Abstract
Background
The majority of prostate cancers harbor gene fusions of the 5′-untranslated region of the androgen-regulated transmembrane protease, serine 2 (TMPRSS2) promoter with erythroblast transformation specific (ETS) transcription factor family members. The common v-ets erythroblastosis virus E26 oncogene homolog [avian] (TMPRSS2–ERG) fusion is associated with a more aggressive clinical phenotype, implying the existence of a distinct subclass of prostate cancer defined by this fusion.
Methods
We used cDNA-mediated annealing, selection, ligation, and extension to determine the expression profiles of 6144 transcriptionally informative genes in archived biopsy samples from 455 prostate cancer patients in the Swedish Watchful Waiting cohort (1987–1999) and the US-based Physicians Health Study cohort (1983–2003). A gene expression signature for prostate cancers with the TMPRSS2-ERG fusion was determined using partitioning and classification models and used in computational functional analysis. Cell proliferation and TMPRSS2-ERG expression in androgen receptor–negative (NCI-H660) and –positive (VCaP-ERβ) prostate cancer cells after treatment with vehicle or estrogenic compounds were assessed by viability assays and quantitative polymerase chain reaction, respectively. All statistical tests were two-sided.
Results
We identified an 87-gene expression signature that distinguishes TMPRSS2-ERG fusion prostate cancer as a discrete molecular entity (area under the curve = 0.80, 95% confidence interval [CI] = 0.792 to 0.81; P<.001). Computational analysis suggested that this fusion signature was associated with estrogen receptor (ER) signaling. Viability of NCI-H660 cells decreased after treatment with estrogen (viability normalized to day 0, estrogen vs vehicle at day 8, mean = 2.04 vs 3.40, difference = 1.36, 95% CI = 1.12 to 1.62) or ERβ agonist (ERβ agonist vs vehicle at day 8, mean = 1.86 vs 3.40, difference = 1.54, 95% CI = 1.39 to 1.69) but increased after ERα agonist treatment (ERα agonist vs vehicle at day 8, mean = 4.36 vs 3.40, difference = 0.96, 95% CI = 0.68 to 1.23). Similarly, expression of TMPRSS2-ERG decreased after ERβ agonist treatment (fold change over internal control, ERβ agonist vs vehicle at 24 hours, NCI H660, mean = 0.57-fold vs 1.0-fold, difference = 0.43, 95% CI = 0.29-fold to 0.57-fold) and increased after ERα agonist treatment (ERα agonist vs vehicle at 24 hours, mean = 5.63-fold vs 1.0-fold, difference = 4.63-fold, 95% CI = 4.34-fold to 4.92-fold).
Conclusions
TMPRSS2-ERG fusion prostate cancer is a distinct molecular subclass. TMPRSS2-ERG expression is regulated by a novel ER-dependent mechanism.
Prostate cancer is a major public health challenge, with an estimated 219,000 new cases diagnosed in 2007 and 27,000 annual deaths expected from the disease in the United States (1). The absence of effective treatment for advanced disease reflects in part the lack of a detailed understanding of the molecular pathogenesis of prostate cancer. A striking recent discovery, however, indicates that 40–70% of prostate cancers harbor an acquired chromosomal translocation that results in the fusion of the promoter region of the transmembrane protease serine 2 (TMPRSS2) gene to the coding region of members of the erythroblast transformation specific (ETS) family of transcription factors, most commonly the v-ets erythroblastosis virus E26 oncogene homolog (avian) (ERG) (2, 3). Prostate cancers with the TMPRSS2-ERG fusion appear to have a more aggressive natural clinical history than other prostate cancers (4). The downstream effects of TMPRSS2-ERG have yet to be identified, and the mechanism by which TMPRSS2-ERG may contribute to the pathogenesis of prostate cancer is entirely unknown.
An important challenge is, therefore, to devise therapeutic strategies to inhibit TMPRSS2-ERG function directly or the critical molecular pathways regulated by the TMPRSS2-ERG fusion. In this study, we used gene expression profiling to identify a gene signature of TMPRSS2-ERG activity in primary prostate cancer specimens. Because it would require more samples than are generally available in tumor banks of frozen prostate cancers to identify a statistically significant gene expression signature of TMPRSS2-ERG-positive tumors, we developed a method to profile the expression levels of 6144 transcriptionally informative genes in routinely collected formalin-fixed paraffin-embedded (FFPE) biopsy samples (5). This method is based on multiplexed locus-specific polymerase chain reaction (PCR) and is amenable to profiling degraded FFPE RNA because the amplified PCR products are extremely short. (Supplementary Figure 1, available online). Using this method, we carried out expression profiling of 455 prostate cancer samples to identify the molecular signature of TMPRSS2-ERG fusion. The signature was further explored using computationally analysis tools to identify molecular pathways associated with the fusion event. The results of this study are presented in the following sections.
Subjects and Methods
Patient Population
Swedish Cohort
The population-based Swedish Watchful Waiting Cohort consists of 1256 men with localized prostate cancer. These men had symptoms of benign prostatic hyperplasia (lower urinary tract symptoms) and were subsequently diagnosed with prostate cancer. All men in this study were determined at the time of diagnosis to have clinical stage T1–T2, Mx, N0, according to the 2002 American Joint Commission Committee Tumor-Node-Metastasis staging system as previously described (6, 7). The prospective follow-up time is now up to 30 years. The regional cohort includes men who were diagnosed at University Hospital in Örebro (1977–1991) (8–10) and at four centers in the southeast region of Sweden: Kalmar, Norrköping, Linköping, and Jonköping (1987–1999) (Table 1). All patients with prostate cancer were recruited through an informed consent process at the respective institutions. This study is compliant with Karolinska and Örebro Ethical Committees. A subset of men from these cohorts (n = 388) were included in the study. Inclusion criteria required the availability of greater than 90% tumor cells compared with surrounding stroma or benign tissue in the diagnostic Trans Urethral Radical Prostatectomy (TURP) biopsy sample. Samples included were derived from an equal proportion of men who died of prostate cancer or developed metastasis and men who lived a minimum of 10 years without clinical recurrence of their disease. Of these 388 patients, only the 354 with reliable TMPRSS2-ERG fusion results were included in the analyses.
Table 1.
Characteristics of the Swedish Watchful Waiting Cohort (with available tissue blocks, N = 1256) by Center
| Characteristic | Örebro, No. (%) | Linköping, No. (%) | Norrköping, No. (%) | Jönköping, No. (%) | Kalmar, No. (%) | Total, No. (%) |
|---|---|---|---|---|---|---|
| Total | 240 | 347 | 168 | 225 | 276 | 1256 |
| Age, y | ||||||
| ≤ 70 | 80 (33%) | 75 (22%) | 40 (24%) | 55 (24%) | 58 (21%) | 308 (25%) |
| > 70 | 160 (67%) | 272 (78%) | 128 (76%) | 170 (76%) | 218 (79%) | 948 (75%) |
| Gleason score | ||||||
| 4–6 | 147 (61%) | 143 (41%) | 64 (38%) | 121 (54%) | 137 (50%) | 612 (49%) |
| 7 | 64 (27%) | 143 (41%) | 56 (33%) | 78 (35%) | 86 (31%) | 427 (34%) |
| 8–10 | 29 (12%) | 61 (18%) | 48 (29%) | 26 (12%) | 53 (19%) | 217 (17%) |
| Cause of death | ||||||
| Prostate cancer | 43 (18%) | 61 (18%) | 44 (26%) | 40 (18%) | 41 (15%) | 229 (18%) |
| Other | 155 (64%) | 208 (60%) | 95 (57%) | 121 (54%) | 163 (59%) | 742 (59%) |
| Still alive | 42 (18%) | 78 (22%) | 29 (17%) | 64 (28%) | 72 (26%) | 285 (23%) |
| Tumor area in biopsy, % | ||||||
| ≤5 (=T1a) | 118 (49%) | 75 (22%) | 55 (33%) | 114 (51%) | 127 (46%) | 489 (39%) |
| 6–25 | 92 (38%) | 178 (51%) | 59 (35%) | 78 (35%) | 101 (36%) | 508 (40%) |
| 26–50 | 13 (5%) | 56 (16%) | 32 (19%) | 11 (5%) | 24 (9%) | 136 (11%) |
| >50 | 17 (7%) | 38 (11%) | 22 (13%) | 22 (10%) | 24 (9%) | 123 (10%) |
| Extreme groups* | ||||||
| Died of prostate cancer | 43 (18%) | 61 (18%) | 44 (26%) | 40 (18%) | 41 (15%) | 229 (18%) |
| Long-term survivors (>10 y) | 90 (37%) | 117 (34%) | 43 (26%) | 87 (39%) | 95 (34%) | 432 (34%) |
| Total | 133 (55%) | 178 (51%) | 87 (52%) | 127(56%) | 136 (49%) | 661 (53%) |
Men who died of prostate cancer or developed metastasis and men who lived a minimum of 10 years without any clinical recurrence of their disease. Percentages may not add to 100% due to rounding error.
