTC CTC CC |
| d | MP98URA5-R | GGG AGG ACA Tga tca agt gcc tgt gtg att aaa agg cg |
| e | URA5MP98-F | CGG TGT TCA GAG GTC TTA Agt gat gtg ctt tga cga agg |
| f | URA5MP98-R | cct tcg tca aag cac atc acT TAA GAC CTC TGA ACA CC G |
| g | NF-1 | tta atc aca cag gcg aa |
| h | NR-5′ | tgc ttg agc tgc ttc cat ct |
| i | NR-5 | tcg ttg agc tgc ttc cat ct |
| j | NF-5 | tgg acc ggt gtc aac tac aa |
| k | MP98-F | tgg cat ctg acc ttg ttg |
| l | URA-R | CC T CGA TGA AGG CAA CTT |
| m | URA-F | AGC TGC AGG AGG TCA GGA A |
| n | MP98-R2 | cag tta cca cca gtc aga |
Lower case designates MP98(CDA2) sequence and upper case URA5 sequence.
Primer designations in Figure 1B.
T cell hybridoma assay.
Strains of Cryptococcus to be tested were grown in liquid YNB medium for 3 d at 30°C with shaking. Cells were collected by centrifugation, suspended in 10 mM Tris, 1 mM EDTA, pH 8 and disrupted with 0.7 mm zirconium beads in a Minibeadbeater-8 (Biospec Products) at 4°C. Supernatants were collected following centrifugation at 20,000 × g for 30 min and stored at −80°C. Protein concentrations were determined by the BCA assay (Pierce). Samples of 10, 2 and 0.4 µg of total protein were assayed for the presence of MP 98 protein as previously described17,28 using the T cell hybridoma cell line, P1D6. This cell line specifically recognizes the MP98 protein when presented by antigen-presenting cells, which were bone marrow dendritic cells from C57Bl/6 mice. Relative amounts of MP98 protein in samples were measured as the induced secretion of Interleukin-2 IL-2) by the P1D6 cells. The IL-2 was quantified in a bioassay using the CTLL-2 cell line.
Immunofluorescence.
All strains were grown in yeast peptone dextrose broth (ypd) or Sabouraud broth (sab) for 3 days at 30°C. For the 3D reconstructions cells were also grown in capsule inducing media.31,34 The cells were washed three times with sterile phosphate-buffered saline (PBS; pH 7.4) and then counted. A suspension of 2 × 106 cells/ml was incubated with 13 µg/ml of MP98 binding mAb 18F2 from hybridoma supernatant in 2% BSA and 0.05% goat serum in PBS for 1 hr at room temperature. Cells were washed three times with 2% BSA and 0.05% goat serum and incubated with 4 µg/ml of goat anti mouse IgG2a-FITC. Cell wall chitin was stained with 10 µg/ml of UVitex 2B (Polysciences Inc., Eppelheim, Germany). To detect additional mannoproteins, 5 µg/ml of concanavalin A (Con A)-Alexa Fluor 594 (Molecular Probes, Eugene, OR) was incubated with C. neoformans cells after incubation with the 18F2 antibody.
Dextran penetration.
C. neoformans strains B3501 and KN99α were grown in minimal media for two days. They were washed with PBS and suspended in tetramethylrhodamine-labeled dextran (MW 10, 40 and 70 kDa) at a concentration of 0.2 mg/ml in PBS.36 Degree of dextran penetration was measured in 40–60 cells at randomly chosen fields of view, then normalized by dividing the penetration (in µm) by the average capsule thickness acquired from 70 cells of each strain.
Confocal microscopy.
Approximately 2 × 106 C. neoformans cells in 100 µl of 2% BSA in PBS with 0.5% goat serum were incubated with 2 µl (10 µg) of mAb 18F2 overnight at 4°C. The cells were washed three times with buffer and incubated with 4 µg/ml of goat anti mouse IgG2a-Alexa Fluor 350 as the secondary antibody for 1 hr at room temperature. The mAb 18B7 was used for GXM staining5,35 as a secondary antibody for detecting mAb 18B7 we used and goat anti mouse IgG1-TRITC. Murine hyperimmune serum was used for GalXM staining31 as a secondary antibody we used goat anti rat IgM-FITC. Stained cells were mounted on glass slides as described above and imaged with a Leica SP2 laser scanning confocal microscope. 3D reconstructions of the captured Z-stacks were made using ImageJ (NIH; http://rsb.info.nih.gov/ij/) and Voxx (Indiana University; www.nephrology.iupui.edu/imaging/voxx/) software.
