Abstract
Insertional mutagenesis screens have provided thousands of mutant alleles for analysing genes of varied functions in Drosophila melanogaster. We here document mechanisms of insertional mutagenesis by a LINE element, the I factor, by determining the molecular structure of RNAs produced from two alleles of the white gene of D.melanogaster, wIR1 and wIR6. These alleles result from insertion of the I factor into introns of the gene. We show that sequences present within the element direct aberrant splicing and termination events. When the I factor is inserted within the white first intron it may lead to the use of a cryptic 3' splice site which does not contain the dinucleotide AG. This splicing gives rise to a chimeric messenger RNA whose synthesis is controlled differently in tissues where the mutated gene is expressed. When the I factor is inserted within the white last intron it induces synthesis of truncated mRNAs. These results provide, for the first time, mechanisms for I factor insertional mutagenesis. They are discussed in the more general context of RNA processing in Drosophila and the evolution of eukaryotic gene introns.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abad P., Vaury C., Pélisson A., Chaboissier M. C., Busseau I., Bucheton A. A long interspersed repetitive element--the I factor of Drosophila teissieri--is able to transpose in different Drosophila species. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8887–8891. doi: 10.1073/pnas.86.22.8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget S. M. Exon recognition in vertebrate splicing. J Biol Chem. 1995 Feb 10;270(6):2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Birchler J. A., Hiebert J. C., Rabinow L. Interaction of the mottler of white with transposable element alleles at the white locus in Drosophila melanogaster. Genes Dev. 1989 Jan;3(1):73–84. doi: 10.1101/gad.3.1.73. [DOI] [PubMed] [Google Scholar]
- Black D. L. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992 May 29;69(5):795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- Bratthauer G. L., Fanning T. G. Active LINE-1 retrotransposons in human testicular cancer. Oncogene. 1992 Mar;7(3):507–510. [PubMed] [Google Scholar]
- Bucheton A. I transposable elements and I-R hybrid dysgenesis in Drosophila. Trends Genet. 1990 Jan;6(1):16–21. doi: 10.1016/0168-9525(90)90044-7. [DOI] [PubMed] [Google Scholar]
- Busseau I., Pelisson A., Bucheton A. I elements of Drosophila melanogaster generate specific chromosomal rearrangements during transposition. Mol Gen Genet. 1989 Aug;218(2):222–228. doi: 10.1007/BF00331272. [DOI] [PubMed] [Google Scholar]
- Chaboissier M. C., Busseau I., Prosser J., Finnegan D. J., Bucheton A. Identification of a potential RNA intermediate for transposition of the LINE-like element I factor in Drosophila melanogaster. EMBO J. 1990 Nov;9(11):3557–3563. doi: 10.1002/j.1460-2075.1990.tb07566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Legrain P., Dujon B., Jacquier A. Interaction between the first and last nucleotides of pre-mRNA introns is a determinant of 3' splice site selection in S. cerevisiae. Nucleic Acids Res. 1994 Jun 11;22(11):1981–1987. doi: 10.1093/nar/22.11.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couderc J. L., Sobrier M. L., Giraud G., Becker J. L., Dastugue B. Actin gene expression is modulated by ecdysterone in a Drosophila cell line. J Mol Biol. 1983 Mar 5;164(3):419–430. doi: 10.1016/0022-2836(83)90059-1. [DOI] [PubMed] [Google Scholar]
- Cáceres J. F., Stamm S., Helfman D. M., Krainer A. R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994 Sep 16;265(5179):1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- Eickbush T. H. Transposing without ends: the non-LTR retrotransposable elements. New Biol. 1992 May;4(5):430–440. [PubMed] [Google Scholar]
- Fanning T. G., Singer M. F. LINE-1: a mammalian transposable element. Biochim Biophys Acta. 1987 Dec 8;910(3):203–212. doi: 10.1016/0167-4781(87)90112-6. [DOI] [PubMed] [Google Scholar]
- Fawcett D. H., Lister C. K., Kellett E., Finnegan D. J. Transposable elements controlling I-R hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell. 1986 Dec 26;47(6):1007–1015. doi: 10.1016/0092-8674(86)90815-9. [DOI] [PubMed] [Google Scholar]
