Summary
13-cis Retinoic acid (13cRA), a stereoisomeric form of retinoic acid, is naturally generated in the body and is also used clinically to treat acute promyelocytic leukaemia, some skin diseases, and cancer. Furthermore, it has been suggested that 13cRA modulates brain neurochemical systems, as increased 13cRA levels are correlated with depression and increased suicidal tendencies. However, the mechanism for the generation of endogenous 13cRA is not well understood. This study identified and characterized a novel enzyme in zebrafish brain, 13-cis isomerohydrolase (13cIMH), that exclusively generated 13-cis retinol, which can be oxidized to 13cRA. 13cIMH shares 74% amino acid sequence identity with human RPE65, an 11-cis isomerohydrolase in the visual cycle, and retains the key residues essential for RPE65’s isomerohydrolase activity. Similar to RPE65, 13cIMH is a membrane-associated protein, requires all-trans retinyl ester as its intrinsic substrate, and its enzymatic activity is dependent on iron. The purified 13cIMH converted atRE exclusively to 13-cis retinol with Km = 2.6 μM and kcat = 4.4 × 10−4s−1. RT-PCR, Western blot analysis and immunohistochemistry detected 13cIMH expression in the brain. These results suggest that 13cIMH may play a key role in the generation of 13cRA and in the modulation of neuronal functions in the brain.
Keywords: brain, retinoic acid, isomerohydrolase, zebrafish, visual cycle, vitamin A
Introduction
Retinoic acids (RA) are a biologically active form of retinoids (vitamin A and its derivatives). The spatiotemporal gradient of RA is essential for the regulation of cell proliferation, differentiation and organ development [1, 2]. Generally, it is considered that endogenous retinoids are stored as all-trans retinyl esters (atRE, Fig. 1, structure 1) in the liver and other tissues [1–3]. As needed, atRE is hydrolyzed to all-trans retinol (atROL, Fig. 1, structure 2), which is subsequently released into the circulation, bound by retinol-binding protein and transported to target cells.
Figure 1. Chemical structures of retinoid derivatives.
All-trans retinyl ester (atRE; structure 1) is major storage form of retinoids, which is hydrolyzed by retinyl ester hydrolase (REH) or generated by lecithin-retinol acyltransferase (LRAT). All-trans retinol (atROL; structure 2) is reversibly oxidized/reduced by retinol dehydrogenase (RDH) to/from all-trans retinal (atRAL; structure 3). atRAL is further oxidized by retinaldehyde dehydrogenase (RALDH) to all-trans retinoic acid (RA; structure 4). Other endogenous stereoisomeric forms of atRA, 9-cis retinoic acid (9cRA; structure 5) and 13-cis retinoic acid (13cRA; structure 6) are presented. Abbreviations in the figure: LRAT, lecithin retinol acyltransferase; REH, retinyl ester hydrolase; RDH, retinol dehydrogenases; RALDH, retinaldehyde dehydrogenases.
In target cells, atROL is converted to atRA through two sequential oxidative reactions (atRAL, Fig. 1, structure 3 and atRA, Fig. 1, structure 4) which are catalyzed by retinol dehydrogenases (RDHs) and retinaldehyde dehydrogenases (RALDHs) [1, 2]. In target cells, the generated RA exerts its functions through binding to the nuclear retinoic acid receptors (RARα/β/γ) and retinoid x-receptors (RXRα/β/γ) [2, 4]. Moreover, in vitro studies have demonstrated that RARs and RXRs form either homodimers or heterodimers that bind to the retinoic acid response element (RARE) in the promoter regions of the target gene, activating target gene transcription in a ligand (RA)-dependent manner [2, 4].
There are three stereoisomeric forms of RA, all-trans retinoic acid (atRA), 9-cis retinoic acid (9cRA, Fig. 1, structure 5) and 13-cis retinoic acid (13cRA or isotretinoin, Fig. 1, structure 6), which show different binding affinities to the retinoic acid receptors. atRA is known to bind exclusively to RARs, whereas 9cRA binds to both RARs and RXRs [2, 4]. In contrast, 13cRA does not exhibit specific binding to RXRs, and has a 100-fold lower affinity to RARs than atRA or 9cRA [5–7]. Thus, 13cRA’s mechanism of action is unclear. There are four possible mechanisms for the physiological function of 13cRA: [i] 13cRA may modulate gene expression through an RAR- and RXR-independent pathway by binding to an unidentified nuclear receptor; [ii] 13cRA may be first isomerized to atRA or 9cRA either enzymatically [8] or spontaneously, and then modulate target gene transcription through atRA or 9cRA [9]; [iii] 13cRA may enhance the translation of target gene mRNA or its protein stability [10]; [iv] 13cRA may directly inhibit retinoid-processing enzymes [11, 12]. The inhibition of the enzymes by RA may be a negative feedback regulation of RA signaling to decrease RA production.
RA signaling is highly sensitive to abnormal changes of RA concentration. It has been shown that either too low or too high concentrations of RA in specific target tissues may cause disruption of tissue patterning and cell differentiation, or result in abnormal development (malformations) of embryos [13–15]. Zebrafish is a commonly used model for research in genetics and pharmacology, vertebrate embryogenesis and vision. Zebrafish models have been used to study the teratogenic effects of RA and its derivatives [15] and deficiencies of retinoid-processing enzymes [16]. Interestingly, excessive doses of 13cRA cause fewer developmental malformations than that of atRA and 9cRA, suggesting that 13cRA may not directly regulate retinoic acid receptor signaling in embryogenesis [15].
AtRA, 13cRA (isotretinoin) and other synthetic retinoids are used clinically for the treatment of acute promyelocytic leukaemia (APL), some skin diseases (e.g. acne, psoriasis and photoaging) and some tumors (e.g. prostate cancer or neuroblastoma) with encouraging outcomes [17, 18]. 13cRA exhibits a longer half-life and higher peak plasma concentrations than other RA isomers in the body, and thus it is considered a storage form of biologically active atRA or 9cRA [18–20]. Due to these features, 13cRA is believed to be more suitable for chemoprevention or chemotherapy, compared to the other RA isoforms [5, 19, 20]. However, RA has a variety of side effects on brain neurochemistry, possibly by regulating neurotransmitter (e.g. dopamine, serotonin and norepinephrine) signaling genes [10, 19, 21, 22]. It has been reported that treatment with 13cRA (isotretinoin) is associated with neurological side effects, such as depression and suicidal tendencies [19, 21, 22], although the molecular mechanisms for these side effects remains obscure. Substantial amounts of RA are present in the mouse brain (34 pmol/g brain tissue [3]), and RARs and RXRs also show broad expression in the brain [23]. Furthermore, there is evidence that RA signaling is essential for neuronal cell phenotypic maintenance in the brain [19, 22, 24]. Therefore, treatment with 13cRA at high concentrations may cause an imbalance of RA in the brain, which may subsequently lead to depression, elevated anxiety and irritability.