Physician Health Study (PHS) Prostatectomy Confirmation Cohort
This cohort included 116 US men who were diagnosed with prostate cancer between 1983 and 2003, and were treated by radical prostatectomy as primary therapy (11) (Table 2). The men were participants in an ongoing randomized trial in the primary prevention of cancer and cardiovascular disease. This study was approved by the Harvard School of Public Health Institutional Review Board, and all patients provided written informed consent at time of initial enrollment. Only the 101 patients with reliable TMPRSS2-ERG fusion results were included in the analysis.
Table 2.
Clinical characteristics of Physician Health Study prostate cancer cohort individuals included in the study, 1983–2003*
| Characteristics | Value |
|---|---|
| Total No. | 108 |
| Gleason score, No. (%) | |
| 2–6 | 31 (29) |
| 7 | 47 (44) |
| 8–10 | 30 (28) |
| pT Stage, No. (%) | |
| T2 | 74 (69) |
| T3 | 22 (20) |
| T4/N1 | 12 (11) |
| Cancers diagnosed before PSA screening, No. (%) | 25 (23) |
| Median PSA at diagnosis, ng/mL† | 7.1 |
| Median follow-up, y | 10.9 |
| Cancer deaths or metastases | 34 |
pT=primary tumor; N=node. Percentages may not add to 100% due to rounding error.
Among men who were diagnosed during the prostate-specific antigen (PSA) screening era.
cDNA-mediated Annealing, Selection, Ligation, and Extension Array Design
We designed a set of four cDNA-mediated annealing, selection, ligation, and extension (DASL) Assay Panels (DAPs) for the discovery of molecular signatures relevant to prostate cancer (Hoshida Y, Setlur SR, Perner S, Camargo A, Gupta S, Moore J, Reich M, Gabriel S, Rubin MA, Golub TR, in preparation). We prioritized informative genes, i.e., genes showing differential expression across samples in previously generated microarray data sets (the datasets are at http://www.broad.mit.edu/cancer/pub/HCC), which included 24 studies, 2149 samples, and 15 tissue types. The top-ranked transcriptionally informative genes that showed the largest variation in expression across the different datasets comprised genes in most of the known biological pathways. To ensure that prostate cancer–related genes were included in the DAP, we performed a meta-analysis of previous microarray datasets from the Oncomine Database (12) and included from that a list of genes that were transcriptionally regulated in prostate cancer. The final array consisted of 6144 genes (6K DAP).
Sample Processing and cDNA-mediated Annealing, Selection, Ligation, and Extension
Foci highly enriched for prostate cancer (>90%) were identified by microscopic examination of the tissue sections by the study pathologists (MAR, SP). Three 0.6-μm biopsy cores per patient were taken from these enriched areas and were placed in one well of a 96-well plate for high-throughput RNA extraction. The CyBi-Well liquid handling system (CyBio AG, Jenna, Germany) was used for high-throughput extraction. Cores were first deparaffinized by incubation with 800 μL Citrisolv (Fisher Scientific, USA) at 60°C for 20 minutes and then with 1.2 mL Citrisolv:absolute alcohol (2:1) at room temperature for 10 minutes. Cores were then washed with absolute alcohol, dried at 55°C, and incubated overnight at 45°C in 300 μL lysis buffer (10 mM NaCl, 500 mM Tris pH 7.6, 20 mM EDTA, 1% SDS) containing 1 mg/mL proteinase K (Ambion, Austin, TX). RNA was extracted from the lysate using the TRIzol LS reagent (Invitrogen, Carlsbad, CA). TRIzol LS reagent (900 μL) was added to the cell lysate, followed by 240 μL of chloroform (Sigma-Aldrich, St. Louis, MO). The samples were mixed thoroughly and centrifuged at 4°C, 5600g for 40 minutes (the same centrifugation settings were used for the rest of the protocol). After centrifugation, the aqueous phase was transferred to a new plate, and the RNA was precipitated by incubation with 620 μL of isopropanol (Sigma-Aldrich) at room temperature for 10 minutes. Glycogen (20 μg; Invitrogen) was added as a carrier. The samples were centrifuged as above, and the pellet was washed with 80% ethanol (Sigma-Aldrich), air dried, and dissolved in RNase-free water. The RNA was quantified using a NanoDrop spectrophotometer (NanoDrop technologies, Wilmington, DE).
SYBR green (QIAGEN Inc., Valencia, CA) quantitative polymerase chain reaction (qPCR) assay for a housekeeping gene, ribosomal protein L13a (RPL13A), was used to estimate RNA quality (RNA with crossover threshold, Ct, of less than 30 cycles was considered to be good quality). Primer sequences for RPL13A were as follows: RPL13A-FWD, GTACGCTGTGAAGGCATCAA, and RPL13A-REV, GTTGGTGTTCATCCGCTT (GenBank accession NM_012423.2). DASL expression assay (Illumina Inc., San Diego, CA) was performed using 50 ng of cDNA according to manufacturer’s instructions.
Cell Lines and Transfection
The prostate cancer cell lines NCI-H660, VCaP, PC3, DU145, and 22Rv1 were obtained from American Tissue Culture Collection (ATCC, Manassas, VA). Cells were maintained according to the supplier’s instructions.
VCaP cells were transiently transfected with an ERβ-containing plasmid (kindly provided by M. Lupien) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Transfection medium was removed after 6 hours, cells were washed in phosphate-buffered saline (PBS, 137 mM NaCl, 10 mM phosphate, 2.7 mM KCl, pH of 7.4) twice, and phenol red–free DMEM (Cellgro Mediatech, Herndon, VA) supplemented with 5% charcoal/dextran-treated-fetal bovine serum (CDT-FBS) (Invitrogen) was added. ERβ mRNA expression levels were determined after transfection by qPCR using the following primers: ERβ-FWD, AAGAAGATTCCCGGCTTTGT and ERβ-REV, TCTACGCATTTCCCCTCATC (GenBank accession code, NM_001437.2).
NCI-H660 cells were transiently transfected with smart pool small-interfering RNA (siRNA) against ERβ or anti-LUC control siRNA (both from Dharmacon Inc., Chicago, IL), both at a concentration of 25 nM, using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Hormone Treatment
17β-Estradiol (E2, Sigma-Aldrich), the ERα agonist propylpyrazole triol (PPT, Tocris Bioscience, Ellisville, MO), and the ERβ agonist diarylpropionitrile (DPN, Tocris Bioscience, Ellisville, MO) were each dissolved in 100% ethanol. Raloxifene, tamoxifen, and fulvestrant (Sigma-Aldrich) were each dissolved in dimethyl sulfoxide (DMSO). All reagents were used at a final concentration of 10nM.
NCI-H660 and VCaP cells were hormone deprived by culture in their respective phenol red–free media (for NCI-H660 without E2 and hydrocortisone) supplemented with 5% CDT-FBS (Invitrogen), for 48 hours (VCaP) or for 72 hours (NCI-H660). Transfected cells were treated with hormones or vehicle 24 hours after transfection.
NCI-H660 cells and transfected VCaP-ERβ cells were treated with the following compounds: E2, DPN, PPT, raloxifene, fulvestrant, or tamoxifen; all at 10 nM final concentration; or vehicle for 12, 24, or 48 hours. Untransfected VCaP cells were treated for 12 or 24 hours with E2, DPN, raloxifene, or fulvestrant (all 10 nM final concentration).