- Fields C. Information content of Caenorhabditis elegans splice site sequences varies with intron length. Nucleic Acids Res. 1990 Mar 25;18(6):1509–1512. doi: 10.1093/nar/18.6.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjose A., Polito L. C., Weber U., Gehring W. J. Developmental expression of the white locus of Drosophila melanogaster. EMBO J. 1984 Sep;3(9):2087–2094. doi: 10.1002/j.1460-2075.1984.tb02095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridell R. A., Pret A. M., Searles L. L. A retrotransposon 412 insertion within an exon of the Drosophila melanogaster vermilion gene is spliced from the precursor RNA. Genes Dev. 1990 Apr;4(4):559–566. doi: 10.1101/gad.4.4.559. [DOI] [PubMed] [Google Scholar]
- Helfman D. M., Ricci W. M. Branch point selection in alternative splicing of tropomyosin pre-mRNAs. Nucleic Acids Res. 1989 Jul 25;17(14):5633–5650. doi: 10.1093/nar/17.14.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz H., Berg C. A. Aberrant splicing and transcription termination caused by P element insertion into the intron of a Drosophila gene. Genetics. 1995 Jan;139(1):327–335. doi: 10.1093/genetics/139.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet F., Ruiz C., Richards G. Puffs and PCR: the in vivo dynamics of early gene expression during ecdysone responses in Drosophila. Development. 1993 Jun;118(2):613–627. doi: 10.1242/dev.118.2.613. [DOI] [PubMed] [Google Scholar]
- Jackson I. J. A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res. 1991 Jul 25;19(14):3795–3798. doi: 10.1093/nar/19.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S., Heidmann T. An indicator gene for detection of germline retrotransposition in transgenic Drosophila demonstrates RNA-mediated transposition of the LINE I element. EMBO J. 1991 Jul;10(7):1927–1937. doi: 10.1002/j.1460-2075.1991.tb07719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H. H., Jr, Wong C., Youssoufian H., Scott A. F., Phillips D. G., Antonarakis S. E. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988 Mar 10;332(6160):164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- Lavigueur A., La Branche H., Kornblihtt A. R., Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993 Dec;7(12A):2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- Levis R., O'Hare K., Rubin G. M. Effects of transposable element insertions on RNA encoded by the white gene of Drosophila. Cell. 1984 Sep;38(2):471–481. doi: 10.1016/0092-8674(84)90502-6. [DOI] [PubMed] [Google Scholar]
- Lynch K. W., Maniatis T. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 1995 Feb 1;9(3):284–293. doi: 10.1101/gad.9.3.284. [DOI] [PubMed] [Google Scholar]
- McCullough A. J., Schuler M. A. AU-rich intronic elements affect pre-mRNA 5' splice site selection in Drosophila melanogaster. Mol Cell Biol. 1993 Dec;13(12):7689–7697. doi: 10.1128/mcb.13.12.7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y., Nishisho I., Horii A., Miyoshi Y., Utsunomiya J., Kinzler K. W., Vogelstein B., Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992 Feb 1;52(3):643–645. [PubMed] [Google Scholar]
- Modolell J., Bender W., Meselson M. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse B., Rotherg P. G., South V. J., Spandorfer J. M., Astrin S. M. Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature. 1988 May 5;333(6168):87–90. doi: 10.1038/333087a0. [DOI] [PubMed] [Google Scholar]
- Mount S. M., Burks C., Hertz G., Stormo G. D., White O., Fields C. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992 Aug 25;20(16):4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M., Green M. M., Rubin G. M. Partial revertants of the transposable element-associated suppressible allele white-apricot in Drosophila melanogaster: structures and responsiveness to genetic modifiers. Genetics. 1988 Feb;118(2):221–234. doi: 10.1093/genetics/118.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita N., Nishio H., Kitoh Y., Ishikawa Y., Ishikawa Y., Minami R., Nakamura H., Matsuo M. Insertion of a 5' truncated L1 element into the 3' end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J Clin Invest. 1993 May;91(5):1862–1867. doi: 10.1172/JCI116402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hare K., Murphy C., Levis R., Rubin G. M. DNA sequence of the white locus of Drosophila melanogaster. J Mol Biol. 1984 Dec 15;180(3):437–455. doi: 10.1016/0022-2836(84)90021-4. [DOI] [PubMed] [Google Scholar]
