Abstract
Hereditary spastic paraplegia (HSP) represents a large group of neurological disorders characterized by progressive spasticity of the lower limbs. One subtype of HSP shows an autosomal recessive form of inheritance with this corpus callosum (ARHSP-TCC), and displays genetic heterogeneity with four known loci. We identified a consanguineous Egyptian family with five affected individuals with ARHSP-TCC. We found linkage to the SPG11 locus and identified a novel homozygous p.Q498X stop codon mutation in exon 7 in the SPG11 gene encoding Spatacsin. Cognitive impairment and polyneuropathy, reported as frequent in SPG11, were not evident. This family supports the importance of SPG11 as a frequent cause for ARHSP-TCC, and expands the clinic SPG11 spectrum.
Keywords: Hereditary spastic paraparesis, Thin corpus callosum, SPG11, Spatacsin, Linkage analysis, Autosomal recessive
1. Introduction
Hereditary spastic paraplegia (HSP) is a clinically and genetically heterogeneous disorder characterized by progressive weakness and spasticity of the lower extremities followed by a similar decline in the upper extremities [1]. HSP can be classified based on the mode of inheritance, to include autosomal recessive, autosomal dominant and X-linked patterns. Clinically, HSP can also be divided into a pure form variant, of which spasticity is the major presenting feature, and a complex variant, which exhibits additional notable symptoms including ataxia, mental retardation, peripheral neuropathy, muscle atrophy or dysarthria [2].
Autosomal recessive hereditary spastic paraplegia with thin corpus callosum (ARHSP-TCC) is one of the more common forms of complicated recessive HSP [3]. ARHSP-TCC is characterized a corpus callosum that is thin but otherwise of normal structure, and complex features including mild mental retardation with learning difficulties and axonal motor or sensorimotor peripheral neuropathy [4]. The ARHSP-TCC phenotype is commonly due to mutations in at least four loci, SPG11, SPG15, SPG18, and SPG46, with two genes identified to date, Spatascin at SPG11 and Spastizin at SPG15 [5, 6]. Additionally, TCC is occasionally observed in association with SPG4 and SPG7 [7, 8]. This genetic heterogeneity makes it difficult to determine a priori which gene is likely to be mutated in a given family with ARHSP-TCC.
SPG11 gene encodes a 40 exon, 8-kb mRNA, expressed broadly in the adult cerebellum, cerebral cortex, hippocampus and pineal gland. The Spatascin protein of 2443 amino acids is of unknown function. It has been suggested that Spatacsin must play a vital biological role due to its high conservation among species [9]. Diverse clinical features have been reported with SPG11 mutations, including diversity in presentation and clinical course of HSP as well as other types of disease. These include Kjellin syndrome (ARHSP-TCC with retinal degeneration) [10], juvenile amyotrophic lateral sclerosis [11] and late-onset L-dopa-responsive parkinsonism [12]. The variability in presentations suggests the presence of genetic or environmental modifiers, and that Spatascin plays diverse roles in the brain.
Disease onset usually manifests itself during childhood or puberty and is marked by progressive spastic paraparesis, axonal or demyelinating polyneuropathy, bladder dysfunction, and remarkable thinning of the corpus callosum as evidenced on magnetic resonance imaging [4, 13]. Mental retardation has also been reported to be a common feature of ARHSP-TCC [9, 13]. Additional symptoms, though less common, include extrapyramidal signs, seizures, cerebellar ataxia and skeletal deformity.
In the current study we ascertained a consanguineous Egyptian family with ARHSP-TCC. Genome-wide microarray SNP scan identified a single linkage peak at the SPG11 locus. The novel mutation is predicted to result in premature protein truncation. The phenotype of the presented family is consistent with previous findings regarding spasticity, dysarthria and thin corpus callosum, but there was no notable mental impairment in the affected members. Additionally, a cerebellar arachnoid cyst was identified in one affected child, although the significance of this finding is uncertain.
