SUMMARY
Biochemical functions of proteins in cells frequently involve interactions with various ligands. Proteomic methods for identification of proteins that interact with specific ligands such as metabolites, signaling molecules, and drugs are valuable in investigating the regulatory mechanisms of cellular metabolism, annotating proteins with unknown functions, and also elucidating pharmacological mechanisms. Here we report an energetics-based target identification method in which target proteins in a cell lysate are identified by exploiting the effect of ligand binding on their stabilities. Urea-induced unfolding of proteins in cell lysates is probed by a short pulse of proteolysis, and the effect of a ligand on the amount of remaining folded protein is monitored on a proteomic scale. As a proof of the principle, we identified proteins that interact with ATP in the E. coli proteome. Literature and database mining confirmed a majority of the identified proteins are indeed ATP-binding proteins. Four identified proteins that were previously not known to interact with ATP were cloned and expressed to validate the result. Except for one protein, the effects of ATP on urea-induced unfolding were confirmed. Analyses of the protein sequences and structure models were also employed to predict potential ATP binding sites in the identified proteins. Our results demonstrate that this energetics-based target identification approach is a facile method to identify proteins that interact with specific ligands on a proteomic scale.
Keywords: protein-ligand interaction, target identification, protein stability, proteolysis, ATP
INTRODUCTION
Biochemical functions of proteins commonly involve interactions with small molecules, which act as substrates, signaling molecules, and allosteric regulators. Discovery of novel interactions between proteins and metabolites provides valuable information on the function and regulation of proteins as well as biochemical roles of the metabolites.1 Bioactive small molecules are frequently discovered from phenotype-based assays without knowing their molecular targets, and it is critical to identify their targets to decipher the mode of actions.2–4 Additionally, the identification of off-targets is essential to understand the mechanisms of side effects of drugs.5 This “target identification problem” or “target deconvolution problem” is actually a significant bottleneck in drug discovery.6–9
Protein-ligand interactions have been conventionally investigated with individual proteins by performing hypothesis-driven biochemical and biophysical assays, which are typically labor-intensive and low-throughput. Recent advances in genomics and proteomics, however, prompt the development of high-throughput approaches to discover novel protein-ligand interactions on a systems level. One of the most common methods employed to identify proteins interacting with a ligand is to capture the binding proteins by affinity-based separation such as affinity chromatography.5,10–12 Although well-established and popular, affinity-based separation still has many technical limitations.6 Small molecules need to be attached to a solid matrix or a detection tag for the separation of the binding proteins. This chemical modification may result in altered affinity or specificity of the molecule. Also, the requirement of chemical modification limits the application of affinity-based separations only to molecules with reactive functional groups for attachment. Another issue with affinity based separation is that the control of stringency is not feasible. Proteins bound nonspecifically are removed by extensive washing. However, this process frequently separate proteins by their dissociation kinetics, not by their affinity.6
Genetic approaches using mutant libraries or arrays have recently gained popularity as a drug target identification method.13–15 When a mutant has a distinct drug response from the wild type, it is likely that the product of the mutated gene is involved in the action of the drug in the cell somehow. Because of the ease of high-throughput applications, genetic approaches appear to be a promising alternative to biochemical approaches. However, genetic approaches also have shortcomings. Construction of mutant libraries is costly and time-consuming. Mutant libraries are mostly available only for lower organisms, and drug targets in humans must be inferred from experimental results obtained with these model organisms. Genetic approaches are not useful for the drugs with subtle pharmacological effects because the inhibition of the growth is the typical readout in genetic screens. Also, genetic approaches are limited to xenobiotic drug-like molecules and cannot be used for metabolites. Finally, identified genes from genetic screens do not necessarily interact with the drug directly. There are many possible indirect mechanisms through which mutants have distinct drug responses even when the direct drug targets are still functional.13
Due to the pros and cons of current approaches, it is important to develop various tools to identify target proteins for small molecules. Target identification methods based on different principles are complementary to each other, and the combined use of several different approaches may provide a more complete picture of system-wide interactions between proteins and small molecules. Energetics-based target identification is a promising new strategy for this purpose. When a protein forms a complex with a ligand, the bound conformation is stabilized according to the dissociation free energy of the complex at a given ligand concentration. This stabilization of the complex results in changes in the energetic properties of the target protein, such as the increase in thermodynamic stability, the decrease in unfolding rates, and the change in the dynamics of the native proteins. Energetics-based target identification exploits these changes in the energetic properties of the target proteins to identify proteins that interact with test molecules in a mixture of proteins, such as a cell lysate.16,17
Here we report an energetics-based target identification method using ‘pulse proteolysis’.18 Pulse proteolysis determines the fraction of folded proteins under a given condition by a brief incubation with a protease. We have shown that this method is a reliable quantitative approach to determine thermodynamic stability and unfolding kinetics of proteins under various circumstances.18–21 The use of proteolysis as a structural probe enables us to monitor urea-induced unfolding of a multitude of proteins in a cell lysate simultaneously without isolating individual proteins for biophysical characterization. By comparing the remaining amount of proteins after pulse proteolysis in the presence and absence of a ligand, targets stabilized by ligand binding can be identified from the mixture of proteins.
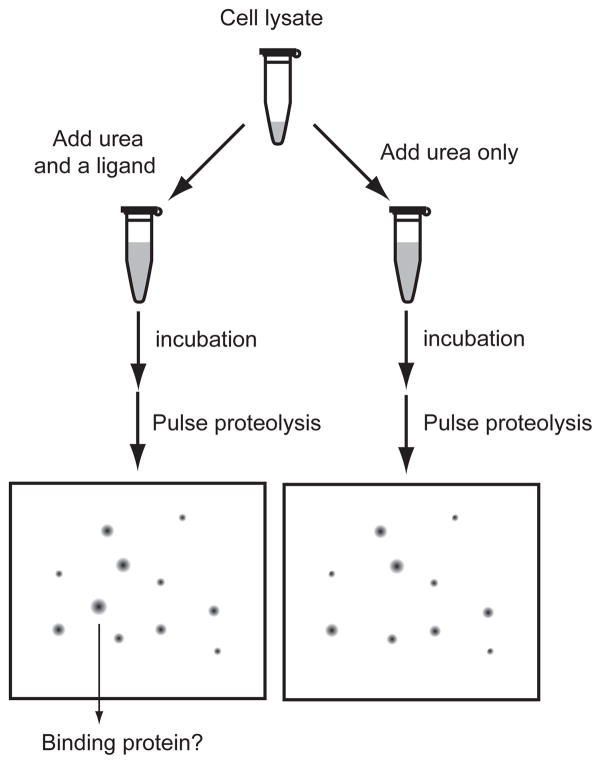
As a proof of principle, we identified ATP-binding proteins in the E. coli proteome by combining pulse proteolysis and two-dimensional (2-D) gel electrophoresis. The schematic diagram of the experimental procedure is shown in Figure 1. A cell lysate is incubated with a ligand in urea, while a control sample is incubated under identical conditions without the ligand. After the incubation, pulse proteolysis is performed in an identical manner for both samples. The remaining proteins are analyzed by 2-D gel electrophoresis. The comparison of the two 2-D gels reveals spots whose intensities are influenced by the presence of the ligand. With this approach, we identified known ATP-binding proteins as well as proteins whose affinity to ATP has not been reported. We characterized several of the identified proteins and confirmed that the biophysical principle of the methodology is valid. This result demonstrates that our energetics-based target identification by pulse proteolysis is a facile and powerful method to identify proteins that interact with specific ligands.
Figure 1.
Energetics-based target identification by pulse proteolysis. A cell lysate is incubated with a ligand and urea. A control reaction is prepared in the identical way except without the ligand. After incubation, unfolded proteins are digested by pulse proteolysis and remaining proteins are analyzed by 2-D gel electrophoresis. Proteins in the spots showing differential intensities on the 2-D gels of the two reactions are identified and characterized further.
RESULTS
Identification of ATP binding proteins
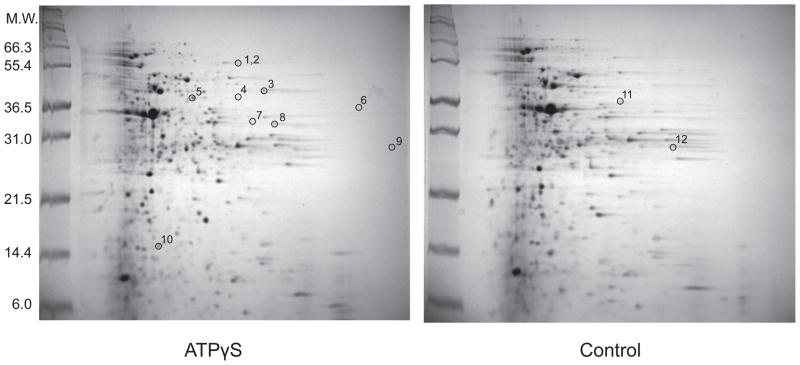
To identify ATP-binding proteins, we incubated an E. coli lysate for 2 hrs in a buffer containing 3.0 M urea and 1.0 mM ATPγS. We also incubated a control without ATPγS under the identical condition. ATPγS was used instead of ATP to minimize the loss of the ligand by enzymatic hydrolysis during the incubation. After the incubation, the reactions were treated with 0.20 mg/mL thermolysin for 1 min (pulse proteolysis), and the resulting proteins were analyzed by 2-D gel electrophoresis. Figure 2 shows a representative pair of the resulting 2-D gels. Overall the two gels look quite similar, indicating that ATPγS did not affect most proteins shown on the gels. Still, a careful comparison revealed spots that show different intensities between the two gels. To rule out possible false positives resulting from the variability in sample preparations and 2-D electrophoresis, we performed three replicate experiments. For further analysis, we selected only the spots showing obvious and consistent changes in intensities from the three pairs of 2-D gels. This screen was not to search exhaustively for ATP-binding proteins, but to prove the principle of the approach. Through this selection process, we chose twelve spots for protein identification. Ten spots had higher intensities on the gels of ATPγS-treated samples than the control gels without ATPγS (spots 1 to 10 in Figure 2), and the other two spots had higher intensities on the control gels than the gels of ATPγS-treated samples (spots 11 and 12 in Figure 2). A spot present only in the control gels without ATPγS may occur when conformational changes associated with ATP binding make a protein susceptible to pulse proteolysis or when the spot contains a partially cleaved protein produced in the absence of ATPγS.