Results were analyzed using GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego CA).
Determination of TMPRSS2-ERG Fusion Status in Biopsy Samples and Expression in Hormone-Treated Prostate Cancer Cell Lines
TMPRSS2-ERG fusion status was determined by ERG break-apart fluorescence in situ hybridization (FISH) assay (13)(n=362) and qPCR (for cases not assessable by FISH (n=98). An aliquot of the RNA used for DASL was used for qPCR. cDNA was synthesized as above using the Illumina kit (Illumina Inc., San Diego, CA). The TMPRSS2-ERG fusion product was detected using SYBR green assay (QIAGEN) with TMPRSS2-ERG_f and TMPRSS2-ERG_r primers (GenBank accession code NM_DQ204772.1) (3). RPL13A was used for normalization. RNA from NCI-H660 cells, which express TMPRSS2-ERG (14), was used as a positive control and a calibrator for quantification. Relative quantification was carried out using the comparative ΔΔCt method (15).
The same protocol was used to quantify the TMPRSS2-ERG fusion product after treatment of NCI-H660 and VCaP prostate cancer cell lines with estrogenic and antiestrogenic compounds. In this case, cDNA was synthesized using Omniscript RT kit (QIAGEN Inc), and a housekeeping gene, hydoxymethylbilane synthase (HMBS) was used for normalization. The primer sequences for HMBS are as follows: HMBS-FWD, CCATCATCCTGGCAACAGCT and HMBS-REV, GCATTCCTCAGGGTGCAGG (Gen Bank accession code NM_000190.3). Two independent experiments were performed, with each sample in triplicate.
Cell Viability Assays
NCI-H660 cells were seeded in 96-well plates (approximately 5 × 103 cells per well) and treated for 8 days with hormone (E2, PPT, or DPN) or vehicle alone as above. Relative cell number was determined before (time 0 used for normalization) and 2, 3, 6, and 8 days after treatment using the Cell Titer-Glo luminescent assay (Promega Corporation, Madison, WI) according to the manufacturer’s instructions. Two independent experiments were performed, both in octuplicate.
Expression of Estrogen Receptor α and Estrogen Receptor β in Prostate Cancer Cell Lines
Quantitative Polymerase Chain Reaction
Total RNA was extracted from the VCaP, NCI-H660, LNCaP, PC3, DU145, and 22Rv1 cell lines. The RNA was reverse transcribed (RT), and 50 μg of the resultant cDNAs was subjected to PCR analysis. RT-PCR was carried out using primers for ERα(16) (GenBank accession code, NM_000125.2) and ERβ (17) (GenBank accession code, NM_001437.2). cDNA was synthesized using the Omniscript RT kit (QIAGEN Inc.), and HMBS was used for normalization. Two independent experiments were performed with each sample in triplicate.
Western Blotting
Expression of ERαand ERβ in was assessed in NCI-H660 cells and in untransfected VCaP cells and VCaP-ERβ cells (48 hours after transfection). Whole-cell extracts were prepared in RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 2 mM sodium orthovanadate, 0.1% Nonidet P-40, 0.1% Tween 20) with 1x Complete Protease Inhibitor Cocktail (Roche, Indianapolis, IN). Protein concentration was determined using the Bio-Rad DC protein assay (Bio-Rad Laboratories, Hercules, CA). Equal amounts (20 μg) of total protein were loaded on NuPAGE 4–12% Tris-Bis gels (Invitrogen) and transferred to Immobilon-P polyvinylidene fluoride membranes (Millipore, Billerica, MA). Blots were incubated with primary antibodies (mouse monoclonal anti-ERα[1:100, NeoMarkers, Labvision Corporation, Fremont, CA] or mouse monoclonal anti-ERβ [1:200, clone 14C8, GeneTex Inc., San Antonio, TX]), washed three times with PBS containing 0.1% Triton X-100, and then incubated with peroxidase-conjugated anti-mouse secondary antibody (1:8000, Amersham Biosciences, Piscataway, NJ) for 1 hour. Rabbit polyclonal anti-β-Actin (1:1000, Cell Signaling technology, Danvers, MA) was used as a control for protein loading and transfer. Antibody–protein complexes were detected using the ECL Western Blotting Analysis System (Amersham Biosciences, Piscataway, NJ) according to the manufacturer’s instructions. Three independent experiments were carried out.
Chromatin Immunoprecipitation
ChIP was performed as described by Carroll et al. (18). Briefly, NCI-H660 cells were hormone -deprived by culture for 3 days in phenol red–free medium (Cellgro Mediatech, Herndon, VA) supplemented with 5% CDT-FBS (Invitrogen) lacking E2 and hydrocortisone. Cells were treated with E2 or vehicle for 60 min, and chromatin was crosslinked using 1% formaldehyde (15). Due to the high homology between ERαand ERβ, sites analyzed included ERαrecruitment sites that had previously been identified upstream of the TMPRSS2 gene (ER3429–ER3433) in an unbiased genome-wide ChIP-Chip experiment in the MCF-7 breast cancer cell line (18). In addition, the TMPRSS2 promoter (TMPRSS2_prom) was added to this analysis. Crosslinked chromatin was immunoprecipitated with mouse monoclonal anti-ERβ antibody (1:200, clone 14C8, GeneTex Inc.). The precipitated DNA was amplified using primers spanning the putative ER binding sites (Supplementary Tables 2 and 3, available online). Three independent experiments were performed.
Statistical Analysis
We used several strategies to both identify and evaluate a gene signature of TMPRSS2-ERG prostate cancer. Briefly, we identified candidate genes by t-test statistic via repeated sampling to ensure robustness. We then built different classification models to evaluate prediction performance of the gene signature using both the Swedish and PHS cohorts. Specifically, we first tested the method on the Swedish cohort by means of a hold-out procedure, i.e., two-thirds (n=235) of the samples were used to build the model and one-third (n=119) was used to evaluate it. In each evaluation phase, a different subset of samples from those used to build the models was used, thus ensuring that the final model did not depend on a specific dataset. Second, a model built using the entire Swedish dataset was used to evaluate the PHS dataset.
Gene Selection and Classification Models Built on the Swedish Cohort
A feature selection procedure (identification of candidate genes) was carried out by applying a two-sided t-test on each gene on 10 repeats of 10-fold cross-validation, resulting in 100 t-tests for each gene. A 10-fold cross-validation works as follows: within each iteration, 1/10 of the samples are held out as “test” and a t-test is computed for each gene using the remaining 9/10 of the samples, known as “training” samples. This procedure, called partitioning, is performed to ensure that similar proportions of fusion-positive and -negative cancers are present in the two partitions. Partitioning is then repeated nine times, resulting in 10 disjoint “test” partitions. This approach ensures that each sample is used as a “training” and a “test” set at least once. In addition, to prevent results that are biased to a specific random splitting of the data, the 10-fold cross-validation procedure was repeated another nine times.
Genes were then selected if they were found to be statistically significantly associated with the TMPRSS2-ERG fusion as determined by FISH or qPCR in at least 50% of the iterations. The number of genes selected was not set a priori but rather depended on the data. This procedure was first applied within the Swedish cohort training set (defined as two-thirds of the entire Swedish cohort) using a P value threshold of .0001. The selected genes were then used to build the classification model (see Classification Model) to predict fusion status based on their expression. The resulting classification model was evaluated on the Swedish cohort validation set (one-third of the entire Swedish cohort) to obtain an initial assessment of classifier performance.
To verify whether the molecular signature seen in the Swedish cohort was present in an independent set of cancers (different population), we used the PHS cohort as an additional evaluation dataset. For this procedure, the same feature-selection (i.e selection of genes) was applied within the entire Swedish cohort, using a P value threshold of .00001. The classification model built using this gene signature on the entire Swedish cohort was then evaluated on the independent set of cancers from the PHS cohort. Finally, to determine the sensitivity of the molecular signature model with respect to the number of genes selected, we used the above described iterative procedure to select the top ranking 75%, 50%, 25%, and 10% of genes from the gene signature (ranking is defined by the frequency with which a subset of genes are called “statistically significant” during the iterative procedure) and evaluated the classifier performances on the PHS dataset.