Table 1.
Primers to make CDA1 deletion cassette and confirm gene replacement
| Primer | Sequence1 | |
| a2 | CDA1-F | cac gtt tga ttg cga acg ct |
| b | CDA1-R | ccg acc taa gac gat cat |
| c | CDA1URA5-F | cca gaa gcg gaa ttt cgc cct tga tcA TGT CCT CCC |
| d | CDA1URA5-R | GGG AGG ACA Tga tca agg gcg aaa ttc cgc ttc tgg |
| e | URA5CDA1-F | CGG TGT TCA GAG GTC TTA Atc taa cga tcg atg ggg t |
| f | URA5CDA1-R | Acc cca tcg atc gtt aga TTA AGA CC T CTG AAC ACC G |
| g | CDA1-F4 | atc ggt cga cag tag cat |
| h | CDA1-R5 | gga aga aga gtc aga ggt |
| i | CDA1-TAA | tta ggc gac ata tac cat acc gac cgc a |
| j | CDA1-F5 | ggt gac att gat gac cgt |
| k | CDA1-F3 | Cca ggc ata cgt tca agt |
| l | URA-R | CC T CGA TGA AGG CAA CTT |
| m | URA-F | AGC TGC AGG AGG TCA GGA A |
| n | CDA1-R3 | ctg tcc aca gtg cat caa |
Lower case designates CDA1 sequence and upper case URA5 sequence.
Primer locations are the same or similar to those designated in Figure 1B if the CDA1 gene was substituted for MP98(CDA2).
Acknowledgements
We thank Susan Buhl for sharing her expertise during the generation of the hybridoma cell lines. We also thank Stuart Chaskes for his technical help in culturing different C. neoformans strains. The data in this paper are from a thesis submitted by Magdia De Jesus in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Sue Golding Graduate Division of Medical Science, Albert Einstein College of Medicine, Yeshiva University, Bronx, NY.
Footnotes
Previously published online: www.landesbioscience.com/journals/virulence/article/13451
Financial Disclosure
This work was supported by NIH grants AI33774, AI33142 and HL59842-01 to A.C. and AI025780 and AI066087 to S.M.L. M.D. was supported by NCI/NIH training grant 2T32CA009173-31.
References
- 1.Yauch LE, Mansour MK, Levitz SM. Receptor-mediated clearance of Cryptococcus neoformans capsular polysaccharide in vivo. Infect Immun. 2005;73:8429–8432. doi: 10.1128/IAI.73.12.8429-8432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell TG, Perfect JR. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rivera J, Mukherjee J, Weiss LM, Casadevall A. Antibody efficacy in murine pulmonary Cryptococcus neoformans infection: a role for nitric oxide. J Immunol. 2002;168:3419–3427. doi: 10.4049/jimmunol.168.7.3419. [DOI] [PubMed] [Google Scholar]
- 4.Schop J. Protective immunity against Cryptococcus neoformans infection. Mcgill J Med. 2007;10:35–43. [PMC free article] [PubMed] [Google Scholar]
- 5.Mukherjee J, Casadevall A, Scharff MD. Molecular characterization of the humoral responses to Cryptococcus neoformans infection and glucuronoxylomannan-tetanus toxoid conjugate immunization. J Exp Med. 1993;177:1105–1116. doi: 10.1084/jem.177.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McFadden D, Zaragoza O, Casadevall A. The capsular dynamics of Cryptococcus neoformans. Trends Microbiol. 2006;14:497–505. doi: 10.1016/j.tim.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Vaishnav VV, Bacon BE, O'Neill M, Cherniak R. Structural characterization of the galactoxylomannan of Cryptococcus neoformans Cap67. Carbohydr Res. 1998;306:315–330. doi: 10.1016/s0008-6215(97)10058-1. [DOI] [PubMed] [Google Scholar]
- 8.Levitz SM, Specht CA. The molecular basis for the immunogenicity of Cryptococcus neoformans mannoproteins. FEMS Yeast Res. 2006;6:513–524. doi: 10.1111/j.1567-1364.2006.00071.x. [DOI] [PubMed] [Google Scholar]