Several lines of evidence have demonstrated that 13cRA is generated endogenously [3, 25], although the mechanism(s) has not been elucidated. It has been suggested that 13cRA could be non-enzymatically generated by spontaneous thermal isomerization from atRA or 9cRA [9]. However, the amount of endogenous 13cRA exceeds the level that could be generated by spontaneous isomerization alone [3, 25]. Furthermore, studies have shown that following liver consumption, there is a 10-fold increase in 13cRA levels compared to atRA in human plasma, which strongly suggests that 13cRA is a physiological metabolite of vitamin A [26]. Likewise, it has been reported that rabbit tracheal epithelial (RTE) cells and HepG2 cells generate and secrete 13cRA [27, 28]. Therefore, there is ample evidence to suggest that unidentified enzymes catalyze the generation of 13-cis retinoids. Recently, Redmond et al showed that RPE65, an isomerohydrolase in the visual cycle in the retinal pigment epithelium (RPE), converts all-trans retinyl ester (atRE) into 11-cis retinol (11cROL) and 13-cis retinol (13cROL) [29]. Therefore, we hypothesize that a homolog of RPE65 could be responsible for generation of 13-cis retinoids in the brain.
In the present study, we identified and characterized an enzyme, 13-cis isomerohydrolase (13cIMH), which is expressed predominantly in the brain and exclusively generates 13cROL from all-trans retinyl ester.
Results
Cloning and amino acid sequence analyses of zebrafish 13cIMH
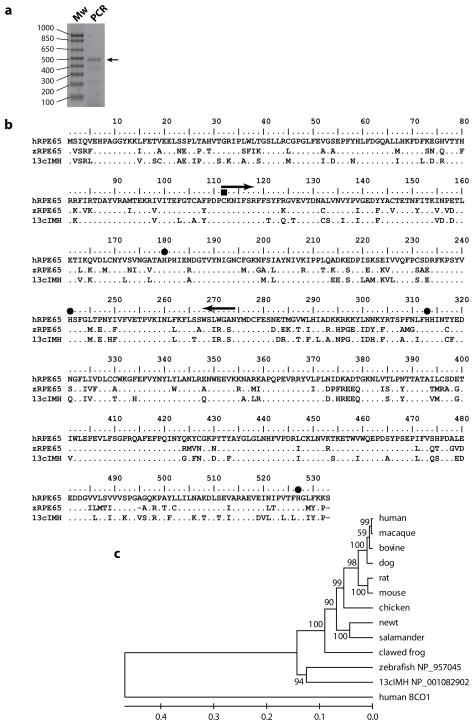
Since RPE65 has been reported to generate both 11-cis and 13-cis retinol from atRE [29], we performed PCR using degenerate primers based on RPE65 sequence and the zebrafish brain cDNA. The products of the degenerate PCR with the expected size were cloned and sequenced (Fig. 2a). One deduced amino acid sequence from the cloned PCR products showed 100% identity to retinal pigment epithelium-specific protein b (accession number in GenBank NP_001082902) of zebrafish and showed 76.5% and 79.2% sequence identities to human and zebrafish RPE65, respectively. We named this cloned gene 13-cis isomerohydrolase (13cIMH) due to the enzymatic activity of its protein product as shown in this manuscript. The full-length zebrafish 13cIMH showed 77.3 % sequence identity to zebrafish RPE65 and 74.1 % to human RPE65 at the amino acid levels, suggesting possible functional similarities to RPE65. Based on the sequence alignment and comparison with human RPE65 and zebrafish RPE65 (Fig. 2b), the 13cIMH conserved the key residues known to be essential for the enzymatic activity of RPE65, such as four His residues forming iron binding site [30, 31] and a Cys residue of palmitylation site for the membrane association of RPE65 protein [32] (Fig. 2b). Phylogenetic tree analysis suggested that the zebrafish 13cIMH gene may be generated by gene duplication prior to diverging to the ancestral amphibian (Fig. 2c). Based on information from GenBank, the zebrafish RPE65 gene is located in chromosome 18, while the 13cIMH gene is in chromosome 8 in zebrafish, suggesting that they are distinct genes.
Figure 2. Identification and analysis of the amino acid sequences of 13cIMH.
(a) The degenerate PCR products amplified from zebrafish brain cDNA were confirmed by 1.2% agarose gel electrophoresis. The arrow indicates the expected size of the PCR product. Mw, DNA size marker; PCR, PCR product using degenerative primers. (b) Alignment of human RPE65 (hRPE65), zebrafish RPE65 (zRPE65) and 13cIMH sequences. The amino acid residues identical to human RPE65 are indicated by dots “.”. The four histidine residues (His180, 241, 313 and 527) that are required for iron-binding [30, 31] and a palmitylated cysteine residue (Cys112) for membrane association [32, 52] in human RPE65 were indicated by filled circles “●” and a filled rectangle“■”, respectively. The locations of degenerate PCR primers were indicated by arrows. (c) A phylogenetic tree was constructed by the UPGMA method in MEGA application version 4.02 [63]. The numbers on the branches indicate the mean of clustering probabilities from 1000 bootstrap re-samplings.
13cIMH is a 13-cis retinoid-specific isomerohydrolase
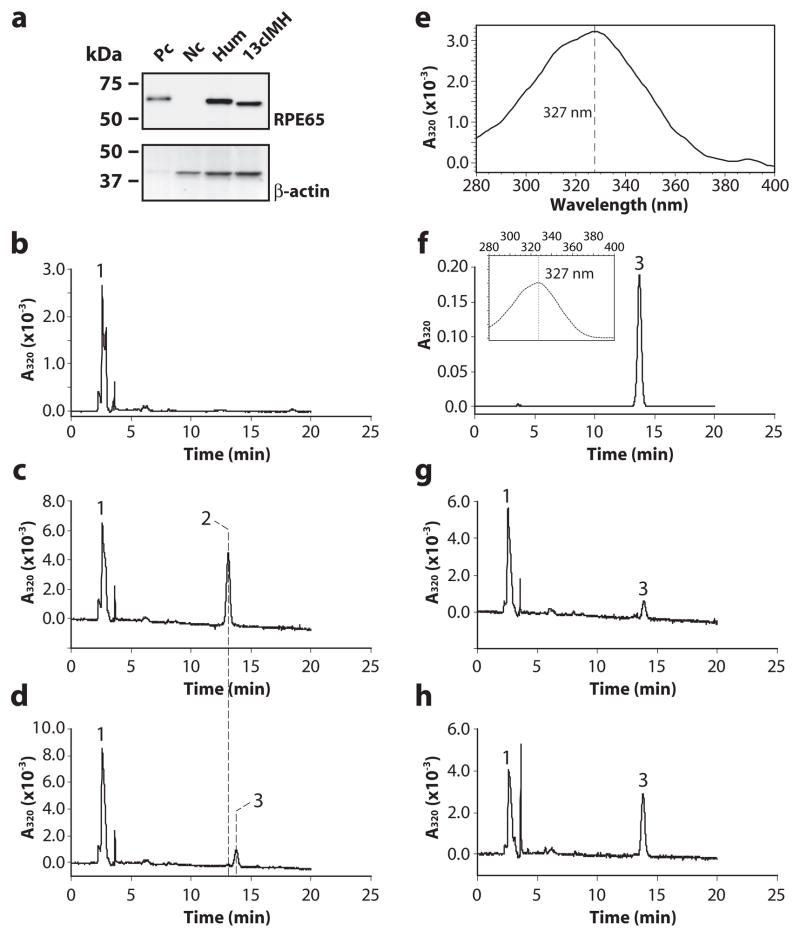