Classification Models
Several classification models based on gene expression values were generated. Support vector machines (SVMs) (19) using both radial basis function and polynomial kernels (degrees equal to 1, 2, or 3) with different costs (cost equal to 0.01, 0.1, 1, or 10) were used. The area under the receiving operator characteristic (ROC) curve (AUC) was used as a performance measure for SVM classification models. The 10 repeats of 10-fold cross-validation previously described were also used to select the best SVM parameters. Each iteration included genes with a P value of .00001or less that had been selected by two-sided t-test. The genes selected here were used only for the purpose of selecting the best SVM model. The best SVM parameters were identified as the ones giving highest mean AUC computed on the test sets. The mean AUC was computed on the total number of iterations, namely 100.
The AUC P value was evaluated via a randomization approach, specifically by counting how many times, out of 1000 iterations, randomly obtained class predictions outperformed the actual classification model. A binomial distribution was used to generate random predictions by setting the probability of positive class according to the frequency of the SVM-predicted positive cancers. Finally, the 95% confidence interval [CI] of the AUC was estimated from the 10 repeats of 10-fold cross-validation within the Swedish training set.
Gene Lists Comparison
To further assess the robustness of the gene signature, we compared the gene list obtained with the iterative procedure described in the previous paragraphs with that obtained by means of a standard two-sided t-test controlled with false discovery rate (FDR, Q value<.01) (20) within the Swedish training set (n=235). In addition, we performed the iterative gene selection on the PHS cohort and measured the overlap with the genes identified in the entire Swedish cohort. A hypergeometric distribution test was computed to assess for the statistical significance of the overlap.
We selected the gene lists based on different thresholds of t-test P values and on the percentage of genes that were statistically significant throughout the repeated cross-validations. We obtained similar results as those presented in this manuscript. Analysis was performed using R 2.4.0 (21).
Computational Functional Analysis
The gene signature identified with the entire Swedish cohort was used for connectivity map (CMAP) (22) and molecular concepts map (MCM) (23, 24) analyses. Connectivity map analyzes the association (ie, the positive or negative correlation) between the given test signature and gene expression profiles of cell lines treated with specific concentrations of various drugs. MCM analyzes the association between the given test signature and various gene sets or “molecular concepts”. Fisher exact two-sided test was used for pairwise comparison of the concepts (MCM, P <.005, odds ratio >1.5).
Results
DASL-Based Expression Array Profiling of Archival Tissue
We developed a novel high-throughput method to profile the expression of 6144 genes in archival tissue specimens. High-quality expression data were obtained from 472 of 504 (93.65%) of the prostate cancer samples (363 from the Swedish cohort and 109 from the PHS cohort). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE8402.
Fusion Status Determination
To define a gene expression signature of TMPRSS2-ERG fusion–expressing prostate cancer, we performed FISH on the 472 prostate cancers for which tissue was available. For samples with inconclusive FISH results, we used qPCR to determine the TMPRSS2-ERG fusion status (455 cancers were successfully annotated, 354 from the Swedish cohort and 101 from the PHS cohort). These experiments indicated that 62 (17.5%) of the prostate tumors of patients in the Swedish Watchful Waiting cohort (diagnosed following transurethral prostate resections for benign prostatic hyperplasia) were positive for the TMPRSS2-ERG fusion. Within the PHS cohort, the majority of cancers (n=83 [82%]) were diagnosed through prostate-specific antigen (PSA) screening, and (n=41, [41%]) of the cancers were positive for TMPRSS2-ERG fusion.
Molecular Signature of TMPRSS2-ERG Fusion
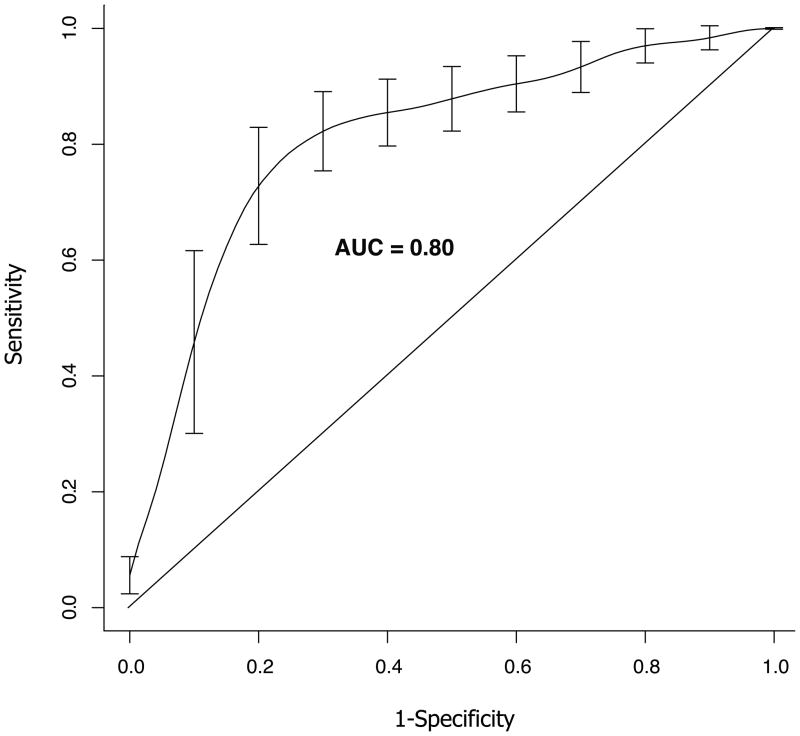
We next asked whether a gene expression signature of TMPRSS2-ERG could be identified. Two-thirds of the Swedish cohort cancers (n = 235) were used as a training set, and the remaining one-third (n = 119) were reserved as a validation set. In the initial analysis, we used the training set to evaluate the number of genes whose expression was statistically significantly correlated with TMPRSS2-ERG fusion status after correcting for multiple hypotheses testing (FDR Q value<.01) (20). One hundred seventy genes were identified, suggesting that TMPRSS2-ERG fusion cancers are indeed molecularly distinct from the fusion-negative cancers. Next, a gene expression–based SVM classifier built on the training set was applied to the 119 cancers in the validation set. The classifier included the genes that were differentially expressed between fusion-positive and fusion-negative cancers (P<.0001) in at least 50% of the 100 resampling iterations within the Swedish cohort training set. The linear kernel SVM (degree = 1) with cost equal to 0.1 was used to build the classification model. The AUC of this predictor was 0.79 (95% CI = 0.782 to 0.80; P<.001) on the validation set, again demonstrating that TMPRSS2-ERG-positive prostate cancers are molecularly distinct from fusion-negative tumors. After this validation step, we combined all 354 cancers in the Swedish Watchful Waiting cohort to build a new SVM model with a new set of 87 genes selected using the same iterative procedure (P<.00001) (Table 3 and Supplementary Figure 2A, available online). This SVM model was applied to the PHS cohort, resulting in an AUC of 0.80 (95% CI = 0.79 to 0.81; P<.001), validating the TMPRSS2-ERG model on the independent PHS cohort (Figure 1 and Supplementary Figure 2B, available online).
Table 3.