Figure 2.
Two-dimensional gel electrophoresis of an E. coli lysate after pulse proteolysis in 3.0 M urea with and without ATPγS. An E. coli cell lysate was incubated with 1.0 mM ATPγS in 3.0 M urea for 2 hrs before pulse proteolysis. A control sample was also prepared under the identical condition except without 1.0 mM ATPγS. Spots with different intensities between the two gels were identified. Spots selected for in-gel digestion are labeled by numbers. Spots 1 and 2 are distinguishable on the actual gel but look as a single spot on this image. Two spots, 11 and 12, only appeared in the control gel. The major spot near 36.5 kDa corresponds to thermolysin.
The identities of the proteins in the selected spots were determined by using in-gel digestion followed by MALDI-TOF-TOF mass spectroscopy (Table 1 and Supplementary Table S1). Ten proteins were successfully identified from the twelve spots (Table 1). The comparison of the molecular weights estimated from the location of the spots with the known molecular weights of the identified proteins suggested that nine spots contain intact proteins and three spots contain fragments (Table 1). Interestingly, intact glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was found only on the control gels. This observation rules out the possibility that GAPDH is digested partially only in the absence of ATPγS.
Table 1.
Identified proteins
| Spot | Gene namea | Descriptiona | ATP binding |
||
|---|---|---|---|---|---|
| Gene Ontologyb | Experimental Evidencec | Kd (μM) | |||
| 1 | atpA | ATP synthase, F1 complex, α subunit | √ | √ | 0.140 |
| 2 | lpd | Lipoamide dehydrogenase (E3 monomer) | |||
| 3d | glnS | Glutaminyl-tRNA synthetase | √ | √ | 19058 |
| 4,7d | pheS | Phenylalanyl-tRNA synthetase, α-chain | √ | √ | 60059 |
| 5 | pfkA | 6-phosphofructokinase-1 | √ | √ | 0.02560 |
| 6 | yncE | Hypothetical protein | √ | ||
| 8 | sucD | Succinyl-CoA synthetase, α subunit | √ | √ | 9061 |
| 9 | mlaC | Periplasmic binding protein of the phospholipid ABC transporter | |||
| 10 | groS | GroES | √ | √44 | |
| 11,12d | gapA | Glyceraldehyde-3-phosphate dehydrogenase-A | |||
The gene name and the description of each protein were collected from EcoCyc, E. coli functional genomics database.
Genes annotated with the GO term ‘ATP binding’ (GO:0005524) in EcoCyc database.
Proteins whose ATP binding is confirmed experimentally.
The spot was found to contain a fragment of the identified protein.
Functions of the identified proteins were surveyed by using the information available on EcoCyc,22 a genomics database of E. coli K12. Seven out of ten proteins are annotated with a Gene Ontology (GO) term, ‘ATP binding’ (GO:0005524). Through literature search, we found experimental evidence of ATP binding to six out of these seven proteins (Table 1). The Kd values for five proteins collected from literature range from 0.025 to 600 μM (Table 1), which are all smaller than the concentration of ATPγS used for the screen (1.0 mM). Apparently, yncE is annotated with ATP binding without experimental evidence based on the existence of P-loop, ATP/GTP-binding site motif (pattern PS00017 in PROSITE database23). The three identified proteins that are not annotated with ATP binding are lipoamide dehydrogenase E3 subunit (E3), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and a periplasmic binding protein (mlaC).
Validation of the results from the proteomic screen
To validate the proteomic screen results, we chose three proteins with no documented ATP binding ability (GAPDH, E3, mlaC) and the hypothetical protein annotated as an ATP-binding protein (yncE). The proteins were over-expressed in E. coli after cloning, and the effects of ATP on their unfolding in urea were monitored by pulse proteolysis. The cell lysates containing the over-expressed proteins were used directly for validation without purification. It is noteworthy that, even in case that the identified protein is a true positive, validation with the over-expressed proteins may not reproduce the observation made with the 2-D gel electrophoresis exactly. In case of a multimeric protein, the apparent stability would be dependent on the protein concentration and can be significantly increased when the protein is overexpressed. Also, if a protein is a component of a protein complex formed with other proteins, overexpression of the single component is not likely to allow the formation of the proper quaternary structure. Therefore, the purpose of the validation with the overexpressed protein is to examine the effect of ATP on the thermodynamics and kinetics of unfolding, not to reproduce the proteomic screen results exactly.
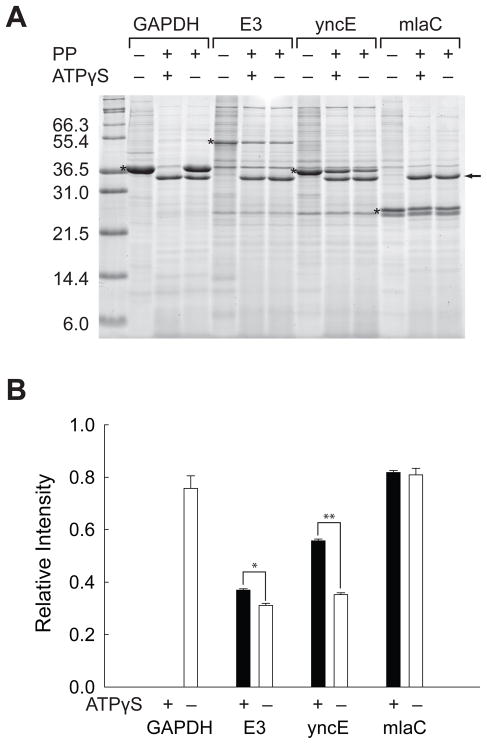
To compare with the proteomic screen, the proteins in crude cell lysates were incubated under the identical condition used for the screen. The cell lysates were incubated in 3 M urea for 2 hrs at 25°C with 1.0 mM ATPγS. Controls were also incubated under the identical condition except without 1.0 mM ATPγS. After pulse proteolysis, the remaining proteins were analyzed by SDS PAGE (Figure 3A). For comparison, the amount of remaining protein after pulse proteolysis was normalized as the relative ratio to the amount of the intact proteins without pulse proteolysis (Figure 3B). The averages of the relative ratios were determined from triplicate experiments. The examination with the over-expressed protein suggests that ATPγS indeed affects the unfolding of GAPDH, E3, and yncE. As observed on the 2-D gels, no detectable amount of GAPDH survives pulse proteolysis when ATPγS is present. E3 shows a small but still statistically meaningful increase in band intensity in the presence of ATPγS (p = 0.0013). The observed difference is somehow much less evident than that observed on the 2-D gels. The fact that E3 is a component of multi-component enzyme systems may explain this marginal effect of ATPγS on the overexpressed protein. YncE shows a statistically significant increase in band intensity in the presence of ATPγS (p < 0.0001), which confirms the proteomic screen result. However, the test with over-expressed mlaC is not consistent with the proteomic screen result. Whether ATPγS is present or not, mlaC is resistant to pulse proteolysis, and the relative intensities are not statistically different. The discrepancy with the proteomic screen result suggests that mlaC may be a false positive or that the experiment with over-expressed mlaC may not truly mimic the experiment performed with the endogenous mlaC due to the reasons described above.
Figure 3.
Validation of the proteomic screen with recombinant proteins. (A) Pulse proteolysis of recombinant GAPDH, E3, yncE and yrbC in E. coli lysates with and without 1.0 mM ATPγS. PP stands for pulse proteolysis. The bands corresponding to over-expressed proteins are marked with asterisks. The bands corresponding to thermolysin are marked with an arrow. (B) Relative intensities of remaining proteins after pulse proteolysis with (black bar) and without (white bar) 1.0 mM ATPγS. The band intensities of over-expressed proteins on the gel shown in Figure 3A were quantified and expressed as ratios to the band intensities of the undigested proteins on the same gel. The error bars indicate the standard deviation of triplicate experiments. *p = 0.0013, **p < 0.0001.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH)
To elucidate the physical origin of the effect of ATP on the susceptibility changes of the identified proteins, we further investigated the effect of ATP on the thermodynamic stability and unfolding kinetics of the proteins. By using pulse proteolysis, we monitored urea-induced unfolding of the over-expressed proteins in cell lysates without purification.