- O'Hare K., Rubin G. M. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983 Aug;34(1):25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- O'hare K., Levis R., Rubin G. M. Transcription of the white locus in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6917–6921. doi: 10.1073/pnas.80.22.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst S. M., Harrison D. A., Remington M. P., Spana C., Kelley R. L., Coyne R. S., Corces V. G. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 1988 Oct;2(10):1205–1215. doi: 10.1101/gad.2.10.1205. [DOI] [PubMed] [Google Scholar]
- Peterson K. M., Davis P. S., Judd B. H. The determined state of white expression in the Drosophila eye is modified by zeste1 in the wzm family of mutants. Mol Gen Genet. 1994 Mar;242(6):717–726. doi: 10.1007/BF00283427. [DOI] [PubMed] [Google Scholar]
- Pirrotta V., Bröckl C. Transcription of the Drosophila white locus and some of its mutants. EMBO J. 1984 Mar;3(3):563–568. doi: 10.1002/j.1460-2075.1984.tb01847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhommeau C., Proust J. I-R hybrid dysgenesis in Drosophila melanogaster: nature and site specificity of induced recessive lethals. Mutat Res. 1990 Jun;230(2):135–157. doi: 10.1016/0027-5107(90)90052-6. [DOI] [PubMed] [Google Scholar]
- Pélisson A., Finnegan D. J., Bucheton A. Evidence for retrotransposition of the I factor, a LINE element of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4907–4910. doi: 10.1073/pnas.88.11.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pélisson A. The I--R system of hybrid dysgenesis in Drosophila melanogaster: are I factor insertions responsible for the mutator effect of the I--R interaction? Mol Gen Genet. 1981;183(1):123–129. doi: 10.1007/BF00270149. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
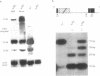
- Sang H. M., Pélisson A., Bucheton A., Finnegan D. J. Molecular lesions associated with white gene mutations induced by I-R hybrid dysgenesis in Drosophila melanogaster. EMBO J. 1984 Dec 20;3(13):3079–3085. doi: 10.1002/j.1460-2075.1984.tb02262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searles L. L., Ruth R. S., Pret A. M., Fridell R. A., Ali A. J. Structure and transcription of the Drosophila melanogaster vermilion gene and several mutant alleles. Mol Cell Biol. 1990 Apr;10(4):1423–1431. doi: 10.1128/mcb.10.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P., Shapiro M. B., Harris N. L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989 Mar 10;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Spikes D., Bingham P. M. Analysis of spliceosome assembly and the structure of a regulated intron in Drosophila in vitro splicing extracts. Nucleic Acids Res. 1992 Nov 11;20(21):5719–5727. doi: 10.1093/nar/20.21.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S., Zhang M. Q., Marr T. G., Helfman D. M. A sequence compilation and comparison of exons that are alternatively spliced in neurons. Nucleic Acids Res. 1994 May 11;22(9):1515–1526. doi: 10.1093/nar/22.9.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talerico M., Berget S. M. Effect of 5' splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990 Dec;10(12):6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Watakabe A., Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol Cell Biol. 1994 Feb;14(2):1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R. A., Graves J., Gibson W., Eisenberg M. Mobile element insertions causing mutations in the Drosophila suppressor of sable locus occur in DNase I hypersensitive subregions of 5'-transcribed nontranslated sequences. Genetics. 1990 Dec;126(4):1071–1082. doi: 10.1093/genetics/126.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachar Z., Chou T. B., Bingham P. M. Evidence that a regulatory gene autoregulates splicing of its transcript. EMBO J. 1987 Dec 20;6(13):4105–4111. doi: 10.1002/j.1460-2075.1987.tb02756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachar Z., Davison D., Garza D., Bingham P. M. A detailed developmental and structural study of the transcriptional effects of insertion of the Copia transposon into the white locus of Drosophila melanogaster. Genetics. 1985 Nov;111(3):495–515. doi: 10.1093/genetics/111.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]