TMPRSS2-ERG 87 Gene Signature*
| Probe_Gene | P | Expression level |
|---|---|---|
| DAP1_2383_RAB27A | 6.46 × 10−8 | decreased |
| DAP1_2857_ALOX15B | 2.95 × 10−8 | decreased |
| DAP4_2027_CSDC2 | 7.27 × 10−8 | decreased |
| DAP4_2259_FOXF2 | 1.79 × 10−9 | decreased |
| DAP4_2818_KLHL21 | 5.88 × 10−9 | decreased |
| DAP4_3051_GRK1 | 1.74 × 10−8 | decreased |
| DAP4_3833_PNLIPRP2 | 1.37 × 10−7 | decreased |
| DAP4_5633_AMELX | 2.17 × 10−8 | decreased |
| DAP2_1182_HPS1 | 6.18 × 10−7 | decreased |
| DAP4_3730_NEUROD1 | 6.43 × 10−7 | decreased |
| DAP4_0050_AADAC | 7.86 × 10−7 | decreased |
| DAP4_1375_AQP2 | 6.64 × 10−7 | decreased |
| DAP4_5366_RGS7 | 1.03 × 10−6 | decreased |
| DAP3_1676_METTL7A | 1.53 × 10−6 | decreased |
| DAP4_2868_CCR1 | 1.66 × 10−6 | decreased |
| DAP4_5915_ITGAD | 1.22 × 10−6 | decreased |
| DAP4_1650_TRO | 1.48 × 10−6 | decreased |
| DAP3_4133_PROM1 | 1.18 × 10−6 | decreased |
| DAP3_1407_ADH5 | 2.56 × 10−6 | decreased |
| DAP4_4205_MPPED2 | 2.23 × 10−6 | decreased |
| DAP1_0743_RAB30 | 3.67 × 10−9 | increased |
| DAP1_1759_PLA2G7 | 2.75 × 10−8 | increased |
| DAP1_2097_ZNF3 | 8.81 × 10−11 | increased |
| DAP1_3022_TPP2 | 9.56 × 10−10 | increased |
| DAP1_5293_DDEF2 | 2.20 × 10−9 | increased |
| DAP1_6021_XRCC5 | 1.18 × 10−9 | increased |
| DAP1_6038_TLE1 | 4.30 × 10−10 | increased |
| DAP2_0721_TWIST1 | 4.77 × 10−8 | increased |
| DAP2_5076_ABCC8 | 1.11 × 10−9 | increased |
| DAP2_5229_ERG | 2.53 × 10−12 | increased |
| DAP3_1883_ECE1 | 4.90 × 10−11 | increased |
| DAP3_2016_CACNA1D | 1.66 × 10−8 | increased |
| DAP3_2857_GP1BB | 5.23 × 10−8 | increased |
| DAP3_3075_RFX1 | 3.99 × 10−11 | increased |
| DAP3_3399_EIF4G3 | 1.74 × 10−9 | increased |
| DAP3_4951_MAP7 | 2.07 × 10−8 | increased |
| DAP3_6085_CADPS | 9.28 × 10−8 | increased |
| DAP4_0208_ARHGAP29 | 1.25 × 10−9 | increased |
| DAP4_0791_DBN1 | 3.05 × 10−9 | increased |
| DAP4_0822_KCNN2 | 4.58 × 10−11 | increased |
| DAP4_1042_HDAC1 | 9.48 × 10−11 | increased |
| DAP4_1360_KHDRBS3 | 2.77 × 10−9 | increased |
| DAP4_1577_SH3YL1 | 1.51 × 10−11 | increased |
| DAP4_2291_EIF5 | 2.30 × 10−8 | increased |
| DAP4_2534_SIPA1L1 | 1.16 × 10−8 | increased |
| DAP4_3958_AMPD3 | 2.83 × 10−8 | increased |
| DAP4_4250_PTPRK | 7.00 × 10−8 | increased |
| DAP4_5710_RPP38 | 2.35 × 10−9 | increased |
| DAP4_5801_PCDHGB7 | 1.23 × 10−7 | increased |
| DAP4_6117_CPSF6 | 1.70 × 10−8 | increased |
| DAP3_1617_PEX10 | 1.10 x10−7 | increased |
| DAP4_0109_SEPT9 | 9.80 × 10−8 | increased |
| DAP4_5082_ALDH18A1 | 9.04 × 10−8 | increased |
| DAP3_1688_TFDP1 | 2.41 × 10−7 | increased |
| DAP4_5322_PSMD13 | 1.71 × 10−7 | increased |
| DAP4_2051_CRISP3 | 2.40 × 10−7 | increased |
| DAP4_2867_PFTK1 | 2.33 × 10−7 | increased |
| DAP3_5801_PDE9A | 4.07 × 10−7 | increased |
| DAP4_0691_RAGE | 3.42 × 10−7 | increased |
| DAP1_0943_BAG5 | 3.02 × 10−7 | increased |
| DAP4_2721_LRP1 | 4.06 × 10−7 | increased |
| DAP4_2225_THUMPD1 | 6.73 × 10−7 | increased |
| DAP4_3740_SAFB | 5.38 × 10−7 | increased |
| DAP3_3625_GHR | 3.88 × 10−7 | increased |
| DAP3_5906_BMPR1B | 4.51 × 10−7 | increased |
| DAP2_0361_COL9A2 | 6.78 × 10−7 | increased |
| DAP3_4969_MYO6 | 4.08 × 10−7 | increased |
| DAP4_5715_ARHGDIB | 6.41 × 10−7 | increased |
| DAP1_5104_PRKAR1B | 9.16 × 10−7 | increased |
| DAP2_1324_KNS2 | 9.95 × 10−7 | increased |
| DAP3_3482_PTK7 | 1.11 × 10−6 | increased |
| DAP1_2954_OCLN | 9.79 × 10−7 | increased |
| DAP2_3975_MLXIP | 6.97 × 10−7 | increased |
| DAP4_0758_KIAA0247 | 1.56 × 10−6 | increased |
| DAP4_5057_RGS10 | 1.59 × 10−6 | increased |
| DAP4_0233_UBE2G1 | 2.19 × 10−6 | increased |
| DAP2_3392_PRKCBP1 | 1.59 × 10−6 | increased |
| DAP3_3105_MAP3K5 | 2.24 × 10−6 | increased |
| DAP4_1178_MAP2K5 | 2.35 × 10−6 | increased |
| DAP4_0028_PGD | 1.85 × 10−6 | increased |
| DAP4_5943_TBP | 1.72 × 10−6 | increased |
| DAP2_1138_NCOA1 | 1.63 × 10−6 | increased |
| DAP3_1276_MTA1 | 2.00 × 10−6 | increased |
| DAP4_1949_SMARCD1 | 2.02 × 10−6 | increased |
| DAP1_2297_SNRPB2 | 2.65 × 10−6 | increased |
| DAP2_5056_UGDH | 3.31 × 10−6 | increased |
| DAP4_1759_NDUFS5 | 3.20 × 10−6 | increased |
P values were calculated using Student two-sided t-test.
Figure 1. Validation of the 87-gene molecular signature of transmembrane protease, serine 2—v-ets erythroblastosis virus E26 oncogene homolog (avian) (TMPRSS2-ERG) gene fusion prostate cancer on the Physician Health Study Cohort.
The 87-gene signature identified in the Swedish Watchful Waiting cohort (n = 354) was validated on the Physician Health Study cohort (n = 101) by using a Support Vector Machine (SVM) model, which showed an area under the receiver operator characteristics curve (AUC) of 0.80 (95% confidence interval = 0.79 to 0.81; P<.001 (two-sided, Student t-test). Error bars represent 95% confidence intervals.
Furthermore, the sensitivity analysis showed that by reducing the number of genes by approximately 50%, to 43 genes, the performance of the classifier on the PHS cohort remained as high as that with 87 genes. However, the performance on the Swedish cohort (which was used to build the classifier) was slightly lower (Supplementary Figure 3, available online). Although the AUCs were similar on the PHS data for the model containing fewer genes, the 87-gene SVM model achieved the best trade-off between sensitivity and specificity. Indeed, the closest point to the perfect solution, i.e., point (0.1) on the ROC curve, belongs to this model.
To further analyze the robustness of the signature, we compared the different gene lists obtained with different approaches as described in the Methods section. The overlap between the 87-gene list (iteration methods) and the 170-gene list (standard t-test with FDR correction) included 73 genes (73/87 = 84%). The overlap between the two lists was highly statistically significant (P= 2.3 × 10−6, hypergeometric test). Moreover, when selecting genes on the PHS cohort with the iterative methods, 44 genes were identified, yielding an overlap of 15 genes (15/44=34%). Again, this overlap was statistically significant (P = 9.6 × 10−18, hypergeometric test), demonstrating the reliability of the gene signature.
We also performed unsupervised analysis using the 87 genes on four other prostate cancer expression datasets that had been generated using different experimental platforms (Supplementary Figure 4, available online). The results suggest the presence of this gene signature in these cohorts.