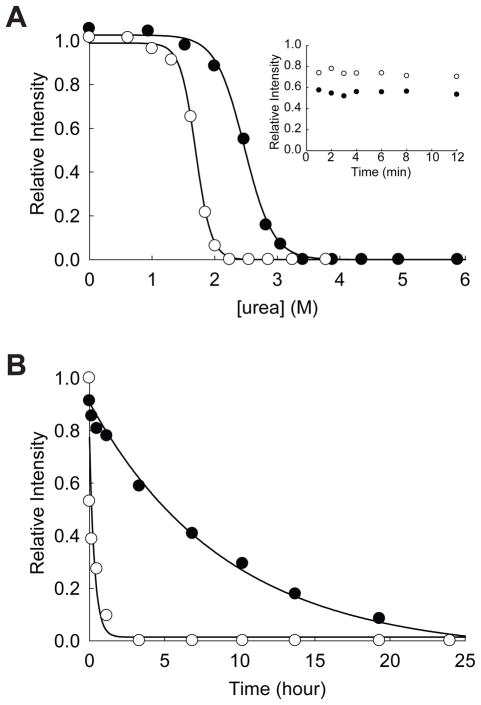
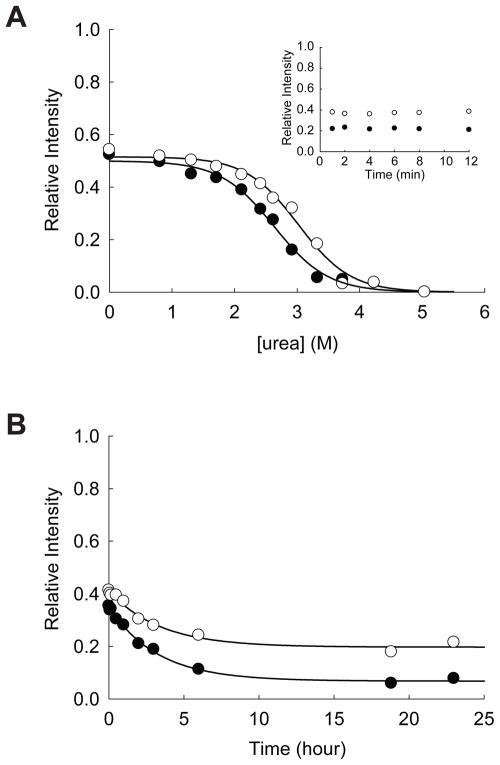
GAPDH is a glycolytic enzyme catalyzing the reduction of glyceraldehyde-3-phosphate using NAD+ as a cofactor. ATP binding to this enzyme has not been previously known. Unfolding of GAPDH in varying concentration of urea for 24 hrs clearly showed a significant decrease in the apparent Cm value (the midpoint of the unfolding transition) from 2.47 ± 0.02 M in the absence of ATPγS to 1.70 ± 0.01 in the presence of 1.0 mM ATPγS (Figure 4A). As a tetrameric protein, the thermodynamic stability of GAPDH is likely to be dependent on the protein concentration. These Cm values are, therefore, apparent Cm values under the given condition.24 Interestingly, both Cm values are lower than 3.0 M urea, which is the urea concentration used for the proteomic screen. The comparison of the unfolding of GAPDH in 3.0 M urea for 2 hrs (~20%, Figure 3) and 24 hrs (~90%, Figure 4A) indicates that the protein does not reach its conformational equilibrium under the condition employed for the proteomic screen. Another interesting observation is that under native conditions GAPDH is resistant to pulse proteolysis even in the absence of ATPγS. This result rules out the possibility that binding of ATPγS induces a conformational change in GAPDH, which makes the protein susceptible to pulse proteolysis.
Figure 4.
Effect of ATP on the stability of GAPDH. (A) Equilibrium unfolding of GAPDH in urea probed by pulse proteolysis with (○) and without (●) 1.0 mM ATPγS. Relative intensities are the ratio of the band intensities of remaining proteins after pulse proteolysis to the band intensity of undigested protein. The apparent Cm values were determined by fitting the relative intensities to Eq. 1. INSET: Prolonged incubation of GAPDH with 0.2 mg/mL thermolysin in 2.7 M urea without ATPγS (●) and in 1.8 M urea with 1.0 mM ATPγS (○). (B) Unfolding of GAPDH in 3 M urea with (○) and without (●) 1.0 mM ATPγS monitored by pulse proteolysis. Unfolding kinetic constants were determined by fitting the relative intensities to a first-order rate equation.
Reliable determination of the fraction of folded proteins (fFold) by pulse proteolysis requires that folded protein is not digested during the 1-min pulse. To confirm that folded protein is not susceptible to pulse proteolysis, proteolysis at a urea concentration near the apparent Cm are typically monitored beyond 1 min. The inset in Figure 4A shows that folded GAPDH is resistant to proteolysis near its apparent Cm, whether ATPγS is present or not. Therefore, pulse proteolysis is a valid probe to determine fFold of this protein. This result also suggests that even in the transition zone ATPγS does not make the folded GAPDH susceptible to pulse proteolysis. Instead, ATPγS seems to decrease the population of native GAPDH in urea by an unknown mechanism. One possible explanation for this unusual destabilizing effect of ATP is that ATP binds and accumulates a non-native form of GAPDH, which is susceptible to proteolysis. This hypothesis is currently being tested in our laboratory.
Unfolding kinetics of GAPDH also demonstrates that ATPγS increases the unfolding rate of this protein significantly (Figure 4B). The unfolding rate constant of GAPDH in 3.0 M urea is increased by ~30-fold from (3.0 ± 0.5) × 10−5 s−1 to (8 ± 3) × 10−4 s−1 by 1.0 mM ATPγS. Typically, ligand binding stabilizes native conformations of proteins and slows down protein unfolding. This increase in the unfolding rate by a ligand is quite unusual and may suggest the stabilization of the unfolding transition state, not the native form of GAPDH, by ATPγS. Also, the unfolding kinetics of GAPDH corroborates that in the absence of ATPγS the conformational equilibrium of this protein in 3.0 M urea cannot be achieved in 2 hrs (Figure 4B).
Dihydrolipoamide dehydrogenase (E3)
Dihydrolipoamide dehydrogenase (E3) is a component of three multicomponent enzyme complexes: pyruvate dehydrogenase multienzyme complex, 2-oxoglutarate dehydrogenase complex, and the glycine cleavage system.25–27 These multicomponent enzymes are large protein complexes composed of 12 – 24 monomeric units of several enzymes. For example, pyruvate dehydrogenase multienzyme complex contains twelve E1 dimers, a 24-subunit E2 core, and six E3 dimers. When isolated, E3 exists as a dimeric form bound to FAD.28 E3 uses FAD as cofactor for an electron transfer to the final acceptor, NAD+. From our literature search, we could not find any report showing ATP binding to this enzyme. Because E3 functions as a component of these large enzyme complexes, it is plausible that our proteomic screen have monitored the effect of ATPγS on the stability of the complexes, not that of the dimeric form of E3. Therefore, as seen in Figure 3, the energetic properties observed with the overexpressed E3 dimer can be different from the property we observed in the proteomic screen.
Unfolding of E3 probed by pulse proteolysis after 24-hr incubation shows an evident increase in Cm from 2.63 ± 0.05 M to 3.02 ± 0.05 M by 1.0 mM ATPγS (Figure 5A). This increase in Cm by ATPγS is consistent with the increase in band intensity after pulse proteolysis upon incubation with ATPγS in 3.0 M urea (Figure 3B). Prolonged incubation with the protease near Cm beyond 1 min confirmed that folded E3 is resistant to pulse proteolysis and that fFold is determined faithfully by pulse proteolysis (Inset of Figure 5A).
Figure 5.
Effect of ATP on the stability of E3. (A) Equilibrium unfolding of E3 in urea probed by pulse proteolysis with (○) and without (●) 1.0 mM ATPγS. Relative intensities are the ratio of the band intensities of remaining proteins after pulse proteolysis to the band intensity of undigested protein. The apparent Cm values were determined by fitting the relative intensities to Eq. 1. INSET: Prolonged incubation of E3 with 0.2 mg/mL thermolysin in 2.8 M urea with (○) and without (●) 1.0 mM ATPγS. (B) Unfolding of E3 in 3 M urea with (○) and without (●) 1.0 mM ATPγS monitored by pulse proteolysis. Unfolding kinetic constants were determined by fitting the relative intensities to a first-order rate equation.
To demonstrate that this change in Cm of ~0.4 M is meaningful, we performed control experiments with proteins that do not interact with ATP, E. coli maltose binding protein and E. coli dihydrofolate reductase (Supplementary Figure S1). In both cases, the differences in Cm values in the presence and absence of ATPγS were less than 0.1 M. This small variation in Cm values is consistent with the typical standard errors in triplicate measurements of Cm by pulse proteolysis. This result clearly shows that the change in Cm value of E3 by ATPγS is statistically significant.