2. Material and methods
Patients
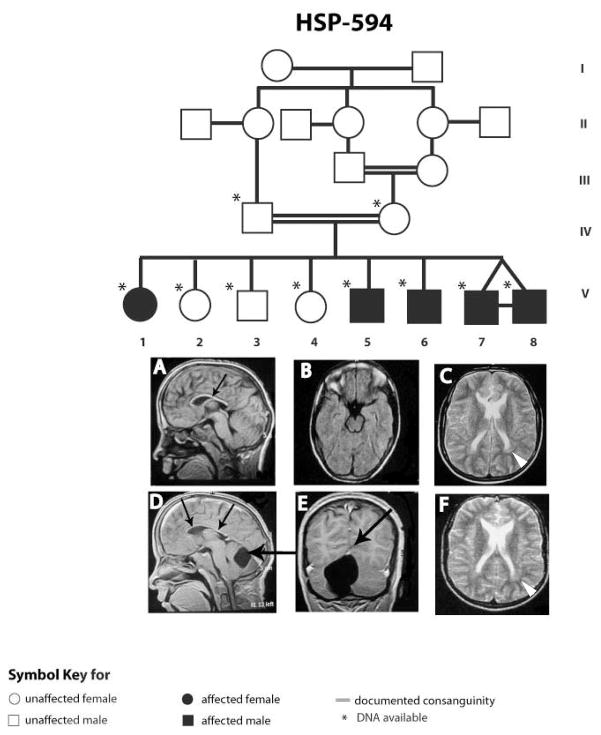
The family HSP-594 was recruited in order to perform molecular diagnosis. Parents were first cousins living in a small village in Upper (i.e. Southern) Egypt with five affected and three unaffected children (Fig. 1). All of the children were assessed by one of the authors (M.Z.). DNA from all genetically informative individuals was genotyped using the Human Linkage-12 Genotyping Bead Chip (Illumina, San Diego, CA). Because V-7 and V-8 were identical twins, only one was genotyped. Statistical analysis of the genotypes was performed using easy Linkage Plus 5.08 running Allegro v1.2 for two-point and multi-point linkage analysis [14]. An autosomal recessive inheritance of the disease was assumed due to multiple affected children, horizontal transmission and parental consanguinity.
Figure 1.
Family pedigree with MRI scans on two identical twins, individual 7 (A, B, C), individual 8 (D, E, F). Both individuals evidence thin corpus callosum (small arrows) and peritrigonal T2 hyperintensity (white arrowheads). Individual 8 shows cerebellar infravermian arachnoid cyst (large arrows).
Primers were designed using Primer 3 to amplify the coding exons for the SPG11 gene (GenBank accession number: NM_025137). PCR products were purified using Exonuclease I (Fermentas, Inc, Glen Burnie, MD) and Shrimp Alkaline Phosphatase (Promega, Madison, WI), and sequenced bi-directionally using Big Dye Terminator Cycle Sequencing (Applied Biosystems, Foster City, CA) on an automated ABI 3100 sequencer. Sequence chromatograms were analyzed using Sequencer 4.8 software (Genecodes, Inc, Ann Arbor, MI).
3. Results
Linkage and clinical findings
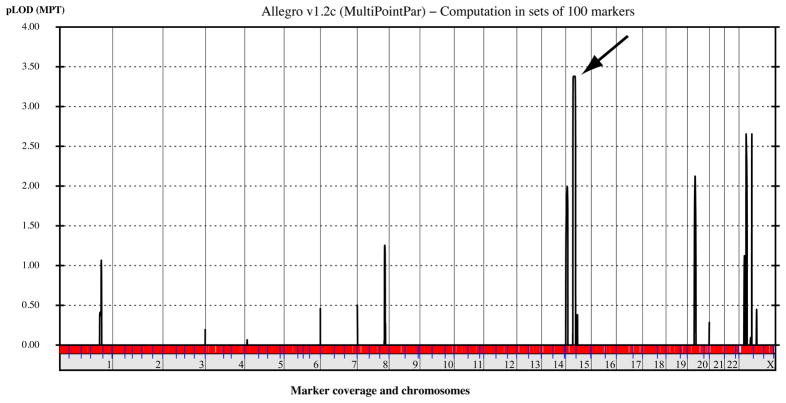
Due to the genetic heterogeneity of ARHSP-TCC, we decided to pursue a whole genome SNP-based linkage analysis to determine whether the cause of disease in this family represented a known or established cause. Analysis of the SNP genotyping results identified a single linkage peak at chromosome 15q21.1 at genome-wide significance with a LOD score of 3.7, and minimal candidate interval containing the SPG11 locus (Fig. 2).
Figure 2.
Whole-genome analysis of linkage results with chromosomal position (x axis) and multipoint LOD scores (y axis) showing a significant LOD score (>3) on chr. 15 (arrow). No other peaks reached this threshold for significance.
The onset age for all affecteds was about 14 years old per parents’ report; the age at clinical evaluation ranged from 17 to 33 years old (Table 1). School performance prior to presentation was not significantly different from the unaffected children. All affected presented with slowly evolving spasticity, which then progressed to dysarthria and inability to walk. They had borderline mental impairment with a score of 85 and 87 on the Full Scale IQ test. Moreover, affected older siblings were wheelchair-bound but did not show signs of cognitive impairment. Urinary incontinence was not reported. Magnetic resonance imaging was performed only on the twins and it showed evidence of a thin corpus callosum and high signal on T2 imaging around the occipital horns as has been reported [4, 15]. In one of the affected males, there was a cerebellar infravermian arachnoid cyst, which was not present in the other twin, and which has not been reported previously in SPG11. The older three affected siblings manifested moderate muscle wasting and joint deformities from the prolonged spasticity. EMG and NCV were only performed for the twin patients and were normal. Two of the affecteds underwent tendon release operations with inadequate results.