Pathway Analysis of the TMPRSS2-ERG Signature
The next challenge was to understand the nature of the final 87-gene TMPRSS2-ERG signature. To do this, we used two computational strategies: 1) the connectivity map (CMAP) (22), which is an approach for identifying correlations between gene signatures of interest and the gene expression consequence of treatment with small-molecule drugs; and 2) the molecular concepts map (MCM) (23, 24), which is a system for comparing a gene signature with a database of protein–protein interaction networks, microarray profiles, and other genomic information. The MCM and CMAP are hypothesis-generating tools requiring independent validation.
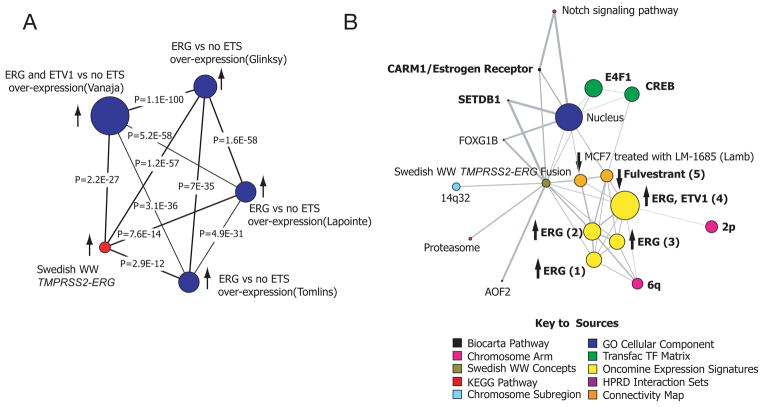
A common hypothesis emerged from both of these analyses, namely, a relationship between the gene signature of TMPRSS2-ERG cancers and estrogen receptor signaling. The MCM and the CMAP showed evidence of anticorrelation between the tumor tissue–derived 87-gene TMPRSS2-ERG signature and the gene expression profile of MCF7 cells treated with the antiestrogen fulvestrant (1 μM). The genes whose expression decreased after treatment were enriched in the sublist of genes in the 87-gene signature, and their expression was increased, suggesting that fulvestrant could potentially reverse the signature induced by TMPRSS2-ERG. In addition, CMAP showed a similar anticorrelation between the TMPRSS2-ERG signature and the expression profile induced by ERβ agonists resveratrol and genistein (Supplementary Table 4, available online). MCM analysis using the signature of genes with increased expression showed a strong association with genes whose expression was positively associated with ERG overexpression in several studies from the Oncomine database (Figure 2A), consistent with the enrichment that was observed in our analysis (Supplementary Figure 4, available online). The MCM also identified several estrogen-related gene sets or “concepts” (23, 24), including the concept for fulvestrant detailed in the CMAP analysis (Figure 2B). The MAP kinase interacting kinase 2 (MKNK2)–human protein reference database interaction set concept was enriched (Supplementary Table 5). MKNK2 selectively associates with the ligand-binding domain of ERβ and is believed to activate ERβ through phosphorylation in a ligand-independent manner (25). Hence, these computational analyses suggested a possible connection between TMPRSS2-ERG fusion and estrogen signaling, a hypothesis that could be tested functionally.
Figure 2. The transcriptional program regulated by transmembrane protease, serine 2—v-ets erythroblastosis virus E26 oncogene homolog (avian) (TMPRSS2-ERG) fusion and target drugs that appear to reverse its molecular profile.
Analysis of TMPRSS2-ERG gene signature through the molecular concepts map (MCM), a compendium of biologically related gene sets or “molecular concepts”. A) A network of molecular concepts related to high expression of erythroblast transformation specific (ETS) genes. Gene expression studies that have reported profiles that include ETS genes were individually explored for signatures associated with expression of ERG and ETV1. The results show statistically significant associations between the TMPRSS2-ERG fusion signature and ERG/ETV1 overexpression–related signatures. B) A broader evaluation of expression data, the Connectivity Map, and other molecular databases (see key to sources) demonstrates that the MCM analysis revealed estrogen receptor (ER) related concepts associated with the TMPRSS2-ERG gene signature. In addition, estrogenic signatures were associated with the TMPRSS2-ERG fusion signature. These estrogenic signatures included the top 5% underexpressed genes in MCF7 cells treated with fulvestrant at 1μM [Fulvestrant (5). Some of the other key signatures to emerge include the overexpression of ERG or ETV1 in prostate cancer expression array studies. These signatures include the top 10% overexpressed genes associated with ERG in different expression array datasets - Tomlins [ERG (1)], Glinsky [ERG (2)], Lapointe [ERG (4)], and the top 20% overexpressed with ERG and ETV1 in Vanaja [ERG, ETV1 (4)]. See “Results” for details. The arrows indicate the direction of expression with respect to the TMPRSS2-ERG fusion signature. The maps demonstrate related concepts linked by lines. The size of the nodes is related to the number of genes that make up the concept.
Validation of the Role of Estrogens in TMPRPSS2-ERG Fusion
Therefore, to investigate the selective role of estrogen and ER-mediated pathways in TMPRSS2-ERG fusion tumors, we performed a series of functional studies in vitro. The NCI-H660 prostate cancer cell line harbors a transcriptionally active homozygous TMPRSS2-ERG fusion (14) and expresses both ERαand ERβ (Supplementary Figure 5, available online). Androgen receptor (AR), which normally regulates wild-type TMPRSS2 expression (26), is absent in this cell line. Consequently, ERG expression is not androgen-regulated in this cell line (14). In contrast, in the androgen-sensitive VCaP cell line, androgen treatment increases ERG expression (3, 14). VCaP cells express AR and low levels of ERβ but do not express ERα(Supplementary Figure 5, available online).
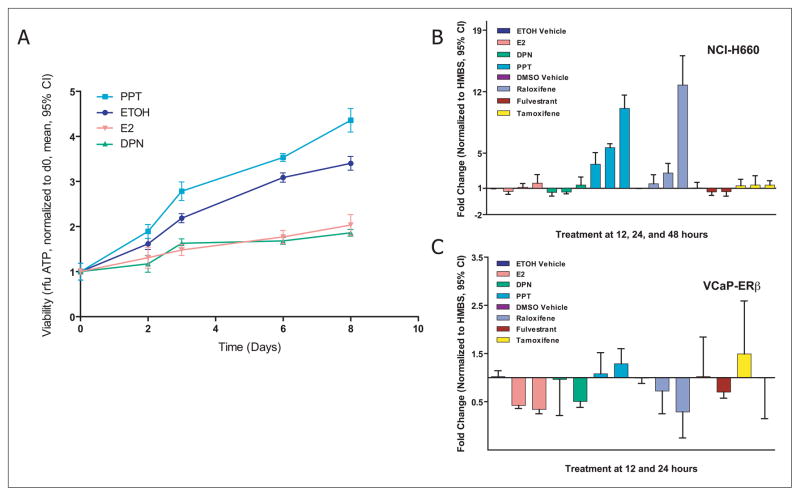
We hypothesized that the observed connections between estrogen signaling and the TMPRSS2-ERG gene signature might be explained by estrogen regulation of the fusion transcript. To test this hypothesis, we analyzed the effect of estrogen (E2) on growth of NCI-H660 prostate cancer cells. NCI-H660 cell growth was inhibited by E2 (viability normalized to day 0, E2 vs ethanol [EtOH] at day 8, mean 2.04 vs 3.40, difference =1.36, 95% CI = 1.12 to 1.62) (Figure 3A), suggesting a growth inhibitory effect that was mediated either by ERαor ERβ. Treatment with an ERα-selective agonist (PPT) resulted in sustained growth (PPT vs EtOH at day 8, mean = 4.36 vs 3.40, difference = 0.96, 95% CI = 0.68 to 1.23), whereas treatment with an ERβ-selective agonist (DPN) reduced viability (DPN vs EtOH at day 8, mean = 1.86 vs 3.40, difference = 1.54, 95% CI = 1.39 to 1.69) (Figure 3A). Consistent with the CMAP and MCM results, the ERαagonist PPT increased TMPRSS2-ERG expression (fold change over internal control, PPT vs EtOH at 24 hours, mean = 5.63-fold vs 1.0-fold, difference = 4.63-fold, 95% CI = 4.34-fold to 4.92-fold) whereas the ERβ agonist DPN suppressed expression of the fusion transcript (fold change over internal control, DPN vs EtOH vehicle control treated at 24 hours, NCI H660, mean = 0.57-fold vs 1.0-fold, difference = 0.43-fold, 95% CI = 0.29-fold to 0.57-fold) (Figure 3B). The antiestrogen fulvestrant reduced TMPRSS2-ERG expression (fulvestrant vs DMSO at 24 hours, NCI H660, mean = 0.58-fold vs 1.0-fold, difference = 0.42-fold, 95% CI = 0.16-fold to 0.68-fold) (Figure 3B). Consistent with these findings, knock-down of ERβ expression by RNA interference increased TMPRSS2-ERG expression (fold change over internal control, ERβ siRNA vs luciferase control at 24 hours, TMPRSS2-ERG expression, mean = 3.62-fold vs 0.75-fold, difference = 2.87-fold, 95% CI = 1.84-fold to 3.90-fold) (Supplementary Figure 5B, available online). Taken together, these results indicate that the TMPRSS2-ERG fusion can be regulated by estrogen receptor action, and that ERβ agonism leads to reduced TMPRSS2-ERG transcript expression, resulting in growth suppression. These results explain the alternate estrogen dependent mechanism by which, expression of TMPRSS2-ERG is regulated in an AR-negative cell line.