The relaxation kinetics of E3 under the proteomic screen condition is not affected by the presence of ATPγS. The relaxation rate constants of E3 in 3.0 M urea are (8.7 ± 0.7) × 10−5 s−1 and (8.3 ± 1.3) × 10−5 s−1 in the absence and presence of 1.0 mM ATPγS. This result indicates that the observed ATPγS effect does not result from the relaxation kinetics but from the increase in fFold. As for GAPDH, the 2-hour incubation for the proteomic screen was not long enough for E3 to reach its conformational equilibrium in 3.0 M urea, but still produce discernable differences in fFold (Figure 5B).
Interestingly, though folded E3 is resistant to pulse proteolysis (Inset in Figure 5A), about 50% of E3 in the cell lysates is readily digested by pulse proteolysis even under native conditions (Figure 5A). The nature of this fraction of E3 susceptible to pulse proteolysis is not clear. One possible explanation is that this susceptible population is misfolded E3 without FAD. FAD is known to be necessary for maturation of E3 into a stable dimer.28 However, incubation of the over-expressed E3 in the cell lysate with added FAD for two hours did not rescue the susceptible fraction (Data not shown).
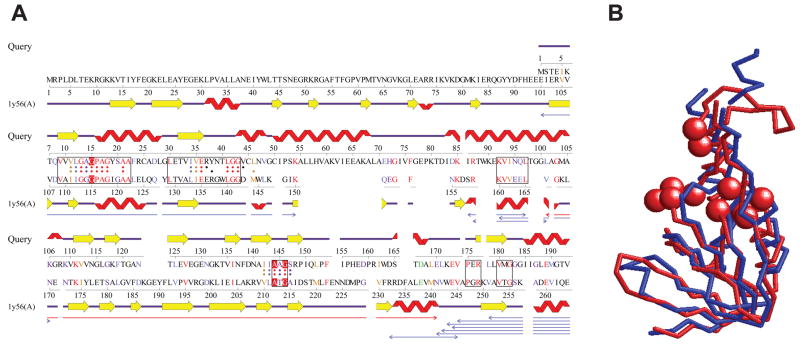
Encouraged by the experimental result suggesting interaction between E3 and ATP, we investigated if E3 has any structural motif for ATP binding. Although the structure of E3 has not been solved, a structure model is available in ModBase29, which is based on a homologous protein structure from P. putida (PDB code: 1lvl). For this structural model, the ProFunc server,30 which matches structural templates of functional sites of known proteins in a query structure, identified a match with an ATP binding site of P. horikoshii L-proline dehydrogenase (PDB code: 1y56) at residues around 10–20 and 145 (Figure 6A). ProFunc identified 22 identical residues and 15 similar residues between the two proteins at the local sites. Indeed, the structure of the whole N-terminal domain (1–156) of the model of E3 overlaps well with a domain (100–213) in the α subunit of L-proline dehydrogenase with a root mean square deviation of 3.5Å (Figure 6B). This structural analysis as well as the observed stabilization by ATP suggests that E3 may have a potential ATP binding site. It is notable that this potential ATP binding site in E3 is part of the tentative FAD binding site; according to the structure of the homologous protein from P. putida, the identified ATP binding site overlaps with the binding site of the ADP moiety of FAD. This model suggests that ATP may compete with FAD for the same binding site and interfere with the regular function of the protein.
Figure 6.
Structure similarity between the N-terminal domain of E3 and L-proline dehydrogenase α subunit. (A) Structural alignment between the N-terminal domain of the model of E3 (Query) and L-proline dehydrogenase α subunit (PDB CODE: 1y56A) computed by the ProFunc server.30 The homology model of E3 was built with ModBase,29 based on a template structure (PDB CODE: 1lvl). The boxes on the sequences of the two proteins represent segments where the sequence identity exceeds 35%. Dots assigned to each sequence indicate residues within 10 Å to the center of the template protein (1y56A) and hence considered in computing the alignment. The three residues in red comprise of the ATP-binding site of the template, which are initially matched in the ProFunc search. The thin arrows below alignments are structurally similar regions between the two proteins, which can be superimposed within a root mean square deviation (RMSD) of 3.0 Å. The red arrow shows the longest such segment. For more details, see the ProFunc website (http://www.ebi.ac.uk/thornton-srv/databases/profunc/). (B) Structural superimposition of the N-terminal domain of the model of E3 (blue, residue 1–156) and the homologous region of L-proline dehydrogenase (red, residue 100–213). Spheres indicate ATP-binding residues of L-proline dehydrogenase. The RMSD of the two structures is 3.48Å.
yncE
yncE is a hypothetical protein with no known function. Still, this protein is annotated as an ATP-binding protein in EcoCyc due to the presence of a P-loop, an ATP-binding sequence motif at the position 320 – 327. A preliminary crystal structure of the protein has been reported in the literature, 31 but the coordinate has not been deposited to the Protein Data Bank.32 Still using homology models of yncE available at the EcoliProteins database,33 we found that the ATP-binding motif region is located at an exposed loop. The location of the binding motif on the surface of the protein confirms that the P-loop is likely involved in ATP binding.
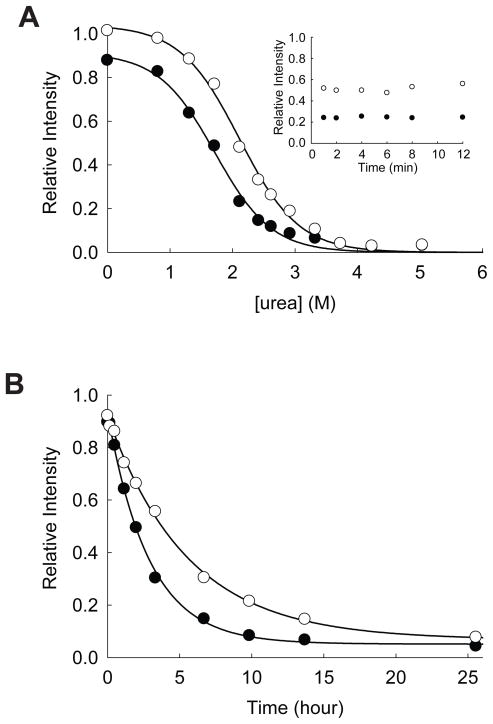
Our experimental validation also supports that yncE is indeed an ATP-binding protein. Equilibrium unfolding of yncE in urea shows that ATPγS increases the Cm of this protein from 1.72 ± 0.06 M to 2.11 ± 0.04 M (Figure 7A). Without ATPγS, a small fraction of yncE (~10%) is digested by pulse proteolysis even under native conditions. However, prolonged incubation with the protease near Cm beyond 1 min confirmed that folded yncE is resistant to pulse proteolysis whether ATPγS is present or not (Inset of Figure 7A). The digestion of the small fraction of the protein under native conditions may suggest that some fraction of the protein is in non-native conformations susceptible to proteolysis, and ATPγS may have converted this fraction of protein into the native conformation resistant to proteolysis. ATPγS slows the unfolding of yncE. The unfolding rate constant of yncE in 3.0 M urea is decreased from (9.7 ± 0.5) × 10−5 s−1 to (5.0 ± 0.2) × 10−5 s−1 by 1.0 mM ATPγS (Figure 7B). The relaxation of this protein is relatively slow; even in the absence of ATPγS the relaxation of yncE in 3 M urea is incomplete after 2-hr incubation. This incomplete relaxation explains the significant amount of remaining protein after pulse proteolysis in 3 M urea (Figure 3), which is actually greater than its Cm values whether ATPγS is present or not.
Figure 7.
Effect of ATP on the stability of yncE. (A) Equilibrium unfolding of yncE in urea probed by pulse proteolysis with (○) and without (●) 1.0 mM ATPγS. Relative intensities are the ratio of the band intensities of remaining proteins after pulse proteolysis to the band intensity of undigested protein. The apparent Cm values were determined by fitting the relative intensities to Eq. 1. INSET: Prolonged incubation of yncE with 0.2 mg/mL thermolysin in 1.9 M urea with (○) and without (●) 1.0 mM ATPγS. (B) Unfolding of yncE in 3 M urea with (○) and without (●) 1.0 mM ATPγS monitored by pulse proteolysis. Unfolding kinetic constants were determined by fitting the relative intensities to a first-order rate equation.
DISCUSSION
Energetics-based discovery of ATP-binding proteins
Using ATPγS as a test molecule, we demonstrated the validity of our energetics-based target identification approach. Out of ten identified positives, six proteins are already known to interact with ATP based on experimental evidence in literature. To validate the ATP binding of the remaining four proteins, we investigated the effect of ATP on their thermodynamic stability and unfolding kinetics individually. Except mlaC, we confirmed that ATP indeed affects their energetic properties (Figures 3–7). This result clearly demonstrates that our energetics-based target identification approach is a reliable way to excavate putative ATP-binding proteins. Though recombinant mlaC was not confirmed to interact with ATP directly, it is still possible that this protein is stabilized by forming a complex with ATP-binding proteins when the protein exists in a stoichiometric amount to its binding partners.