Table 1.
Clincial features of affected individuals in family HSP-594
| ID number | Sex | Age | Age at onset (years) | First symptom | Severity | Dysarthria | Mental Retardation | Sphincter disturbances | Cerebral MRI | EMG/NCV findings | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IV-1 | F | 33 | ~14 | Spasticity | WB | + | not reported | − | n/a | n/a | Musle wasting and skeletal deformities |
| IV-5 | M | 25 | ~14 | Spasticity | WB | + | not reported | − | n/a | n/a | Muscle wasting, skeletal deformities and tendon release operation |
| IV-6 | M | 22 | ~14 | Dysarthria | WB | + | not reported | − | n/a | n/a | Muscle wasting, skeletal deformities and tendon release operation |
| IV-7 | M | 17 | ~14 | Spasticity | AG | mild | not reported | − | Thin corpus callosum, High signal of white matter around occipital horn | Normal | − |
| IV-8 | M | 17 | ~14 | Spasticity | AG | mild | not reported | − | Thin corpus callosum, High signal of white matter around occipital horn; retrocerebellar cyst | Normal | − |
WB- wheelchair-bound
AG- abnormal gait
Genotyping findings
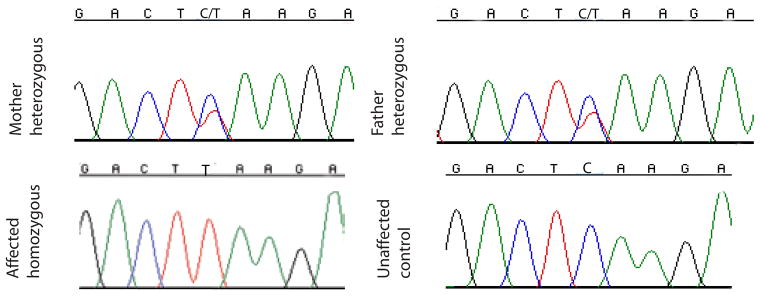
Based on the linkage data indicating the association with chromosome 15, and in conjunction with the phenotype findings, we tested samples from the family for mutation in the SPG11 gene. Polymerase chain reaction was performed on genomic DNA from one affected individual for all 40 coding exons, with one unaffected individual set as a control. We identified a single c.C1492T base substitution in exon 7. The mutation was present in the homozygous state in all affecteds. Additional family members were sequenced to verify that the c.C1492T mutation segregated only with the disease. We found that parents as well as two of the three unaffected siblings were carriers of the mutation (Fig. 3), whereas none of the unaffected members were homozygous for this mutation. This missense nucleotide change results in amino acid position p.Q498X premature stop codon in the first part of the protein.
Figure 3.
Sequence chromatograms illustrating mutation in SPG11gene; affected individual displaying a homozygous single base substitution c.C1492T in exon 7, leading to a premature stop codon p.Q498X in the first third of the protein. Mother and father are heterozygous for this mutation. Control DNA sample shows the normal base sequence.
4. Discussion
In the current study we identified a novel mutation on exon 7 associated with ARHSP-TCC in a consanguineous family from Upper Egypt. The phenotype and age of onset of the affected individuals is consistent with the characteristics of previously reported SPG11 mutations [4, 16]. The family did not display evidence of ataxia or neuropathy and there were also no symptoms of sphincter dysfunction, which are not uncommonly observed in patients with SPG11 mutations [3]. Additionally, contrary to multiple reports of moderate to severe mental retardation among individuals with ARHSP-TCC, within the current family only the two twin boys evidenced only borderline cognitive dysfunction. Other affected individuals were reported to be able to carry on meaningful conversations and to lead functional lives, however detailed IQ testing was not performed on all affecteds. It has been suggested that the TCC may result from progressive atrophy (as evidence by abnormal white matter signal on brain MRI) rather than hypoplasia, and follow up MRI studies in this family might more precisely clarify these changes over time.
Because of genetic heterogeneity of ARHSP-TCC, with as many as 7 or more causative loci, it is challenging to determine an optimal screening strategy for individual families to arrive at genetic diagnosis. Diseases such as HSP, with variable phenotypic features and multiple causative genes, are particularly challenging because rational screening strategies are not apparent. In this instance, we decided to perform whole genome SNP analysis, which led to the identification of the SPG11 locus as the cause in this family, but an alternative strategy might be to test polymorphic markers around each of the five common ARHSP-TCC loci to exclude specific genes, and then sequence only the genes that cannot be excluded. Since SPG11 mutations may be identified in 50–80% of patients with ARHSP-TCC, an alternative strategy would have been to sequence this candidate gene. However, higher-throughput methodology, including whole exome or whole genome sequencing strategies are achieving wider-spread use, especially as costs drop rapidly [17, 18].