Figure 3. Cell viability and expression of transmembrane protease serine 2 v-ets erythroblastosis virus E26 oncogene homolog (avian)(TMPRSS2-ERG) in response to estrogenic and antiestrogenic compounds.
A) NCI-H660 cell proliferation after treatment with 17β-estradiol (E2) or specific ERα/β agonists. Cell viability assays were performed using NCI-H660 cells challenged with E2, the estrogen receptor alpha (ERα)-specific agonist propylpyrazole triol (PPT), the estrogen receptor beta (ERβ)-specific agonist diarylpropionitrile (DPN), or vehicle (ethanol, EtOH). Relative cell numbers were determined after 2, 3, 6, and 8 days of stimulation. Means and 95% confidence intervals from one representative experiment performed in octuplicate are shown. B) Expression of TMPRSS2-ERG in androgen receptor (AR) negative NCI-H660 cells after treatment with estrogenic and antiestrogenic compounds. NCI-H660 cells were treated for 12, 24, and 48 hours with E2, ERβ agonist DPN, ERαagonist PPT, raloxifene, fulvestrant, or tamoxifen (10 nM final concentration). NCI-H660 cells treated with vehicle (EtOH or dimethyl sulfoxide [DMSO]) alone were harvested at the same time points and used for normalization. Quantitative polymerase chain reactions were performed using primers spanning the TMPRSS2-ERG gene fusion. Means and 95% confidence intervals from three independent experiments performed in triplicate are shown. C) Expression of TMPRSS2-ERG in AR-positive VCaP cells transfected with ERβ (VCaP-ERβ) after treatment with estrogenic and antiestrogenic compounds. VCaP cells were transiently transfected with ERβ to increase expression of this receptor subtype (Supplementary Figure 5A, available online). VCaP-ERβ cells were treated for 12 or 24 hours with the same compounds. Vehicle-treated (DMSO or EtOH) VCaP-ERβ cells were used for normalization. Means and 95% confidence intervals are shown from two independent experiments performed in triplicate. Results for untransfected VCaP cells are shown in Supplementary Figure 5C (available online).
We then extended these observations to a second TMPRSS2-ERG expressing cell line, VCaP, which expresses AR but lacks high levels of ERαand ERβ expression. Treatment of ERβ-overexpressing VCaP cells with the ERβ agonists DPN and E2 led to reduced expression of the TMPRSS2-ERG fusion transcript compared with the vehicle treated control cells (fold change over internal control, DPN vs EtOH at 24 hours, mean = 0.51-fold vs 1.0-fold, difference = 0.49-fold, 95% CI = 0.38-fold to 0.61-fold; E2 vs EtOH at 24 hours, mean = 0.34-fold vs 1.0-fold, difference = 0.66-fold, 95% CI = 0.56-fold to 0.76-fold) (Figure 3C), whereas this lowered expression was not seen in non–ERβ-overexpressing parental VCaP cells (Supplementary Figure 5A, Supplementary Figure 5C, available online). These results further indicate a role of estrogen receptors in regulating TMPRSS2-ERG expression.
Next, we tested the effect of the selective estrogen receptor modulators (SERMs) raloxifene and tamoxifen on TMPRSS2-ERG expression (Figure 3B and C). Raloxifene has higher affinity for ERαthan ERβ, and led to increased expression of TMPRSS2-ERG in NCI-H660 cells compared with the vehicle treated cells, consistent with an ERαagonist effect (fold change over internal control, raloxifene vs DMSO at 48 hours, mean = 12.75-fold vs 1.0-fold, difference = 11.7-fold, 95% CI = 9.60-fold to 13.9-fold) (Figure 3B). In contrast, raloxifene induced a remarkable decrease in expression of the fusion transcript in ERα–negative VCaP-ERβ cells (fold change over internal control, raloxifene vs DMSO at 24 hours, mean = 0.29-fold vs 1.0-fold, difference = 0.71-fold, 95% CI = 0.36-fold to 1.06-fold) (Figure 3C). From this result, we conclude that although raloxifene can lead to decreased expression of TMPRSS2-ERG when it binds to ERβ, it makes for a poor treatment alternative in TMPRSS2-ERG prostate cancers expressing ERα.
To further confirm the regulation of TMPRSS2-ERG by ERβ, we examined five ER binding sites previously identified in MCF7 cells that are upstream of the TMPRSS2 gene (18) and the TMPRSS2 promoter. Chromatin immunoprecipitation experiments showed that ERβ localized to a previously unrecognized site in the TMPRSS2 promoter in NCI-H660 cells (Supplementary Figure 6, available online). This result suggests that ERβ regulation of TMPRSS2-ERG expression occurs through direct transcriptional regulation of the gene fusion.
Discussion
Our findings that the molecularly distinct TMPRSS2-ERG class of prostate cancer is regulated by estrogen receptor–dependent pathways have potential clinical implications. First, sustained expression of the TMPRSS2-ERG fusion transcript in castration-resistant prostate cancers (27) suggests that the TMPRSS2 promoter may remain active through ERαstimulation (Supplementary Figure 7, available online). Increased expression of ERαhas been found to be associated with prostate cancer progression, metastasis, and the castration-resistant phenotype (28). Therefore, clinical use of SERMs that have ERα-stimulating activity (eg, raloxifene) may favor the progression of TMPRSS2-ERG-dependent prostate cancer. Our data also suggest a mechanism by which ERβ may function as a tumor suppressor – that is, through negative regulation of TMPRSS2-ERG expression (29). The inhibitory effect of ERβ was suggested by the CMAP analysis result, which flagged phytoestrogens (eg, resveratrol and genistein, both of which are known to have ERβ agonistic activity) as yielding a gene expression signature that is inversely correlated with the TMPRSS2-ERG gene signature. Consistent with this observation, we found that activation of ERβ by DPN decreased TMPRSS2-ERG expression. Importantly, loss of ERβ protein expression has been associated with prostate cancer progression (30), and castration-resistant prostate cancers often lack ERβ expression (31). Loss of ERβ expression would be expected to result in increased TMPRSS2-ERG expression, leading to sustained stimulation of tumor cell growth. These results highlight the need to test ERβ-specific agonists in the treatment of prostate cancer and raise a cautionary note regarding the use of therapeutic agents with ERαagonist activity.
We also note that these findings may have relevance for a recently initiated phase III trial testing the ability of the ERαantagonist toremifene to reduce the incidence of clinically significant prostate cancer in a cohort of 16,000 American men (32). Our results raise the possibility that men who have clinically undetected TMPRSS2-ERG fusion prostate cancers may preferentially benefit from toremifene chemopreventive treatment as compared with men who have TMPRSS2-ERG-negative prostate cancers.