According to GO terms in EcoCyc, products of 361 out of 4144 protein-coding genes are annotated as ATP binding proteins (GO:0005524). Because ATP binding to some proteins still may not be known, ATP binding proteins are roughly estimated to be ~10% of the proteome. Considering that typically ~500 proteins are observable by 2-D gel electrophoresis with the staining method employed in our study, we expect ~50 ATP binding proteins are expressed enough to be identified by 2-D gel electrophoresis. Because quite a few proteins still remain folded in 3.0 M urea (The control gel in Figure 2) and ATP binding to these folded proteins would not make any difference upon pulse proteolysis, the upper limit of the number of identifiable ATP-binding proteins under our experimental condition would be 20 – 30 proteins. Although the positive spots were not searched exhaustively, identification of ten ATP-binding proteins from selected spots, which are well isolated and reproducible from triplicate experiments, suggests that the coverage of this approach is still in a reasonable range. The limitation in the coverage seems due to the resolving power of 2-D gel electrophoresis. More quantitative proteomics platforms, such as differential gel electrophoresis (DIGE)34 or various quantitative mass spectrometry approaches such as stable isotope labeling with amino acids in cell culture (SILAC),35,36 may improve the proteome coverage significantly.
The presented result shows that our target identification approach is applicable to the identification of enzymes using a metabolite as their substrates. Four confirmed ATP-binding proteins from our proteomic screen (glutaminyl-tRNA synthetase, phenylalanyl-tRNA synthetase, succinyl-CoA synthetase, and 6-phosphofructokinase) are known to use ATP for their catalytic functions. Recently, a powerful new proteomic method, dubbed ‘activity-based protein profiling’,37,38 has been introduced to identify proteins with specific enzymatic activities by using mechanism-based conjugation reagents. This approach has been shown to be quite effective in identifying novel enzymes and monitoring the changes in enzymatic activities on a proteomic scale. Our target identification provides an alternative way to identify enzyme-substrate interactions without creating mechanism-based conjugation reagents.
Our target screen identifies not only enzymes utilizing the test molecule as a substrate but also proteins interacting with the test molecule through non-catalytic binding sites, which may have regulatory roles. Our screen identified ATP synthase F1-α subunit as an ATP-interacting protein (Table 1). The cytosolic F1 complex of ATP synthase contains two subunits, F1-α and F1-β. The catalytic subunit is F1-β in which ATP is synthesized. Though F1-α is not involved in catalysis, this subunit has been shown to bind ATP with high affinity.39,40 Though the biochemical role is still unknown,41,42 ATP binding by F1-α may have a regulatory function. Our screen also identified GroES (Table 1), which forms a heptameric lid to cap the chaperone protein GroEL.43 GroES has been shown to bind ATP by azido-ATP labeling.44 While GroEL utilizes ATP for its chaperone function, the role of ATP binding by GroES is not yet known. Still, this ATP binding may have a regulatory role. The identification of ATP synthase F1-α subunit and GroES demonstrates that our target identification approach may discover protein–ligand interactions at non-catalytic sites as well as at active sites. It is still noteworthy that F1-α and GroES may be stabilized by ATP indirectly by forming complexes with F1-β and GroEL, respectively.
Our screen identified several novel ATP interactions that have not been reported previously. E3 was not known to interact with ATP. YncE is a hypothetical protein with an unknown function, and its interaction with ATP was suggested only by a sequence analysis. Validation with recombinant proteins corroborates our proteomic identification of E3 and yncE as putative ATP-binding proteins (Figure 3). In addition, a potential ATP binding site was identified in the structure model of E3. These proteins exemplify the utility of our target identification and subsequent validation processes using unfolding energetics and bioinformatics in discovering novel biochemical functions and regulatory roles of metabolites. The putative interactions discovered through this approach would provide highly reliable leads, which is valuable in generating hypotheses for further rigorous biochemical and biophysical investigations.
Interestingly, we also found that GAPDH is apparently destabilized in the presence of ATPγS. Our preliminary characterization suggests that natively folded GAPDH does not bind ATPγS, but nonnative conformations of GAPDH may interact with ATPγS. Through this interaction with nonnative conformations, ATP seems to modulate the energetics of unfolding of GAPDH. This unusual effect of ATP on unfolding of this protein is quite interesting because the ATPγS concentration used in our screen is close to the physiological concentration of ATP in E. coli cytosol.45 It is plausible that ATP may exert a similar effect on this protein in vivo. Recently, the advance of proteomics enables the investigations of the energetic properties of proteins on a proteomic scale, which uncover proteins with exceptional energetic properties such as resistance against proteolysis46 or resistance against denaturation by SDS.47 These studies demonstrate surprising diversity in energetic properties of proteins in cells and provide valuable insights on how the energetic properties are linked with the functions of the proteins. The finding of the effect of ATP on GAPDH exemplifies that our screen not only reports protein–ligand interactions but also reveal their unusual energetic consequences.
Experimental parameters to be considered
Our screen discovers protein–ligand interactions based on the effect of ligand binding on the conformational energies of target proteins. Due to the diversity in thermodynamic stabilities and binding affinities of target proteins in a proteome, a single experimental condition would not guarantee the identification of every target. However, by choosing experimental conditions tactfully one may maximize the efficiency of the screen. Several experimental parameters are worth a careful consideration.
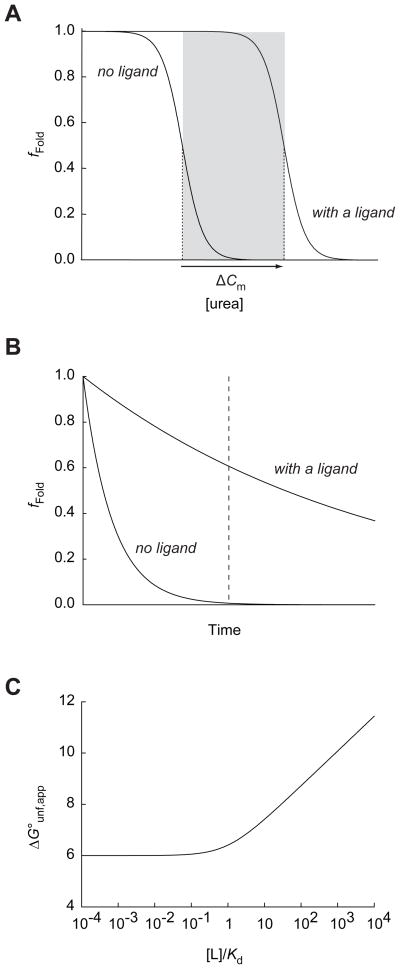
First, the concentration of urea is an important parameter that determines the experimental window of the proteome coverage. To be identified under a given experimental condition, a target protein needs to be unfolded in the absence of the ligand but to be folded in the presence of the ligand (Figure 8A). This window of the urea concentration for identification is unique to each target. If the urea concentration is outside of this window, the target protein would not show significant difference in fFold upon ligand binding. As a proof-of-principle experiment, we performed our screen at a single urea concentration (3.0 M). Performing screens at several different urea concentrations may increase the coverage of the proteome further. The exceptional stability of thermolysin allows pulse proteolysis to be performed in up to 8 M urea.18
Figure 8.
Experimental parameters in energetics-based target identification. (A) The window of urea concentrations allowing the detection of ligand binding by energetics-based target identification. The grey box indicates the range of urea concentrations between the Cm value of a protein in the presence and absence of a certain concentration of a ligand. In this range the target would show a significant difference in the fraction of folded protein (fFold), which is probed by pulse proteolysis. (B) Incubation time as an experimental parameter. Even when the system does not reach equilibrium at the time of pulse proteolysis (indicated by a dashed line), the target may show a significant difference in fFold due to the slow unfolding in the presence of the ligand. For the demonstration of the principle in this example, the unfolding rate constant is decreased by 10-fold in the presence of ligand. (C) Ligand concentration as an experimental parameter. The apparent thermodynamic stability (ΔG°unf,app) of a protein with the global stability (ΔGunf°) of 6.0 kcal/mol is calculated with varying ratios of a ligand concentration to a dissociation equilibrium constant ([L]/Kd) by using Eq. 2.
Second, the duration of the incubation in urea before pulse proteolysis is also a parameter affecting the screening results. Our screen does not require a complete equilibration of the proteins under a given condition. Ligand binding frequently slows unfolding of target proteins. Even before conformational equilibrium is achieved, target proteins may show difference in fFold in the presence of the ligand due to the effect on unfolding kinetics (Figure 8B). YncE is an example of this case. This protein does not reach its conformational equilibrium in 2 hrs under our experimental condition. However, the decreased unfolding rate of the protein in the presence of ATP still results in difference in fFold of the protein (Figure 7B). Actually a prolonged incubation of this protein under the experimental condition would lead to complete unfolding of the protein regardless of the presence of the ligand, and no yncE would be detectable after pulse proteolysis even in the presence of ATP. Moreover, a prolonged incubation of a cell lysate at room temperature may cause adverse effects on the integrity of the proteins in the lysate. Considering these factors, we chose to incubate the lysate for 2 hrs. Again, this duration of the incubation can be adjusted according to the conformational energetic characteristics of proteins in a given proteome.