To study the enzymatic activity of 13cIMH, a plasmid expressing human RPE65 [32] and that expressing 13cIMH were separately transfected into 293A-LRAT cells [31]; an expression plasmid expressing red fluorescent protein (RFP) was used as the negative control. Forty-eight hours post-transfection, protein expression was confirmed by Western blot analysis (Fig. 3a). Due to the highly hydrophobic feature of atRE, we employed a novel in vitro isomerohydrolase activity assay, which was recently developed in our laboratory and utilizes atRE incorporated in the liposomes as substrate [33] to evaluate its isomerohydrolase activity. As shown by HPLC analysis, the RFP expressing cell lysate did not produce detectable 11cROL (Fig. 3b), whereas the cell lysate expressing RPE65 generated significant amounts of 11cROL from atRE (Fig. 3c). Under the same assay conditions, the 13cIMH cell lysate exclusively generated 13cROL after incubation with the liposome containing atRE, without any detectable product of 11cROL (Fig. 3d). The generated 13cROL has a characteristic retention time 13.8 min, which is distinct from that of 11cROL (13.1 min) in the HPLC profile (Fig. 3d). Furthermore, the UV-Vis absorption spectrum of the peak 3 with retention time of 13.8 min showed a λmax of 327 nm (Fig. 3e), identical to the characteristic λmax (Fig. 3f, inset) and retention time (Fig. 3f) of 13cROL standard. For further confirmation, the 13cROL standard (Fig. 3f) was spiked into the reaction products generated by 13cIMH (Fig. 3g, before spike and 3h, after spike). The retention time of the 13cIMH-generated peak was identical to that of the 13cROL standard, indicating that 13cIMH is a unique and novel isomerohydrolase, converting atRE exclusively to 13cROL (Fig. 3g, h). Our assays showed that the activity of 13cIMH was higher when it was expressed in 293A cells without LRAT (Fig. S1). Therefore, all further experiments were performed with 293T cells (without LRAT, same as 293A cell), unless specified.
Figure 3. Zebrafish 13cIMH is a 13-cis specific isomerohydrolase.
The expression plasmids of human RPE65, zebrafish 13cIMH and red fluorescent protein (RFP, negative control) were separately transfected into 293A-LRAT cells. (a) Protein expression was confirmed by Western blot analyses. (b-d) Equal amounts of total cellular proteins from the cells (125 μg) expressing RFP (b), human RPE65 (c) and 13cIMH (d) were incubated with liposomes containing atRE (250 μM lipids, 3.3 μM atRE) for 1 hour at 37°C, and the generated retinoids were analyzed by HPLC. (e-f) A peak 3 in panel (d) was identified as the generated 13cROL based on retention time (d) and the absorption spectrum (e) compared to retention time (f) and absorption spectrum (inset) of the 13cROL standard. The x-axis of inset in panel (f) represents wavelength (nm). (g, h) For further confirmation of the identity of generated 13cROL, the 13cROL standard was spiked into the reaction products. The 13cROL peaks before (g) and after (h) the spike. The peaks were identified as follows: 1, retinyl esters; 2, 11cROL; 3, 13cROL.
All-trans retinyl ester is the substrate of 13cIMH
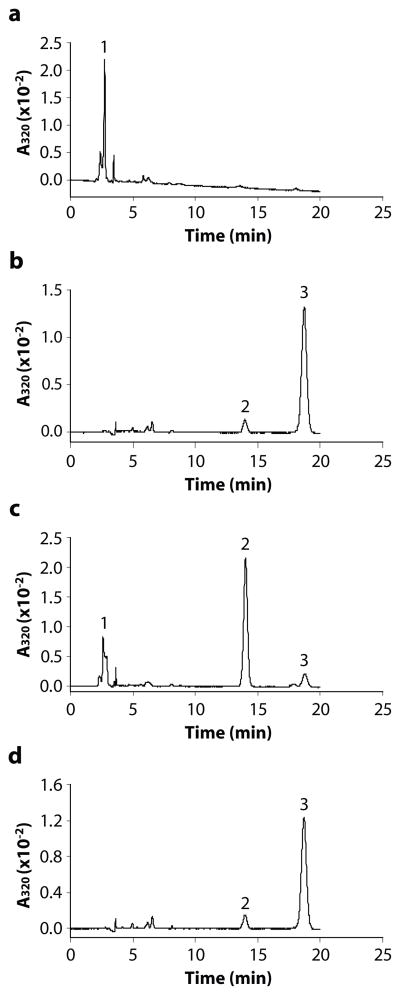
Previously, we reported that atRE is the direct substrate of RPE65 in the generation of 11cROL [34]. To verify whether 13cIMH also requires atRE as a direct substrate to generate 13cROL, we incubated 13cIMH with liposomes containing either atROL or atRE as substrate. The RFP cell lysates incubated with atRE did not produce detectable 13cROL (Fig. 4a); likewise, the RFP cell lysates incubated with atROL showed a major peak of exogenous atROL and only a minor peak of 13cROL (Fig. 4b). In contrast, a substantial amount of 13cROL was generated when the 13cIMH-expressing cell lysate was incubated with atRE (Fig. 4c), while only a small amount of 13cROL was generated when the same cell lysate was incubated with atROL (Fig. 4d). This minor peak of 13cROL was likely generated by spontaneous thermal isomerization from atROL, as this peak was also observed in negative control cell lysates lacking 13cIMH expression (Fig. 4b). The results suggested that similar to RPE65, 13cIMH requires atRE as its specific substrate.
Figure 4. Retinyl ester is the substrate of 13cIMH.
The cell lysate expressing RFP was incubated with liposomes containing atRE (a) or atROL (b). The cell lysate expressing 13cIMH was incubated with liposomes containing atRE (c) or atROL (d). Generated retinoids were extracted and analyzed by HPLC. The peaks were identified as follows: 1, retinyl esters; 2, 13cROL; 3, atROL.
13cIMH is an iron-dependent enzyme
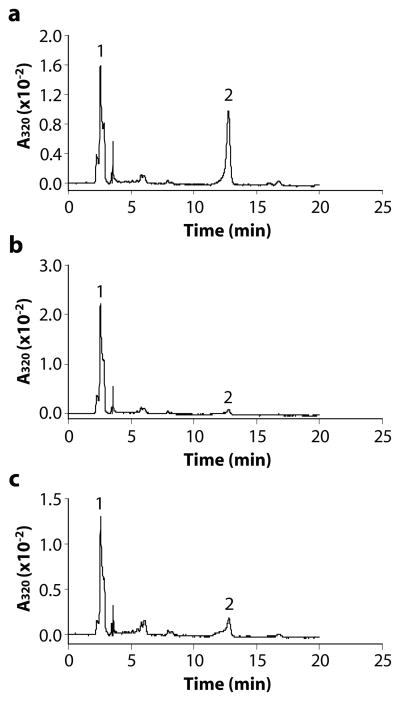
We have previously shown that RPE65 is an iron (II)-dependent enzyme [35]. We predicted that 13cIMH is also an iron-dependent enzyme, as it retains the four His residues known to coordinate iron in RPE65. To determine if the enzymatic activity of 13cIMH is dependent on iron, the 13cIMH cell lysate was incubated with the atRE-liposomes and a metal chelator, bypiridine. The HPLC profile of the extracted reaction showed a significant 13cROL peak in the absence of the metal chelator (Fig. 5a). In contrast, the generation of 13cROL from atRE was almost completely abolished when the reaction was incubated with the iron chelator (Fig. 5b). Addition of 6 mM FeSO4 into the iron chelator reaction restored partial 13cIMH activity (Fig. 5c), suggesting that 13cIMH is an iron-dependent enzyme, similar to RPE65.