- 9.Heiss C, Klutts JS, Wang Z, Doering TL, Azadi P. The structure of Cryptococcus neoformans galactoxylomannan contains beta-D-glucuronic acid. Carbohydr Res. 2009;344:915–920. doi: 10.1016/j.carres.2009.03.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Jesus M, Chow SK, Cordero RJ, Frases S, Casadevall A. Galactoxylomannans from Cryptococcus neoformans varieties neoformans and grubii are structurally and antigenically variable. Eukaryot Cell. 9:1018–1028. doi: 10.1128/EC.00268-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frases S, Nimrichter L, Viana NB, Nakouzi A, Casadevall A. Cryptococcus neoformans capsular polysaccharide and exopolysaccharide fractions manifest physical, chemical and antigenic differences. Eukaryot Cell. 2008;7:319–327. doi: 10.1128/EC.00378-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.James PG, Cherniak R, Jones RG, Stortz CA, Reiss E. Cell-wall glucans of Cryptococcus neoformans Cap 67. Carbohydr Res. 1990;198:23–38. doi: 10.1016/0008-6215(90)84273-w. [DOI] [PubMed] [Google Scholar]
- 13.Vartivarian SE, Reyes GH, Jacobson ES, James PG, Cherniak R, Mumaw VR, Tingler MJ. Localization of mannoprotein in Cryptococcus neoformans. J Bacteriol. 1989;171:6850–6852. doi: 10.1128/jb.171.12.6850-6852.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy JW. Influence of cryptococcal antigens on cellmediated immunity. Rev Infect Dis. 1988;10:432–435. doi: 10.1093/cid/10.supplement_2.s432. [DOI] [PubMed] [Google Scholar]
- 15.Reiss E, Huppert M, Cherniak R. Characterization of protein and mannan polysaccharide antigens of yeasts, moulds and actinomycetes. Curr Top Med Mycol. 1985;1:172–207. doi: 10.1007/978-1-4613-9547-8_7. [DOI] [PubMed] [Google Scholar]
- 16.Eigenheer RA, Jin Lee Y, Blumwald E, Phinney BS, Gelli A. Extracellular glycosylphosphatidylinositol-anchored mannoproteins and proteases of Cryptococcus neoformans. FEMS Yeast Res. 2007;7:499–510. doi: 10.1111/j.1567-1364.2006.00198.x. [DOI] [PubMed] [Google Scholar]
- 17.Specht CA, Nong S, Dan JM, Lee CK, Levitz SM. Contribution of glycosylation to T cell responses stimulated by recombinant Cryptococcus neoformans mannoprotein. J Infect Dis. 2007;196:796–800. doi: 10.1086/520536. [DOI] [PubMed] [Google Scholar]
- 18.Chaka W, Verheul AF, Vaishnav VV, Cherniak R, Scharringa J, Verhoef J, et al. Induction of TNFalpha in human peripheral blood mononuclear cells by the mannoprotein of Cryptococcus neoformans involves human mannose binding protein. J Immunol. 1997;159:2979–2985. [PubMed] [Google Scholar]
- 19.Murphy JW, Mosley RL, Cherniak R, Reyes GH, Kozel TR, Reiss E. Serological, electrophoretic and biological properties of Cryptococcus neoformans antigens. Infect Immun. 1988;56:424–431. doi: 10.1128/iai.56.2.424-431.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delfino D, Cianci L, Lupis E, Celeste A, Petrelli ML, Curro F, et al. Interleukin-6 production by human monocytes stimulated with Cryptococcus neoformans components. Infect Immun. 1997;65:2454–2456. doi: 10.1128/iai.65.6.2454-2456.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pitzurra L, Perito S, Baldelli F, Bistoni F, Vecchiarelli A. Humoral response against Cryptococcus neoformans mannoprotein antigens in HIV-infected patients. Clin Exp Immunol. 2003;133:91–96. doi: 10.1046/j.1365-2249.2003.02181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pietrella D, Mazzolla R, Lupo P, Pitzurra L, Gomez MJ, Cherniak R, et al. Mannoprotein from Cryptococcus neoformans promotes T-helper type 1 anticandidal responses in mice. Infect Immun. 2002;70:6621–6627. doi: 10.1128/IAI.70.12.6621-6627.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pietrella D, Cherniak R, Strappini C, Perito S, Mosci P, Bistoni F, et al. Role of mannoprotein in induction and regulation of immunity to Cryptococcus neoformans. Infect Immun. 2001;69:2808–2814. doi: 10.1128/IAI.69.5.2808-2814.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vecchiarelli A. Immunoregulation by capsular components of Cryptococcus neoformans. Med Mycol. 2000;38:407–417. doi: 10.1080/mmy.38.6.407.417. [DOI] [PubMed] [Google Scholar]