Third, the concentration of the ligand is another important parameter determining the stringency of the screen. When a target protein binds a ligand through its native conformation, the thermodynamic stabilization of the protein is a function of the ligand concentration as shown below:
| (2) |
where ΔG°unf, app is the apparent thermodynamic stability of the protein in the presence of the ligand, ΔG°unf is the thermodynamic stability of the protein in the absence of the ligand, [L] is the ligand concentration, and Kd is the dissociation equilibrium constant of the complex. This quantitative relationship between ΔG°unf, app and [L] is shown in Figure 8C. When [L] ≫ Kd, the apparent stability of the target protein is linearly proportional to ln[L]. As the target protein is stabilized further, the effective window of the experimental parameters for identification (the shaded area in Figure 8A) is broadened. The higher ligand concentration is desirable to maximize the coverage of the screen. However, as the ligand concentration is increased, the chance of observing nonspecific interactions may also increase. Figure 8C also demonstrates how the stringency of the screen can be controlled by the ligand concentration. A target with a Kd value much smaller than the ligand concentration ([L]/Kd ≫ 1) would have a better chance for identification due to its greater stabilization with the given concentration of the ligand. Targets with Kd values greater than the ligand concentration ([L]/Kd < 1) do not experience any significant stabilization. Therefore, a suitable ligand concentration should be determined based on the desired stringency of the screen.
Comparison with alternative approaches
Our energetics-based target identification approach by pulse proteolysis overcomes several limitations of traditional target identification methods. First, this method is performed without modifying the test molecules, and possible adverse effects of the modification on binding are eliminated. The direct use of test molecules in target identification significantly reduces the experimental time and cost. Second, the stringency of the screen can be modulated by changing the ligand concentration in our screen. This modulation of the stringency is quite valuable in minimizing nonspecific binding. Also, because the identification is based on the effect of ligand binding on the conformational energies of the target proteins, the method is universally applicable to any protein–ligand interactions regardless of the functional consequences of the binding events. Our result suggests that the method can be used to identify not only the interactions with substrates but also the interactions with regulatory molecules, signaling molecules, inhibitors, and drugs.
The concept of energetics-based target identification has been demonstrated recently by other laboratories also by identifying drug targets from the changes in proteolytic susceptibility16 and methionine oxidation17 of target proteins upon the addition of a drug. Our approach using pulse proteolysis is based on physical and chemical principles distinct from these two approaches. Though proteolysis is employed as a probe, our approach differs from the one based on the change in proteolytic susceptibility16 in several aspects. Proteolytic susceptibility is determined by the accessibility to a non-native high energy conformation, frequently partially unfolded forms, under native conditions.48,49 Our approach is not strongly dependent on the changes in proteolytic susceptibility due to the brief incubation (1 min) with the protease. As shown with over-expressed proteins, folded GAPDH, E3, and yncE are not digested through pulse proteolysis even in the absence of ATP (Insets of Figures 4, 5, and 7). The observed changes in the amount of remaining proteins after proteolysis are not from the difference in the susceptibility but the difference in thermodynamics and kinetics of unfolding. Moreover, ligand binding may not result in the change in proteolytic susceptibility. Our previous study on E. coli maltose binding protein clearly showed that maltose does not affect the proteolytic susceptibility of this protein.50 The proteolysis of this protein occurs through partial unfolding that does not involve the maltose binding site. The protein is digested without releasing the bound maltose, and the rate of proteolysis is identical in the presence and absence of maltose. Still, the thermodynamic stability and unfolding kinetics of maltose binding protein is strongly dependent on the ligand concentration.18,19 Therefore, our method based on the changes in unfolding behaviors in urea may identify the targets that the susceptibility-based methods cannot identify.
The target identification method based on methionine oxidation17 is similar to our approach in that the effect of ligand binding is monitored by the change in fFold of the target proteins in chemical denaturants. By employing quantitative mass spectrometry, the change in methionine oxidation was monitored at multiple denaturant concentrations, while only a single urea concentration was used in our approach with 2-D gel electrophoresis. Also, their mass spectrometry approach using isobaric mass tags allowed a quantitative comparison between the samples with and without the ligand. However, this approach covers only the proteins containing buried methionine residues whose susceptibility to oxidation increases significantly upon unfolding. Our approach uses nonspecific proteases, which can digest any unfolded proteins regardless of their amino acid compositions or sequences.
As any other proteomic methods, our approach cannot cover the complete proteome. ~20% of E. coli soluble proteins are susceptible to pulse proteolysis even under native conditions (Na and Park, unpublished result). Unless the association with the test molecule confers the proteins resistance against pulse proteolysis, our method would not identify these proteins. Also, quite a few E. coli proteins, possibly including membrane proteins, remain folded even in 8 M urea (Na and Park, unpublished result). Our method would not be able to identify targets in this category of proteins either. Therefore, these energetics-based target identification approaches (proteolytic susceptibility, methionine oxidation, and pulse proteolysis) are actually complementary to each other. The application of two or more of these approaches may increase the proteome coverage significantly.
Here we demonstrate the principle of our energetics-based target identification approach by employing pulse proteolysis and 2-D gel electrophoresis. The results clearly show that this approach is valid and powerful in discovering unknown interactions between proteins and ligands (metabolites, signaling molecules, drug, and so forth) on a proteomic scale. When combined with more powerful proteomics tools, the proteome coverage of the approach would be improved significantly. Though we demonstrated its feasibility by using E. coli lysates in this study, we expect that this approach is also applicable to other proteomes including human, in which many medically significant questions can be pursued with an unprecedented efficiency.
MATERIAL AND METHODS
Escherichia coli lysate preparation
E. coli K-12 (MG1655) cells were grown at 37°C to reach log phase (OD600 = 0.6) and harvested by centrifugation. The cell pellet was washed with ice-cold water and resuspended in 20 mM Tris-HCl buffer (pH 8.0) containing 10 mM EDTA and 1 mM DTT. The resuspended cells were lysed on ice by sonication. Cell debris was removed by centrifugation. To remove metabolites, which may interfere with the following procedures, the lysates were first dialyzed against 20 mM Tris-HCl (pH 8.0) containing 1.0 M NaCl and 0.1% β-mercaptoethanol, and then dialyzed against 20 mM Tris-HCl buffer (pH 8.0) containing 50 mM NaCl and 0.1% β-mercaptoethanol. Finally, the lysate was dialyzed against 20 mM Tris-HCl buffer (pH 8.0) containing 50 mM NaCl and 1 mM DTT. Dialysis was performed at 4°C. The total protein concentration was estimated spectrophotometrically by using absorbance at 260 nm and 280 nm to subtract the contribution of nucleic acid.51,52 The resulting cell lysate was aliquoted and stored at −80°C for further experiments.
Pulse proteolysis
To identify proteins stabilized by ATP binding, the cell lysate was incubated at 25°C for 2 hrs in 20 mM Tris-HCl buffer (pH 8.0) containing 1.0 mM ATPγS (Sigma-Aldrich, San Louis, MO), 50 mM NaCl, 5 mM MgCl2, 1 mM TCEP, and 3.0 M urea. To prevent the ligand from being hydrolyzed by endogenous enzymes, we used ATPγS, a non-hydrolysable ATP analogue. A control reaction was prepared in an identical manner except without ATPγS. After the incubation, pulse proteolysis was performed as described previously.18,53 Briefly, thermolysin was added to the final concentration of 0.20 mg/mL, and the reaction was quenched after 1 min by 16 μM phosphoramidon and then again by 10 mM EDTA. Phosphoramidon, a competitive inhibitor for thermolysin, was used to suppress autolysis of thermolysin,54 which complicates 2-D gel analysis by producing multiple fragments of the protease. EDTA inactivates thermolysin by chelating Ca2+ ion, which is essential for the structural integrity of the enzyme.55
2-D gel electrophoresis
To remove salts prior to 2-D gel electrophoresis, the buffer of the quenched reaction was exchanged with 8.0 M urea by Protein Desalting Spin Column (Pierce, Rockford, IL). The resulting solution in 8.0 M urea was then mixed with an equivalent volume of a two-fold concentrated stock solution for isoeletric focusing to make the final solution of 8.0 M urea, 2% CHAPS, 20 mM DTT, 0.002% bromophenol blue, and 0.5% IPG strip buffer (3–10 NL) (GE Healthcare, Pittsburgh, PA). The solution was then centrifuged to remove any precipitants. 250 μL of the resulting solution (~500 μg of total protein) was used to rehydrate 13-cm 3–10 NL DryStrip (GE Healthcare, Pittsburgh, PA). The hydrated strip was focused with Ettan IPGphor II (GE Healthcare, Pittsburgh, PA) according to the manufacturer’s instruction. Separation on the second dimension was performed in 15% (w/v) continuous SDS PAGE gel with SE 600 Ruby Complete (GE Healthcare, Pittsburgh, PA). The gels were stained with Colloidal Coomassie staining solution.56 Spots showing different intensities on gels of a sample and its control were identified by visual inspection. To rule out the false positives from artifacts in 2-D gel electrophoresis, the same experiment was performed in triplicate. Spots that were identified consistently in the repeated experiments were selected for further characterization.