Figure 5. Zebrafish 13cIMH is an iron-dependent enzyme.
The 293T cell lysate expressing 13cIMH was incubated with liposomes containing atRE (a), liposome containing atRE in the presence of 1 mM bypiridine (b) and liposome containing atRE, in the presence of 1 mM bypiridine and 6 mM FeSO4 (c). Generated retinoids were analyzed by HPLC. The peaks were identified as follows: 1, retinyl esters; 2, 13cROL.
Characterization of the kinetic parameters for 13cIMH’s enzymatic activity
To determine the steady-state kinetics of the enzymatic activity of 13cIMH, the assay conditions were optimized to ensure that all of the measurements were taken within the linear range. First, we plotted the time course of 13cROL generation after incubation of the atRE-liposomes with 125 μg of total cell lysate expressing 13cIMH. The time course of 13cROL production appeared linear in its initial phase (Fig. 6a), therefore, all of the further experiments in this study were conducted within this range. Secondly, to establish the dependence of 13cROL production on the level of 13cIMH protein, increasing amounts of the 13cIMH expression plasmid (0.5 to 6 μg DNA) were transfected into 293T cells. Western blot analysis confirmed that 13cIMH expression levels increased as greater amounts of the 13cIMH expression plasmid was used for transfection (Fig. 6b). The cell lysates with increasing 13cIMH expression levels were incubated with liposomes containing atRE. The production of 13cROL was found to be a linear function to the 13cIMH protein levels, within a specific range of 13cIMH (27 to 514 of arbitrary units) (Fig. 6c). Finally, to measure the kinetic parameters of 13cIMH activity, we constructed an adenovirus expressing 6×His-tagged 13cIMH to achieve higher expression levels for purification and in vitro enzyme assays using the purified enzyme. The purity of 13cIMH was verified by SDS-PAGE (Fig. 6d1) and Western blot analysis (Fig. 6d2, 3). We measured the initial reaction velocity using different concentrations of atRE-liposomes and the purified 13cIMH. Michaelis-Menten analysis of the data yielded the kinetic parameters for the reaction: Michaelis constant (Km) was 2.6 μM, and turnover number (kcat) was 4.4 × 10−4s−1 for purified 13cIMH (Fig. 6e).
Figure 6. Enzymatic parameters of 13cIMH.
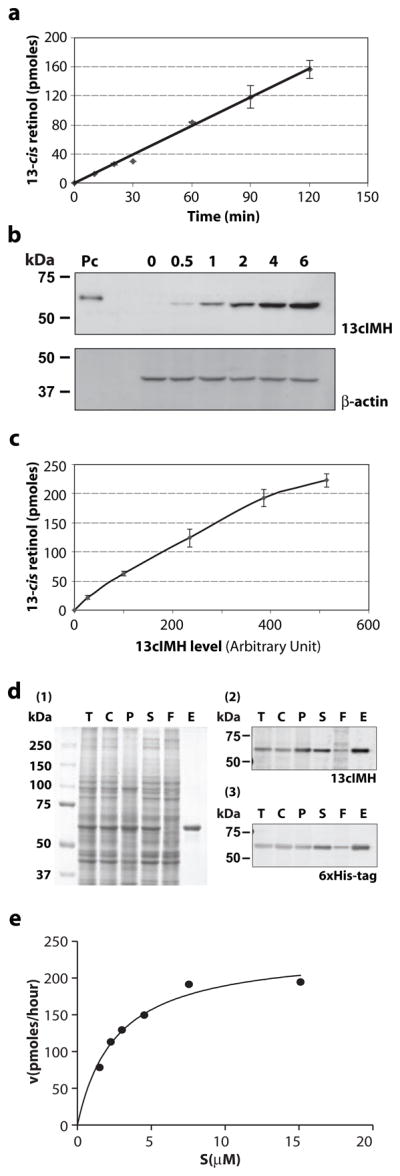
(a) Time course of 13cROL production. Equal amounts of microsomal proteins (125 μg) from 293T cells expressing 13cIMH were incubated with liposome containing atRE for the indicated time intervals. (b) Increasing amounts of the 13cIMH plasmid (0.5–6.0 μg) was transfected into 293T cells, and the expression was confirmed by Western blot analysis. (c) Dependence of production of 13cROL on 13cIMH expression levels. Equal amounts of 293T cellular proteins expressing various levels of 13cIMH were incubated with liposome containing atRE for 1 hour. The produced 13cROL was calculated from the area of the 13cROL peak (mean ± SD, n = 3) and plotted against protein levels of 13cIMH (arbitrary units). (d) 293A-LRAT cells were infected with adenovirus expressing His-tagged 13cIMH at MOI 100 and cultured for 24 hours. Expressed 13cIMH was purified using Ni-NTA resin. SDS-PAGE (d1) and Western blot analysis of the purified 13cIMH. Equal amount of proteins (25 μg) and 0.5 μg of eluted protein were resolved by 8% SDS-PAGE. T; lysed total cellular protein, C; total cellular protein incubated with 0.1% CHAPS for 2 hours, P; insoluble fraction by 0.1% CHAPS, S; solubilized total cellular protein by 0.1% CHAPS, F; flow through from Ni-NTA resin, E; elution. (d2-3) Same order but half amount of proteins were resolved in SDS-PAGE gel and subjected to Western blot analysis with antibodies for RPE65 (d2) and 6×His-tag (d3). (e) Mihaelis-Menten plot of 13cROL generation by the purified 13cIMH. Liposomes with increasing concentrations (s, pico-moles) of atRE were incubated with 9.0 μg of purified 13cIMH. Initial rates (v) of 13cROL generation were calculated based on 13cROL production recorded by HPLC.