- 25.Pitzurra L, Cherniak R, Giammarioli M, Perito S, Bistoni F, Vecchiarelli A. Early induction of interleukin-12 by human monocytes exposed to Cryptococcus neoformans mannoproteins. Infect Immun. 2000;68:558–563. doi: 10.1128/iai.68.2.558-563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dan JM, Wang JP, Lee CK, Levitz SM. Cooperative stimulation of dendritic cells by Cryptococcus neoformans mannoproteins and CpG oligodeoxynucleotides. PLoS ONE. 2008;3:2046. doi: 10.1371/journal.pone.0002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansour MK, Schlesinger LS, Levitz SM. Optimal T cell responses to Cryptococcus neoformans mannoprotein are dependent on recognition of conjugated carbohydrates by mannose receptors. J Immunol. 2002;168:2872–2879. doi: 10.4049/jimmunol.168.6.2872. [DOI] [PubMed] [Google Scholar]
- 28.Levitz SM, Nong S, Mansour MK, Huang C, Specht CA. Molecular characterization of a mannoprotein with homology to chitin deacetylases that stimulates T cell responses to Cryptococcus neoformans. Proc Natl Acad Sci USA. 2001;98:10422–10427. doi: 10.1073/pnas.181331398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker LG, Specht CA, Donlin MJ, Lodge JK. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell. 2007;6:855–867. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casadevall A, Mukherjee J, Devi SJ, Schneerson R, Robbins JB, Scharff MD. Antibodies elicited by a Cryptococcus neoformans-tetanus toxoid conjugate vaccine have the same specificity as those elicited in infection. J Infect Dis. 1992;165:1086–1093. doi: 10.1093/infdis/165.6.1086. [DOI] [PubMed] [Google Scholar]
- 31.De Jesus M, Nicola AM, Rodrigues ML, Janbon G, Casadevall A. Capsular localization of the Cryptococcus neoformans polysaccharide component galactoxylomannan. Eukaryot Cell. 2009;8:96–103. doi: 10.1128/EC.00331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orendi JM, Verheul AF, De Vos NM, Visser MR, Snippe H, Cherniak R, et al. Mannoproteins of Cryptococcus neoformans induce proliferative response in human peripheral blood mononuclear cells (PBMC) and enhance HIV-1 replication. Clin Exp Immunol. 1997;107:293–299. doi: 10.1111/j.1365-2249.1997.283-ce1169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner SH, Cherniak R, Reiss E. Fractionation and characterization of galactoxylomannan from Cryptococcus neoformans. Carbohydr Res. 1984;125:343–349. doi: 10.1016/0008-6215(84)85172-1. [DOI] [PubMed] [Google Scholar]
- 34.Zaragoza O, Fries BC, Casadevall A. Induction of capsule growth in Cryptococcus neoformans by mammalian serum and CO(2) Infect Immun. 2003;71:6155–6164. doi: 10.1128/IAI.71.11.6155-6164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casadevall A, Cleare W, Feldmesser M, Glatman-Freedman A, Goldman DL, Kozel TR, et al. Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob Agents Chemother. 1998;42:1437–1446. doi: 10.1128/aac.42.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mondon P, Chang YC, Varma A, Kwon-Chung KJ. A novel episomal shuttle vector for transformation of Cryptococcus neoformans with the ccdB gene as a positive selection marker in bacteria. FEMS Microbiol Lett. 2000;187:41–45. doi: 10.1111/j.1574-6968.2000.tb09134.x. [DOI] [PubMed] [Google Scholar]
- 37.Wickes BL, Edman U, Edman JC. The Cryptococcus neoformans STE12alpha gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol Microbiol. 1997;26:951–960. doi: 10.1046/j.1365-2958.1997.6322001.x. [DOI] [PubMed] [Google Scholar]
- 38.Wickes BL, Edman JC. The Cryptococcus neoformans GAL7 gene and its use as an inducible promoter. Mol Microbiol. 1995;16:1099–1109. doi: 10.1111/j.1365-2958.1995.tb02335.x. [DOI] [PubMed] [Google Scholar]