In-gel digestion
In-gel digestion was performed with selected spots on 2-D gels as described previously.46 Peptides extracted from gel pieces were desalted using Ziptipμ-C18 (Millipore; Bedford, MA). 1 μL of the desalted eluant was mixed with an equal volume of 5 mg/mL α-cyano-4-hydroxy cinnamic acid in 60% acetonitrile containing 0.1% trifluoroacetic acid and allowed to dry on a MALDI target plate. Mass spectra were acquired on a 4800 Plus MALDI TOF/TOF™ Analyzer (Applied Biosystems; Foster City, CA). MS/MS analysis of each sample was performed on the top ten peaks from each mass spectrum. Proteins were identified from Swiss-Prot database by using MASCOT database search engine57 in GPS Explorer (Applied Biosystems; Foster City, CA). All identified proteins have significant MASCOT scores (> 100) and GPS Explorer protein confidence index (> 95%).
Confirmation with recombinant proteins
To confirm the results from the proteomic screen, GAPDH, yncE, E3, and mlaC were cloned into pAED4 expression vector by amplifying the corresponding genes from E. coli K-12 genomic DNA by polymerase chain reaction. The cloned proteins were over-expressed in BL21(DE3) pLysS by induction with IPTG. After lysing the cells, supernatants were collected by centrifugation. The effect of ATPγS on the unfolding of the recombinant proteins in urea was determined by pulse proteolysis under the same condition as the proteomic screen with the E. coli cell lysate. The lysates of cells over-expressing the cloned proteins were used for pulse proteolysis without further purification as described previously.18,19 Briefly, the lysates were incubated in 20 mM Tris-HCl buffer (pH 8.0) containing 50 mM NaCl, 5.0 mM MgCl2, 1.0 mM TCEP, and 3.0 M urea with and without 1.0 mM ATPγS at 25°C for 2 hrs. Pulse proteolysis was performed to digest the unfolded protein. The remaining proteins were determined by quantifying the band intensities on SDS PAGE gels by ImageJ, an image analysis software (http://rsbweb.nih.gov/ij/).
Determination of Cm
To determine the influence of ATPγS on the global stability, Cm was determined by pulse proteolysis as described previously.18 Briefly, the recombinant proteins were incubated for 24 hrs at 25°C in 20 mM Tris-HCl buffer (pH 8.0) containing 50 mM NaCl, 5.0 mM MgCl2, 1.0 mM TCEP, and varying concentrations of urea. The lysates of the cells overexpressing the recombinant proteins were used for this experiment without purification. Pulse proteolysis was performed with 0.20 mg/mL thermolysin for 1 min to digest the unfolded protein. The amount of remaining protein in each reaction was determined by quantifying the band intensities of intact proteins on SDS PAGE gels with ImageJ.
The Cm values were determined by fitting the band intensities to the following equation:
| (1) |
where I is the observed band intensity, I0 is the band intensity of the protein digested by pulse proteolysis under native conditions, and m is the dependence of global stability on urea.
Relaxation kinetics
The relaxation kinetics of the recombinant proteins in 3.0 M urea were determined by pulse proteolysis as described previously.19 The lysates of the cells overexpressing the recombinant proteins were used for this experiment without purification. Unfolding was initiated by adding urea to the cell lysates with the recombinant proteins. The resulting unfolding condition was 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 5.0 mM MgCl2, 1.0 mM TCEP, and 3.0 M urea. Progress of unfolding was monitored by performing pulse proteolysis with aliquots of the reaction at designated time points. The amounts of remaining proteins after pulse proteolysis were determined by quantifying band intensities on SDS PAGE gels with ImageJ. The relaxation kinetic constants were determined by fitting the band intensities to the first-order rate equation.
Supplementary Material
Acknowledgments
We thank Joseph R. Kasper for recombinant cysteine-free DHFR and Youngil Chang for recombinant MBP. We also thank Jonathan P. Schlebach, Joseph R. Kasper, and Mark W. Hinzman for helpful comments on this manuscript. The work was partly funded by National Institutes of Health (R01 GM075004 to D. K.) and National Science Foundation (EF0850009, IIS0915801, DMS800568 to D. K.).
Abbreviations used
- 2-D
two-dimensional
- ADP
Adenosine 5′-diphosphate
- ATP
Adenosine 5′-triphosphate
- ATPγS
Adenosine 5′-[γ-thio]triphosphate
- CHAPS
3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate
- DIGE
differential gel electrophoresis
- DTT
dithiothreitol
- E3
Dihydrolipoamide dehydrogenase E3 subunit
- EDTA
Ethylenediaminetetraacetic acid
- FAD
flavin adenine dinucleotide
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GO
Gene Ontology
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- MALDI
Matrix-assisted laser desorption/ionization
- NAD+
nicotinamide adenine dinucleotide
- SDS
sodium dodecyl sulfate
- PAGE
polyacrylamide gel electrophoresis
- TCEP
tris(2-carboxyethyl)phosphine
- Tris
tris(hydroxymethyl)aminomethane
- TOF
time-of-flight
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mercier KA, Baran M, Ramanathan V, Revesz P, Xiao R, Montelione GT, Powers R. FAST-NMR: functional annotation screening technology using NMR spectroscopy. J Am Chem Soc. 2006;128:15292–15299. doi: 10.1021/ja0651759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luesch H, Chanda SK, Raya RM, DeJesus PD, Orth AP, Walker JR, Izpisua Belmonte JC, Schultz PG. A functional genomics approach to the mode of action of apratoxin A. Nat Chem Biol. 2006;2:158–167. doi: 10.1038/nchembio769. [DOI] [PubMed] [Google Scholar]
- 3.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 4.Hantschel O, Rix U, Schmidt U, Burckstummer T, Kneidinger M, Schutze G, Colinge J, Bennett KL, Ellmeier W, Valent P, Superti-Furga G. The Btk tyrosine kinase is a major target of the Bcr-Abl inhibitor dasatinib. Proc Natl Acad Sci USA. 2007;104:13283–13288. doi: 10.1073/pnas.0702654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Missner E, Bahr I, Badock V, Lucking U, Siemeister G, Donner P. Off-target decoding of a multitarget kinase inhibitor by chemical proteomics. ChemBioChem. 2009;10:1163–1174. doi: 10.1002/cbic.200800796. [DOI] [PubMed] [Google Scholar]
- 6.Burdine L, Kodadek T. Target identification in chemical genetics: The (often) missing link. Chem Biol. 2004;11:593–597. doi: 10.1016/j.chembiol.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Terstappen GC, Schlupen C, Raggiaschi R, Gaviraghi G. Target deconvolution strategies in drug discovery. Nat Rev Drug Discov. 2007;6:891–903. doi: 10.1038/nrd2410. [DOI] [PubMed] [Google Scholar]
- 8.Ong SE, Schenone M, Margolin AA, Li X, Do K, Doud MK, Mani DR, Kuai L, Wang X, Wood JL, Tolliday NJ, Koehler AN, Marcaurelle LA, Golub TR, Gould RJ, Schreiber SL, Carr SA. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci USA. 2009;106:4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan JN, Nislow C, Emili A. Recent advances and method development for drug target identification. Trends Pharmacol Sci. 2009;31:82–88. doi: 10.1016/j.tips.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Godl K, Wissing J, Kurtenbach A, Habenberger P, Blencke S, Gutbrod H, Salassidis K, Stein-Gerlach M, Missio A, Cotten M, Daub H. An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc Natl Acad Sci USA. 2003;100:15434–15439. doi: 10.1073/pnas.2535024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat Chem Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 12.Sleno L, Emili A. Proteomic methods for drug target discovery. Curr Opin Chem Biol. 2008;12:46–54. doi: 10.1016/j.cbpa.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 13.Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW. Chemogenomic profiling: Identifying the functional interactions of small molecules in yeast. Proc Natl Acad Sci USA. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, Wang J, Ketela T, Brenner C, Brill JA, Fernandez GE, Lorenz TC, Payne GS, Ishihara S, Ohya Y, Andrews B, Hughes TR, Frey BJ, Graham TR, Andersen RJ, Boone C. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 15.Hoon S, Smith AM, Wallace IM, Suresh S, Miranda M, Fung E, Proctor M, Shokat KM, Zhang C, Davis RW, Giaever G, StOnge RP, Nislow C. An integrated platform of genomic assays reveals small-molecule bioactivities. Nat Chem Biol. 2008;4:498–506. doi: 10.1038/nchembio.100. [DOI] [PubMed] [Google Scholar]
- 16.Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, Wang J, Wu RP, Gomez F, Loo JA, Wohlschlegel JA, Vondriska TM, Pelletier J, Herschman HR, Clardy J, Clarke CF, Huang J. Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. U.S.A; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.West GM, Tucker CL, Xu T, Park SK, Han X, Yates JR, 3rd, Fitzgerald MC. Quantitative proteomics approach for identifying protein-drug interactions in complex mixtures using protein stability measurements. Proc Natl Acad Sci USA. 2010;107:9078–9082. doi: 10.1073/pnas.1000148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park C, Marqusee S. Pulse proteolysis: A simple method for quantitative determination of protein stability and ligand binding. Nat Methods. 2005;2:207–212. doi: 10.1038/nmeth740. [DOI] [PubMed] [Google Scholar]