Tissue distribution and subcellular fractionation of 13cIMH
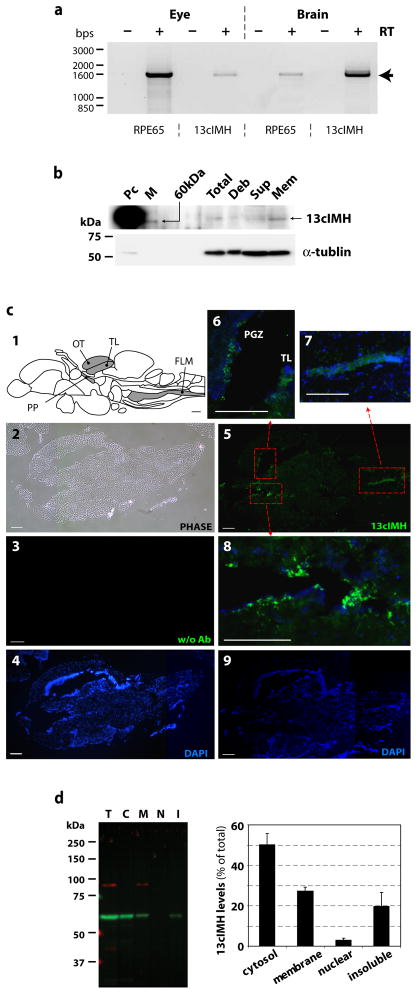
To determine the tissue distribution of 13cIMH, total RNA was extracted from adult zebrafish brain and eye. RT-PCR was performed using primers specific for zebrafish RPE65 and 13cIMH. The results showed that the RPE65 mRNA was predominantly expressed in the eye and at lower levels in the brain. In contrast, the 13cIMH mRNA was detected at high levels in the brain, and low levels in the eye (Fig. 7a). To detect endogenous 13cIMH in the brain, we performed Western blot analysis using whole brain homogenates. We also isolated total membrane fraction from the brain to enrich 13cIMH for Western blot analysis and in vitro enzymatic assay. A faint, yet, single band was observed in both the total brain homogenates and in the membrane fraction of the brain (Fig. 7b). The band showed an apparent molecular weight of 61 kDa, which is identical to that of the recombinant 13cIMH, although the intensity was lower than that of the recombinant protein (Fig. 7b). The molecular weight of the band also matched the calculated molecular weight from the amino acid sequence of 13cIMH. No 13cROL activity was detected in the brain homogenates or in the membrane fraction by HPLC (data not shown). This suggests that 13cIMH may be expressed only in a small region of the brain, and thus, 13cIMH was diluted in the whole brain homogenates or membrane fraction, so that its activity was not detected due to the assay sensitivity.
Figure 7. Localization of zebrafish 13cIMH in the brain and eye.
(a) RT-PCR analysis of RPE65 and 13cIMH using RNA from the zebrafish eye and brain. RT-PCR was performed in the absence (indicated by “−“) and the presence of (“+”) reverse transcriptase (RT) to exclude possible genomic DNA contamination. The arrow indicates the expected product size of 1.6 kbs. (b) Western blot analysis of endogenous 13cIMH in the total membrane fraction of brain. Cellular proteins (2.5 μg) of 293A-LRAT cells expressing 13cIMH were used as a positive control (Pc). Equal amounts (50 μg) of total zebrafish brain homogenates (Total), unbroken cell debris (Deb), supernatants following centrifugation (Sup) and total membrane fraction (Mem) were resolved in 8% SDS-PAGE gel and transferred onto the membrane. The endogenous 13cIMH expression was confirmed by Western blot analysis (upper panel), and then the membrane was stripped and re-blotted with an antibody for tublin (Abcam, Cambridge, MA, lower panel). (c) Immunohisotochemistry of 13cIMH in the zebrafish brain. (c1) The diagram shows a drawing sagittal section of zebrafish brain (modified from Rupp, B. et al. [36]). Gray-colored regions indicate the stained areas by immunohistochemistry. OT; Optic Tectum, TL; torus longitudinalis, PP; periventricular pretectum, FLM; fasciculus longitudinalis medialis. (c2) A phase contrast image of a sagittal section of zebrafish brain. (c3, 4) The brain section was incubated without the primary antibody for 13cIMH (c3; FITC channel, c4; dapi). (c5-9) the brain section was incubated with the primary antibody for 13cIMH. Green fluorescence indicated the signals of 13cIMH in low magnification images (c5; 13cIMH and c9; dapi) and in high magnification images from the boxed areas in c5: PGZ; periventricular grey zone, TL; torus longitudinalis (c6-8). Scale bar = 200 μm. (d) Subcellular localization of 13cIMH in cultured cells. Forty-eight hours post-transfection of the 13cIMH plasmid, the cells were harvested and separated to 4 subcellular fractions by FractionPrepTM kit. Equal amounts of fractionated proteins (25 μg for total protein, 5 μg each fraction) were used for Western blot analyses using an anti-13cIMH antibody. T; total cell lysates, C; cytosolic, M; membrane, N; nuclear fractions and I; detergent-insoluble fraction. The level of 13cIMH in each fraction was quantified by densitometry and expressed as % of total 13cIMH (means ± SEM) from 4 independent experiments.
To determine the location of 13cIMH in the brain, zebrafish brain sections were stained with or without the anti-13cIMH antibody by immunohistochemistry (Fig. 7c 2-4, without primary antibody (negative control), Fig. 7c 5-9, with primary antibody). The sections were missing the forebrain (Fig. 7c 1, 2), but 13cIMH expressions were detected in the periventricular grey zone (PGZ) of optic tectum (OT) and torus longitudinalis (TL) (Fig. 7c 5 and 6), at the fasciculus longitudinalis medialis (FLM) in the medulla oblongata (Fig. 7c 5 and 7), and at the periventricular pretectum (PP), which is a boundary area between brain and ventricles (Fig. 7c 5 and 8) (Referred from [36-38]). Similarly, the cross section of the zebrafish brain at the optic tectum showed that 13cIMH is expressed in the preventricular grey zone (PGZ) of optic tectum (OT) (see Fig. S2). In addition, subcellular fractionation and Western blot analysis of the 293A-LRAT cells expressing 13cIHM showed that 13cIMH was present in both of the cytosolic and membrane fractions (Fig. 7d).
Discussion
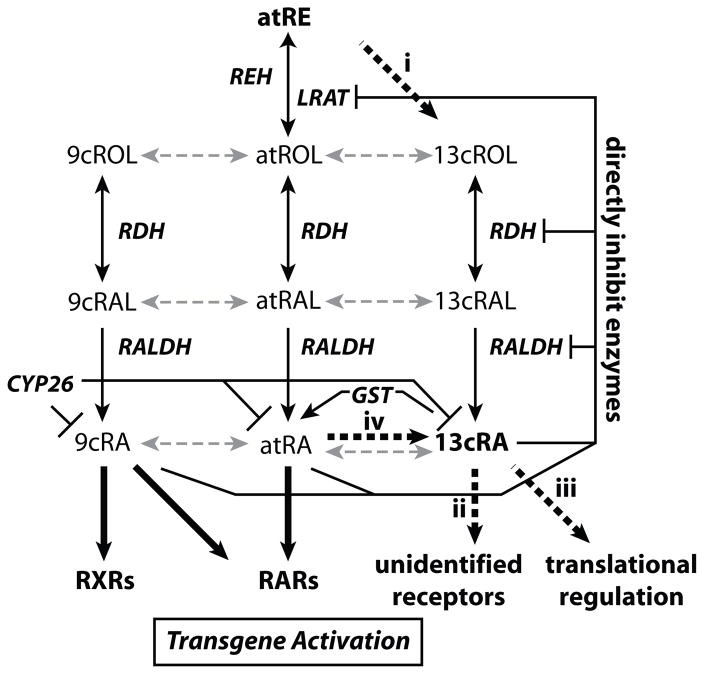
13cRA is an important isoform of RA and has crucial biological functions, especially in the central nervous system [10, 19-22]. High levels of 13cRA in the brain are associated with depression [19, 21]. The actual mechanism for the generation of endogenous 13cRA was unclear, although several possible pathways for generating 13cRA have been speculated (See Fig. 8). Here we identified the first enzyme that specifically generates 13cROL (Fig. 8, line i), a precursor of 13cRA, suggesting a potential role of 13cIMH in the production of 13cRA. This isomerohydrolase is expressed predominantly in the brain, suggesting a neurological-associated function. This novel finding indicates that there is an enzyme-dependent metabolic pathway to generate 13-cis retinoids in neuronal tissue.