However, one limitation of the current study is that we do not how important the estrogenic influence is in vivo, where the androgen receptor is most often intact and functional. We also have not accounted for the potential role of other genomic alterations that may be associated with the gene fusion event (eg, phosphatase and tensin homolog loss). Future work will explore these questions.
Perhaps most importantly, our results suggest a mechanism by which prostate cancers might develop androgen independence from an initial androgen-dependent state. Specifically, the TMPRSS2-ERG oncogene is regulated by estrogen receptors, whereby ERαagonists (eg, endogenous estrogens) can stimulate oncogene expression. These experiments suggest that pharmacologic inhibition of TMPRSS2-ERG expression using drugs that antagonize ERαactivity and function as ERβ agonists may have promise as a new therapeutic strategy for prostate cancer.
Supplementary Material
Acknowledgments
The authors would like to thank Mark Gerstein for substantial conversations regarding analytic methodology. The authors are also grateful to Chungdak Namgyal of the DFHCC TMA core facility, Danielle Cullinane, and Christopher LaFargue for technical support critical to this study.
The sponsors had no role in the study design, data collection and analysis, interpretation of the results, the preparation of the manuscript, or the decision to submit the manuscript for publication.
Francesca Demichelis, Sven Perner, Scott Tomlins, Arul M. Chinnaiyan, and Mark A. Rubin are co-inventors on a patent filed by The University of Michigan and The Brigham and Women’s Hospital covering the diagnostic and therapeutic fields for ETS fusions in prostate cancer.
Funding:
National Institutes of Health, Prostate SPORE at the Dana-Farber/Harvard Cancer Center (NCI P50 CA090381, R01AG21404 to M.A.R., A.M.C.), Swiss Foundation for Medical-Biological Grants SSMBS (SNF 1168 to K.D.M.), Department of Defense Grant (PC050965 to S.R.S and PC61474 to S.P.), and Prostate Cancer Foundation (F.D.).
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66(7):3396–400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 3.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 4.Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26(31):4596–9. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 5.Bibikova M, Talantov D, Chudin E, Yeakley JM, Chen J, Doucet D, et al. Quantitative gene expression profiling in formalin-fixed, paraffin-embedded tissues using universal bead arrays. Am J Pathol. 2004;165(5):1799–807. doi: 10.1016/S0002-9440(10)63435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andren O, Fall K, Franzen L, Andersson SO, Johansson JE, Rubin MA. How well does the Gleason score predict prostate cancer death? A 20-year followup of a population based cohort in Sweden. J Urol. 2006;175(4):1337–40. doi: 10.1016/S0022-5347(05)00734-2. [DOI] [PubMed] [Google Scholar]
- 7.Varenhorst E, Garmo H, Holmberg L, Adolfsson J, Damber JE, Hellstrom M, et al. The National Prostate Cancer Register in Sweden 1998–2002: trends in incidence, treatment and survival. Scand J Urol Nephrol. 2005;39(2):117–23. doi: 10.1080/00365590510007793. [DOI] [PubMed] [Google Scholar]
- 8.Johansson JE, Adami HO, Andersson SO, Bergstrom R, Holmberg L, Krusemo UB. High 10-year survival rate in patients with early, untreated prostatic cancer. Jama. 1992;267(16):2191–6. [PubMed] [Google Scholar]
- 9.Johansson JE, Adami HO, Andersson SO, Bergstrom R, Krusemo UB, Kraaz W. Natural history of localised prostatic cancer. A population-based study in 223 untreated patients. Lancet. 1989;1(8642):799–803. doi: 10.1016/s0140-6736(89)92269-1. [DOI] [PubMed] [Google Scholar]
- 10.Johansson JE, Holmberg L, Johansson S, Bergstrom R, Adami HO. Fifteen-year survival in prostate cancer. A prospective, population-based study in Sweden. Jama. 1997;277(6):467–71. [PubMed] [Google Scholar]
- 11.Gann PH, Hennekens CH, Stampfer MJ. A prospective evaluation of plasma prostate-specific antigen for detection of prostatic cancer. Jama. 1995;273(4):289–94. [PubMed] [Google Scholar]
- 12.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. [accessed December 2007];Neoplasia. 2004 6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. WWW.Oncomine.org. [DOI] [PMC free article] [PubMed]
- 13.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, et al. TMPRSS2:ERG Fusion-Associated Deletions Provide Insight into the Heterogeneity of Prostate Cancer. Cancer Res. 2006;66(17):8337–41. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 14.Mertz KD, Setlur SR, Dhanasekaran SM, Demichelis F, Perner S, Tomlins S, et al. Molecular characterization of TMPRSS2-ERG gene fusion in the NCI-H660 prostate cancer cell line: a new perspective for an old model. Neoplasia. 2007;9(3):200–6. doi: 10.1593/neo.07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Strom A, Hartman J, Foster JS, Kietz S, Wimalasena J, Gustafsson JA. Estrogen receptor beta inhibits 17beta-estradiol-stimulated proliferation of the breast cancer cell line T47D. Proc Natl Acad Sci U S A. 2004;101(6):1566–71. doi: 10.1073/pnas.0308319100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau KM, LaSpina M, Long J, Ho SM. Expression of estrogen receptor (ER)-alpha and ER-beta in normal and malignant prostatic epithelial cells: regulation by methylation and involvement in growth regulation. Cancer Res. 2000;60(12):3175–82. [PubMed] [Google Scholar]
- 18.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38(11):1289–97. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 19.Vapnik VN. Statistical learning theory. New York: Wiley; 1998. [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc B. 1995;57(1):289–300. [Google Scholar]
- 21.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2004. [accessed February 2008]. http://www.R-project.org. [Google Scholar]
- 22.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313(5795):1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 23.Rhodes DR, Kalyana-Sundaram S, Tomlins SA, Mahavisno V, Kasper N, Varambally R, et al. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia. 2007;9(5):443–54. doi: 10.1593/neo.07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39(1):41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 25.Slentz-Kesler K, Moore JT, Lombard M, Zhang J, Hollingsworth R, Weiner MP. Identification of the human Mnk2 gene (MKNK2) through protein interaction with estrogen receptor beta. Genomics. 2000;69(1):63–71. doi: 10.1006/geno.2000.6299. [DOI] [PubMed] [Google Scholar]
- 26.Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, et al. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59(17):4180–4. [PubMed] [Google Scholar]
- 27.Bonkhoff H, Fixemer T, Hunsicker I, Remberger K. Estrogen receptor expression in prostate cancer and premalignant prostatic lesions. Am J Pathol. 1999;155(2):641–7. doi: 10.1016/S0002-9440(10)65160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonkhoff H, Fixemer T, Hunsicker I, Remberger K. Progesterone receptor expression in human prostate cancer: correlation with tumor progression. Prostate. 2001;48(4):285–91. doi: 10.1002/pros.1108. [DOI] [PubMed] [Google Scholar]
- 29.Cheng J, Lee EJ, Madison LD, Lazennec G. Expression of estrogen receptor beta in prostate carcinoma cells inhibits invasion and proliferation and triggers apoptosis. FEBS Lett. 2004;566:169–72. doi: 10.1016/j.febslet.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 30.Pasquali D, Rossi V, Esposito D, Abbondanza C, Puca GA, Bellastella A, et al. Loss of estrogen receptor beta expression in malignant human prostate cells in primary cultures and in prostate cancer tissues. J Clin Endocrinol Metab. 2001;86(5):2051–5. doi: 10.1210/jcem.86.5.7441. [DOI] [PubMed] [Google Scholar]
- 31.Fixemer T, Remberger K, Bonkhoff H. Differential expression of the estrogen receptor beta (ERbeta) in human prostate tissue, premalignant changes, and in primary, metastatic, and recurrent prostatic adenocarcinoma. Prostate. 2003;54(2):79–87. doi: 10.1002/pros.10171. [DOI] [PubMed] [Google Scholar]
- 32.Price D, Stein B, Sieber P, Tutrone R, Bailen J, Goluboff E, et al. Toremifene for the prevention of prostate cancer in men with high grade prostatic intraepithelial neoplasia: results of a double-blind, placebo controlled, phase IIB clinical trial. J Urol. 2006;176:965–70. doi: 10.1016/j.juro.2006.04.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.