Acknowledgments
Center of Inherited Disease Research at The Johns Hopkins University generated SNP allele calls and helped with the analysis. The authors wish to thank Tom Lidner from University of Erlangen-Nurenberg for his contribution in statistical analysis of linkage data. This work was supported by grants from the NIH and from Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Salinas S, et al. Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol. 2008;7(12):1127–38. doi: 10.1016/S1474-4422(08)70258-8. [DOI] [PubMed] [Google Scholar]
- 2.Fink JK. GeneReviews at GeneTests: Medical Genetics Information Resource (database online). Copyright, University of Washington, Seattle. 1997-2008. University of Washington; Seattle: 2009. Hereditary Spastic Paraplegia Overview. GeneReviews at GeneTests: Medical Genetics Information Resource (database online) [Google Scholar]
- 3.Stevanin G, Dürr A, Brice A. GeneReviews at GeneTests: Medical Genetics Information Resource (database online). Copyright, University of Washington, Seattle. 1997-2010. University of Washington; Seattle: 2009. Spastic Paraplegia Type 11. GeneReviews at GeneTests: Medical Genetics Information Resource (database online) [Google Scholar]
- 4.Stevanin G, et al. Mutations in SPG11 are frequent in autosomal recessive spastic paraplegia with thin corpus callosum, cognitive decline and lower motor neuron degeneration. Brain. 2008;131(Pt 3):772–84. doi: 10.1093/brain/awm293. [DOI] [PubMed] [Google Scholar]
- 5.Hanein S, et al. Identification of the SPG15 gene, encoding spastizin, as a frequent cause of complicated autosomal-recessive spastic paraplegia, including Kjellin syndrome. Am J Hum Genet. 2008;82(4):992–1002. doi: 10.1016/j.ajhg.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stevanin G, et al. Mutations in SPG11, encoding spatacsin, are a major cause of spastic paraplegia with thin corpus callosum. Nat Genet. 2007;39(3):366–72. doi: 10.1038/ng1980. [DOI] [PubMed] [Google Scholar]
- 7.Orlacchio A, et al. Hereditary spastic paraplegia: clinical genetic study of 15 families. Arch Neurol. 2004;61(6):849–55. doi: 10.1001/archneur.61.6.849. [DOI] [PubMed] [Google Scholar]
- 8.Coutinho P, et al. Clinical heterogeneity of autosomal recessive spastic paraplegias: analysis of 106 patients in 46 families. Arch Neurol. 1999;56(8):943–9. doi: 10.1001/archneur.56.8.943. [DOI] [PubMed] [Google Scholar]
- 9.Liao SS, et al. Novel mutations of the SPG11 gene in hereditary spastic paraplegia with thin corpus callosum. J Neurol Sci. 2008;275(1–2):92–9. doi: 10.1016/j.jns.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 10.Orlen H, et al. SPG11 mutations cause Kjellin syndrome, a hereditary spastic paraplegia with thin corpus callosum and central retinal degeneration. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(7):984–92. doi: 10.1002/ajmg.b.30928. [DOI] [PubMed] [Google Scholar]
- 11.Orlacchio A, et al. SPATACSIN mutations cause autosomal recessive juvenile amyotrophic lateral sclerosis. Brain. 2010;133(Pt 2):591–8. doi: 10.1093/brain/awp325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anheim M, et al. SPG11 spastic paraplegia. A new cause of juvenile parkinsonism. J Neurol. 2009;256(1):104–8. doi: 10.1007/s00415-009-0083-3. [DOI] [PubMed] [Google Scholar]
- 13.Paisan-Ruiz C, et al. Clinical heterogeneity and genotype-phenotype correlations in hereditary spastic paraplegia because of Spatacsin mutations (SPG11) Eur J Neurol. 2008;15(10):1065–70. doi: 10.1111/j.1468-1331.2008.02247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann K, Lindner TH. easy LINKAGE-Plus--automated linkage analyses using large-scale SNP data. Bioinformatics. 2005;21(17):3565–7. doi: 10.1093/bioinformatics/bti571. [DOI] [PubMed] [Google Scholar]
- 15.Del Bo R, et al. SPG11: a consistent clinical phenotype in a family with homozygous spatacsin truncating mutation. Neurogenetics. 2007;8(4):301–5. doi: 10.1007/s10048-007-0095-z. [DOI] [PubMed] [Google Scholar]
- 16.Schule R, et al. Frequency and phenotype of SPG11 and SPG15 in complicated hereditary spastic paraplegia. J Neurol Neurosurg Psychiatry. 2009;80(12):1402–4. doi: 10.1136/jnnp.2008.167528. [DOI] [PubMed] [Google Scholar]
- 17.Lupski JR, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362(13):1181–91. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi M, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106(45):19096–101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]