Figure 8. Retinoid metabolism and retinoic acid signaling.
Scheme of retinoid metabolism. Solid lines indicate the reversible or irreversible conversion of retinoids by enzymes (indicated by italics). Gray broken lines represent the isomerization of retinoids by spontaneous thermal isomerization. Generally, it is considered that endogenous retinoids are stored as atRE in the liver and other tissues [1–3]. As needed, atRE is hydrolyzed to atROL, which is subsequently released into the circulation, bound by retinol-binding proteins and transported to target cells. Generated atRA binds to RARs, whereas 9cRA binds to both RARs and RXRs and activate target gene regulation. Bold broken lines indicate unidentified mechanisms and pathways related to 13-cis retinoids in the RA signaling: (i) The enzymes or mechanisms to generate 13cROL from atRE as shown in this study; (ii) 13cRA functions through unidentified signaling pathways or receptors; (iii) 13cRA may function by unidentified mechanism to enhance the translation of target gene mRNA or its protein stability [10]; (iv) Generation of 13cRA through the unidentified mechanism in RTE and HepG2 cells [27, 28]. Abbreviations in the figure: REH, retinyl ester hydrolase; LRAT, lecithin retinol acyltransferase; RDH, retinol dehydrogenases; RALDH, retinaldehyde dehydrogenases; GST, glutathione S-transferases.
13cRA might modulate expression of target genes through unidentified retinoic acid receptors (Fig. 8, line ii). Alternatively, it may be first isomerized to atRA or 9cRA, which can regulate gene expression through RARs or RXRs [8, 9]. Previous studies suggested that 13cRA is generated endogenously by an unknown mechanism [3, 25, 27, 28]. Isomerization from atRE to 13cROL is a key step in the generation of 13cRA. It is reported that the short-chain dehydrogenase/reductase family and the alcohol dehydrogenase family belonging to RDH family enzymes are expressed in the brain [39–41] and have ability to oxidize 13cROL to 13cRAL, even though with weaker activity than that of favorable substrates [11, 42, 43]. 13cRAL is further oxidized to 13cRA by ubiquitous retinal dehydrogenases in the brain such as RALDH2 [16, 44]. Our in vitro enzymatic activity assay showed that 13cIMH efficiently converts atRE to 13cROL. Therefore, this newly identified enzyme can catalyze a key reaction in the generation of 13cRA.
The possible physiological function of 13cIMH in the central nervous system may be associated with retinoic acid signaling in the regulation of synaptic plasticity, injury repair, learning and memory behavior [19, 22, 45]. It was reported that exposure to a clinical dose (1 mg/kg/day) or higher (40 mg/kg/day) of 13cRA suppresses hippocampal cell survival, division and proliferation both in vivo and in vitro [45, 46]. The hippocampal cell dysfunction induced by 13cRA has been shown to decrease the learning process, memory and induce depression-related behavior in a mouse model [21, 45, 47]. Likewise, 13cRA has been found to alter cellular morphology and exert non-transcriptionally mediated effects, such as significant increases in serotonin (5-HT1A) receptor and serotonin reuptake transporter (SERT) on cultured serotonergic cells [10, 48]. The decreased synaptic serotonin levels may impair neuronal function and result in improper neural communication. Our results also showed predominant expression of 13cIMH in the brain, further supporting its proposed role in the modulation of neuronal function.
The present study showed that 13cIMH shares high sequence homology with RPE65, an isomerohydrolase which converts atRE to 11cROL, a key step in the visual cycle [30, 49, 50]. RPE65 was considered an orphan gene, as it does not share high sequence homology with any known genes. Previously, only genes from the β-carotene monooxygenase family were found to have limited, but significant sequence homology with RPE65 (36.6% to human β-carotene monooxygenase and 36.7% to β-carotene monooxygenase from fruit fly [51]). 13cIMH represents the first protein identified to have high sequence homology (74% amino acid identity) to RPE65. Furthermore, sequence alignment showed that 13cIMH conserves particular features of RPE65 known to be essential for its enzymatic activity, e.g., four His residues for iron binding [30, 31] and a Cys residue for palmitylation and membrane association [32, 52]. Subcellular fractionation analysis showed that 13cIMH is also a membrane associated protein, similar to RPE65. It has been shown that membrane association of RPE65 is essential for its enzymatic activity [27]. Enzymatically, it also shares common features with RPE65, including the utilization of atRE as its direct substrate [34] and iron-dependent catalytic activity [35]. These structural and enzymatic similarities suggest that 13cIMH is also an isomerohydrolase in retinoid processing.
The catalytic efficiency (kcat/Km) of the purified recombinant 13cIMH was 169 M−1 s−1, which is 4.3-fold higher than that of purified recombinant chicken RPE65 (39 M−1 s−1) under the same assay condition [33]. In addition, we previously reported that recombinant chicken RPE65 exhibited 7.7-fold higher isomerohydrolase activity than that of recombinant human RPE65 [53], suggesting that 13cIMH is 33-fold more active than human RPE65 in isomerohydrolase activity. This higher enzymatic activity of 13cIMH may contribute to rapid synthesis of 13cROL, the precursor of 13cRA, in the limited expression areas of the enzyme in the brain (see Fig. 7c).
Redmond et al. recently reported that RPE65 generates equal amounts of 11cROL and 13cROL using an in-cell assay model [29]. We observed a similar phenomenon under our in vitro assay condition, i.e., RPE65 produced high levels of 11cROL and relatively low levels of 13cROL. However, the 13cROL production was detected only in the absence of LRAT in the reaction mixture (Suppl. Fig. 3b). A possible explanation for the higher 13cROL production in Redmond’s assay system, is that in their “in cell” assay condition, the reaction was allowed at 37 °C for 7 hours. The prolonged incubation period at 37 °C could generate more 13cROL through thermal isomerization of atROL to 13cROL, independent of the RPE65 activity.