- 19.Na YR, Park C. Investigating protein unfolding kinetics by pulse proteolysis. Protein Science. 2009;18:268–276. doi: 10.1002/pro.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim MS, Song J, Park C. Determining protein stability in cell lysates by pulse proteolysis and Western blotting. Protein Science. 2009;18:1051–1059. doi: 10.1002/pro.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlebach JP, Kim M-S, Joh NH, Bowie JU, Park C. Probing membrane protein unfolding with pulse proteolysis. J Mol Biol. 2011 doi: 10.1016/j.jmb.2010.12.018. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD. EcoCyc: A comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJ. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park C, Marqusee S. Analysis of the stability of multimeric proteins by effective ΔG and effective m-values. Protein Sci. 2004;13:2553–2558. doi: 10.1110/ps.04811004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettit FH, Reed LJ. Alpha-keto acid dehydrogenase complexes. 8 Comparison of dihydrolipoyl dehydrogenases from pyruvate and alpha-ketoglutarate dehydrogenase complexes of Escherichia coli. Proc Natl Acad Sci USA. 1967;58:1126–1130. doi: 10.1073/pnas.58.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guest JR, Creaghan IT. Lipoamide dehydrogenase mutants of Escherichia coli K 12. Biochem J. 1972;130:8P–8P. doi: 10.1042/bj1300008p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steiert PS, Stauffer LT, Stauffer GV. The lpd gene product functions as the L protein in the Escherichia coli glycine cleavage enzyme system. J Bacteriol. 1990;172:6142–6144. doi: 10.1128/jb.172.10.6142-6144.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindsay H, Beaumont E, Richards SD, Kelly SM, Sanderson SJ, Price NC, Lindsay JG. FAD insertion is essential for attaining the assembly competence of the dihydrolipoamide dehydrogenase (E3) monomer from Escherichia coli. J Biol Chem. 2000;275:36665–36670. doi: 10.1074/jbc.M004777200. [DOI] [PubMed] [Google Scholar]
- 29.Pieper U, Eswar N, Davis FP, Braberg H, Madhusudhan MS, Rossi A, Marti-Renom M, Karchin R, Webb BM, Eramian D, Shen MY, Kelly L, Melo F, Sali A. MODBASE: A database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskowski RA, Watson JD, Thornton JM. ProFunc: A server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baba-Dikwa A, Thompson D, Spencer NJ, Andrews SC, Watson KA. Overproduction, purification and preliminary X-ray diffraction analysis of YncE, an iron-regulated Sec-dependent periplasmic protein from Escherichia coli. Acta Crystallographica Section F, Structural Biology and Crystallization Communications. 2008;64:966–969. doi: 10.1107/S1744309108029515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang YD, Spratt P, Chen H, Park C, Kihara D. Sub-AQUA: Real-value quality assessment of protein structure models. Protein Eng Des Sel. 23:617–632. doi: 10.1093/protein/gzq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ünlü M, Morgan ME, Minden JS. Difference gel electrophoresis: A single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 35.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 36.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 37.Leung D, Hardouin C, Boger DL, Cravatt BF. Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nat Biotechnol. 2003;21:687–691. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- 38.Evans MJ, Saghatelian A, Sorensen EJ, Cravatt BF. Target discovery in small-molecule cell-based screens by in situ proteome reactivity profiling. Nat Biotechnol. 2005;23:1303–1307. doi: 10.1038/nbt1149. [DOI] [PubMed] [Google Scholar]
- 39.Senda M, Kanazawa H, Tsuchiya T, Futai M. Conformational change of the α subunit of Escherichia coli F1 ATPase: ATP changes the trypsin sensitivity of the subunit. Arch Biochem Biophys. 1983;220:398–404. doi: 10.1016/0003-9861(83)90429-0. [DOI] [PubMed] [Google Scholar]
- 40.Dunn SD, Futai M. Reconstitution of a functional coupling factor from the isolated subunits of Escherichia coli F1 ATPase. J Biol Chem. 1980;255:113–118. [PubMed] [Google Scholar]
- 41.Perlin DS, Latchney LR, Wise JG, Senior AE. Specificity of the proton adenosinetriphosphatase of Escherichia coli for adenine, guanine, and inosine nucleotides in catalysis and binding. Biochemistry. 1984;23:4998–5003. doi: 10.1021/bi00316a026. [DOI] [PubMed] [Google Scholar]
- 42.Myers JA, Boyer PD. Catalytic properties of the ATPase on submitochondrial particles after exchange of tightly bound nucleotides under different steady state conditions. FEBS Lett. 1983;162:277–281. doi: 10.1016/0014-5793(83)80771-6. [DOI] [PubMed] [Google Scholar]
- 43.Hunt JF, Weaver AJ, Landry SJ, Gierasch L, Deisenhofer J. The crystal structure of the GroES co-chaperonin at 2.8 Å resolution. Nature. 1996;379:37–45. doi: 10.1038/379037a0. [DOI] [PubMed] [Google Scholar]
- 44.Martin J, Geromanos S, Tempest P, Hartl FU. Identification of nucleotide-binding regions in the chaperonin proteins GroEL and GroES. Nature. 1993;366:279–282. doi: 10.1038/366279a0. [DOI] [PubMed] [Google Scholar]
- 45.Schneider DA, Gourse RL. Relationship between growth rate and ATP concentration in Escherichia coli: A bioassay for available cellular ATP. J Biol Chem. 2004;279:8262–8268. doi: 10.1074/jbc.M311996200. [DOI] [PubMed] [Google Scholar]
- 46.Park C, Zhou S, Gilmore J, Marqusee S. Energetics-based protein profiling on a proteomic scale: Identification of proteins resistant to proteolysis. J Mol Biol. 2007;368:1426–1437. doi: 10.1016/j.jmb.2007.02.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia K, Manning M, Hesham H, Lin Q, Bystroff C, Colon W. Identifying the subproteome of kinetically stable proteins via diagonal 2D SDS/PAGE. Proc Natl Acad Sci USA. 2007;104:17329–17334. doi: 10.1073/pnas.0705417104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hubbard SJ. The structural aspects of limited proteolysis of native proteins. Biochim Biophys Acta. 1998;1382:191–206. doi: 10.1016/s0167-4838(97)00175-1. [DOI] [PubMed] [Google Scholar]
- 49.Park C, Marqusee S. Probing the high energy states in proteins by proteolysis. J Mol Biol. 2004;343:1467–1476. doi: 10.1016/j.jmb.2004.08.085. [DOI] [PubMed] [Google Scholar]
- 50.Chang Y, Park C. Mapping transient partial unfolding by protein engineering and native-state proteolysis. J Mol Biol. 2009;393:543–556. doi: 10.1016/j.jmb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Grimsley GR, Pace CN. Spectrophotometric determination of protein concentration. Curr Protoc Protein Sci. 2004;(Unit 3):1. doi: 10.1002/0471140864.ps0301s33. [DOI] [PubMed] [Google Scholar]
- 52.Mach H, Middaugh CR, Denslow N. Determining the identity and purity of recombinant proteins by UV absorption spectroscopy. Curr Protoc Protein Sci. 2001;(Unit 7.2) doi: 10.1002/0471140864.ps0702s01. [DOI] [PubMed] [Google Scholar]
- 53.Park C, Marqusee S. Quantitative determination of protein stability and ligand binding by pulse proteolysis. Curr Protoc Protein Sci. 2006;46:20.11.21–20.11.14. doi: 10.1002/0471140864.ps2011s46. [DOI] [PubMed] [Google Scholar]
- 54.Wildes D, Anderson LM, Sabogal A, Marqusee S. Native state energetics of the Src SH2 domain: Evidence for a partially structured state in the denatured ensemble. Protein Sci. 2006;15:1769–1779. doi: 10.1110/ps.062136006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dahlquist FW, Long JW, Bigbee WL. Role of Calcium in the thermal stability of thermolysin. Biochemistry. 1976;15:1103–1111. doi: 10.1021/bi00650a024. [DOI] [PubMed] [Google Scholar]
- 56.Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 57.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 58.Bhattacharyya T, Bhattacharyya A, Roy S. A fluorescence spectroscopic study of glutaminyl-tRNA synthetase from Escherichia coli and its implications for the enzyme mechanism. Eur J Biochem. 1991;200:739–745. doi: 10.1111/j.1432-1033.1991.tb16239.x. [DOI] [PubMed] [Google Scholar]
- 59.Kosakowski HM, Holler E. Phenylalanyl-tRNA synthetase from Escherichia coli K10. Synergistic coupling between the sites for binding of L-phenylalanine and ATP. Eur J Biochem. 1973;38:274–282. doi: 10.1111/j.1432-1033.1973.tb03059.x. [DOI] [PubMed] [Google Scholar]
- 60.Johnson JL, Reinhart GD. MgATP and fructose 6-phosphate interactions with phosphofructokinase from Escherichia coli. Biochemistry. 1992;31:11510–11518. doi: 10.1021/bi00161a032. [DOI] [PubMed] [Google Scholar]
- 61.Joyce MA, Fraser ME, Brownie ER, James MN, Bridger WA, Wolodko WT. Probing the nucleotide-binding site of Escherichia coli succinyl-CoA synthetase. Biochemistry. 1999;38:7273–7283. doi: 10.1021/bi990527s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.