In the presence of LRAT, however, our in vitro assay showed that RPE65 predominantly generated 11cROL (Fig. 3c and Suppl. Fig. 1c, also [31, 32, 50, 53]). This is consistent with our previous results showing that RPE65 predominantly generates 11cROL under our in vitro assay condition (at 37 °C for 1 hr) in the presence of LRAT and CRALBP [31, 32, 50, 53], which are the same protein sets in the experiments of Fig. 3c and Suppl. Fig. 1c in this manuscript. We suggest that CRALBP stabilizes 11cROL generated by RPE65, whereas other free retinoids, including 13cROL, can be re-esterified by LRAT and isomerized again by RPE65. Moreover, it was reported that 11cROL is a poor substrate of LRAT, compared to atROL and 13cROL [54, 55], which may account for the selective accumulation of 11cROL as the major product over 13cROL, although RPE65 has the ability to generate both 11cROL and 13cROL. We speculate that these are the potential reasons for the predominant 11cROL generation by RPE65 in the presence of LRAT under our in vitro assay condition and actual physiological conditions in the RPE which expresses LRAT.
Nonetheless, we noticed a difference in their products, when we compared the ratio of 11cROL to 13cROL produced by these RPE65 and 13cIMH under the same assay conditions. We have never detected any 11cROL generation by 13cIMH under any conditions. Similarly, we have never observed that RPE65 generates exclusively 13cROL under any conditions, indicating the difference between the 13cIMH and RPE65.
We have previously shown that RPE65, a homolog of 13cIMH, uses iron as a cofactor in the iron-binding site consisting of 4 conserved His residues, and existence of iron was confirmed by RPE65 crystal structure analysis [31, 35, 52]. 13cIMH showed 74% of amino acid sequence identity to human RPE65 and retains the four His forming the iron-binding site in RPE65, suggesting that 13cIMH is likely an iron-dependent enzyme. Our results confirmed this notion. It is noteworthy that the enzymatic activity of 13cIMH by supplement of FeSO4 following deprivation of endogenous iron did not recover completely. This observation was consistent with our previous result with recombinant human RPE65 [35], i.e., RPE65 activity after metal chelator incubation was not fully restored by the addition of iron. This partial recovery of the enzymatic activity by the addition of iron may be ascribed to the free radical generation caused by oxidation of ferrous ion to ferric ion (Fenton reaction) [56] and/or concomitant protein modification by these radicals, in the presence of high and possibly toxic concentrations of iron.
Our RT-PCR analysis shows that zebrafish RPE65 is predominantly expressed in the eye, and to lesser extent in the brain. Detection of RPE65 in the brain was not surprising, as there is a phototransduction system in the brain of lower vertebrates that is thought to be involved in regulating light-dependent circadian rhythms [57–61]. In contrast, 13cIMH is predominantly expressed in the brain, and only weakly expressed in the eye. The high level of 13cIMH expression in the brain supports the notion that it may function in the pathway of 13cRA synthesis, and thus 13cIMH may be important for the 13cRA-mediated regulation of neuronal function. The function of 13cIMH in the eye is unclear. Unlike 11cRAL which forms rhodopsin and 9cRAL which forms isorhodopsin, 13cRAL cannot form stable visual pigments [62]. Therefore, 13cIMH is unlikely to participate in the visual cycle. It is more likely that 13cIMH is necessary to generate a small amount of 13cRA in the eye that regulates retinal development and/or the neuronal function.
In summary, this study has identified the first 13-cis retinoid specific isomerohydrolase and will contribute to the understanding of 13cRA generation and its neurological functions.
Materials and Methods
Cloning of RPE65 homolog from the zebrafish brain
The brain were dissected from adult zebrafish, total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA) and further purified by an RNeasy kit (Qiagen, Valencia, CA). The cDNA was synthesized using the TaqMan reverse transcriptase (RT) system (Applied Biosystemes Inc., Foster City, CA) with an oligo-dT primer and random hexamer. PCR was performed at 94 C for 5 min followed by 35 cycles of 94 C for 30 sec, 45 C for 30 sec, and 72 C for 30 sec by using a pair of degenerate primers (DegRPE65-Fwd1; 5’-TGCARRAAYATHTTYTCCAG-3’ and DegRPE65-Rev1; 5’-TTKGMYCCYCWRAKRCTCCA-3’, expected size is 488 base pairs). The sizes of the PCR products were confirmed by 1.2 % of agarose gel electrophoresis and the identities of the products further confirmed by DNA sequencing.
Amino acid sequence comparisons and phylogenetic tree analyses of RPE65
Alignments of human RPE65 (NP_000320), zebrafish RPE65 (RPE65a, NP_957045) and 13cIMH (NP_001082902) were performed using “Clustal-W” program in BioEdit (Ibis Therapeutics, Carlsbad, CA). A phylogenetic tree was constructed using UPGMA method with 1000 times bootstrap re-sampling in MEGA application version 4.02 [63]. The known RPE65 sequences of human, macaque monkey (XP_001095946), bovine (NP_776878), dog (NP_001003176), rat (NP_446014), mouse (NP_084263), chicken (NP_990215), Japanese fireberry newt (BAC41351), tiger salamander (AAD12758), African clawed frog (AAI25978), zebrafish RPE65 and 13cIMH were used for phylogenetic analysis. Human β-carotene 15,15’-monooxygenase (BCO1, NP_059125) was used as the out-group of the tree.
Construction of 13cIMH expression vectors
The full-length cDNA clones were purchased from Open Biosystems (Huntsville, AL). The 13cIMH (NP_001082902) was sub-cloned into the pcDNA3.1(−) expression vector (Invitrogen, Carlsbad, CA) as described previously [32]. Briefly, the gene-specific primers (forward primer containing a NotI site and the Kozak sequence [64], 13cIMH-Fwd; 5’-GCGGCCGCCACCATGGTCAGTCGTCTTGAACAC-3’ and a reverse primer containing a HindIII site, 13cIMH-Rev; 5’-AAGCTTCTAAGGTTTGTAG ATGCCGTGGAG-3’) were used for PCR. PCR was performed with Pfu-Turbo (Stratagene, La Jolla, CA) at 94 C for 5 min followed by 35 cycles of 94 C for 1 min, 58 C for 1 min, and 72 C for 2 min. Following agarose gel electrophoresis, the PCR product was purified and cloned into the pGEM-T easy vector (Promega, Madison, WI). The insert was sequenced using an ABI-3770 automated DNA sequencer (Applied Biosystems Inc., Foster City, CA) from both directions to exclude any mutations. The confirmed 13cIMH cDNA was subcloned into an expression vector, pcDNA3.1(-) (Invitrogen, Carlsbad, CA). Following sequence confirmation, the expression constructs were purified by QIAfilter Maxi Prep kit (Qiagen, Valencia, CA). Further, histidine hexamer tag (6×His-tag) was fused to N-terminus of 13cIMH and cloned into pShuttle-CMV vector (Qbiogen, Montreal, Canada) for construction of adenovirus. Preparations, amplification and titration of recombinant adenovirus were performed as described previously [31, 50].
Western blot analysis
Briefly, total cellular protein concentration was measured by Bradford assay [65]. Equal amounts of protein (20 μg) were resolved by SDS-PAGE and blotted with a 1:1000 dilution of a rabbit polyclonal antibody to human RPE65 [66] which recognizes 13cIMH but not zebrafish RPE65 (see Fig. S3) and a 1:5000 dilution of mouse monoclonal antibody to β-actin (Abcam, Cambridge, MA) as a loading control. The membrane was then incubated for 1.5 hour with 1:25000 dilutions of anti-mouse IgG conjugated with DyLight 549 and anti-rabbit IgG conjugated with DyLight 649 (Pierce, Rockford, IL) and the bands were detected using FluorChem Q imaging system (AlphaInnotech, San Leandro, CA). The bands (intensity × area) were semi-quantified by densitometry using AlphaView Q software (AlphaInnotech, San Leandro, CA), and averaged from at least 3 independent experiments.
In vitro isomerohydrolase activity assay
293A-LRAT and 293T cells were separately transfected with plasmids expressing human RPE65, 13cIMH or RFP. The enzyme activity assays were carried out as described previously [33]. The peak of each retinoid isomer in high performance liquid chromatography (HPLC) profile was identified based on its characteristic retention time and the absorption spectrum of each retinoid standard. The isomerohydrolase activity was calculated from the area of the 11cROL and 13cROL peaks and represented as average ± SEM from 3 independent measurements.
Purification of recombinant 13cIMH
The 293A-LRAT cells were infected by adenoviruses expressing 6×His-tagged 13cIMH at MOI (multiplicity of infection) 100 and cultured for 24 hours. Then, the cells were lysed by sonication in the binding buffer (20 mM Tris, pH. 8.0, 150 mM NaCl, 10% glycerol and 10 mM imidazole), and the membrane associated proteins solubilized by incubation with 0.1% (w/v) of CHAPS (3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate hydrate) for 2 hours with gentle agitation. The CHAPS-solubilized 13cIMH was loaded into the Ni-NTA column (EMD chemicals, Gibbstown, NJ) and washed by the washing buffer (20 mM Tris, pH. 8.0, 150 mM NaCl, 10% glycerol and 30 mM imidazole). Finally, 13cIMH was eluted with elution buffer (20 mM Tris, pH. 8.0, 150 mM NaCl, 10% glycerol and 250 mM imidazole) and imidazole was eliminated by sequential centrifugations (10 mL × 5) using amicon-ultra centrifugation unit (Millipore, Billerica, MA).
RT-PCR analyses
The eye and brain were dissected from adult zebrafish, total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA), and further purified by an RNeasy kit (Qiagen, Valencia, CA). The cDNA was synthesized using the TaqMan reverse transcriptase (RT) system (Applied Biosystemes Inc., Foster City, CA) with an oligo-dT primer and random hexamer. Simultaneously, the same RNAs were used for the reaction without RT enzyme as a negative control (RT minus group). PCR was performed with Pfu-Turbo (Stratagene, La Jolla, CA) at 94 C for 5 min followed by 35 cycles of 94 C for 1 min, 58 C for 1 min, and 72 C for 2 min by using the same primer sets for 13cIMH cloning and gene specific primers of zebrafish RPE65 (zRPE65-Fwd; 5’-GCGGCCGCCACCATGGTCAGCCGTTTTGAACAC-3’ and zRPE65-Rev; 5’-GATATCTTATGGTTTGTACATCCCATGGAAAG-3’). The sizes of the PCR products were confirmed by 0.8 % of agarose gel electrophoresis and the identities of the products further confirmed by DNA sequencing.
Immunohistochemistry
The dissected zebrafish brain was fixed in 100 mM phosphate buffer containing 4% paraformaldehyde. The fixed tissues were used for the frozen sections. Following blocking with 3% bovine serum albumin (BSA) and 10% pre-immuned goat serum, the slides were incubated with a 1:1000 dilution of an anti-human RPE65 monoclonal antibody (Millipore, Billerica, MA) which recognizes 13cIMH, but not zebrafish RPE65 (Fig. S3). After 3 washes, the slides were incubated with a 1:200 dilution of Cy3-labeled anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA). Following 3 washes, the slides were treated with mounting medium containing DAPI (Vectorlab, San Diego, CA). The fluorescent images were captured using a Zeiss LSM-510META laser scanning confocal microscope (Carl Zeiss, Thromwood, NY).
Subcellular fractionation of 13cIMH in cultured cells
The 293A-LRAT cells expressing zebrafish 13cIMH was harvested and washed twice with ice-cold PBS. Subcellular fractionation analyses were performed as described previously [32]. Western blot analyses using the monoclonal antibody to 13cIMH and rabbit polyclonal antibody to calnexin (ER membrane marker, 1:2500 dilution, Abcam, Cambridge, MA) were used to identify the subcellular localization of RPE65 and to verify the membrane preparation. Distribution of RPE65 in each fraction was analyzed by densitometry and expressed as the mean ± SEM from 4 independent experiments.
Supplementary Material
Acknowledgments
We thank Dr. Tomoko Obara (University of Oklahoma Health Sciences Center, Oklahoma City, OK) for providing the zebrafish and Dr. Anne Murray for critical review of the manuscript. This study was supported by NIH grants EY018659, EY012231, EY019309, a grant (P20RR024215) from the National Center For Research Resources, and a grant from OCAST.
Abbreviations
- RA
Retinoic acid
- atRE
all-trans retinyl ester
- atROL
all-trans retinol
- RDH
retinol dehydrogenase
- RALDH
retinaldehyde dehydrogenase
- RAR
retinoic acid receptor
- RXR
retinoid x-receptor
- atRA
all-trans retinoic acid
- 9cRA
9-cis retinoic acid
- 13cRA
13-cis retinoic acid
- APL
acute promyelocytic leukaemia
- RPE65
retinal pigment epithelium specific 65-kDa protein
- RPE
retinal pigment epithelium
- 11cROL
11-cis retinol
- 13cROL
13-cis retinol
- 13cIMH
13-cis isomerohydrolase
- BCO1
β-carotene 15,15’-monooxygenase
- LRAT
lecithin retinol acyltransferase
- RT
reverse transcriptase
- PCR
polymerase chain reaction
- RFP
red fluorescent protein
- HPLC
high performance liquid chromatography
- MOI
multiplicity of infection;
- CHAPS
(3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate hydrate).
References
- 1.Morriss-Kay GM, Ward SJ. Retinoids and mammalian development. Int Rev Cytol. 1999;188:73–131. doi: 10.1016/s0074-7696(08)61566-1. [DOI] [PubMed] [Google Scholar]
- 2.Blomhoff R, Blomhoff HK. Overview of retinoid metabolism and function. J Neurobiol. 2006;66:606–630. doi: 10.1002/neu.20242. [DOI] [PubMed] [Google Scholar]
- 3.Kane MA, Folias AE, Wang C, Napoli JL. Quantitative profiling of endogenous retinoic acid in vivo and in vitro by tandem mass spectrometry. Anal Chem. 2008;80:1702–1708. doi: 10.1021/ac702030f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giguere V. Retinoic acid receptors and cellular retinoid binding proteins: complex interplay in retinoid signaling. Endocr Rev. 1994;15:61–79. doi: 10.1210/edrv-15-1-61. [DOI] [PubMed] [Google Scholar]
- 5.Idres N, Marill J, Flexor MA, Chabot GG. Activation of retinoic acid receptor-dependent transcription by all-trans-retinoic acid metabolites and isomers. J Biol Chem. 2002;277:31491–31498. doi: 10.1074/jbc.M205016200. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong JL, Redfern CP, Veal GJ. 13-cis retinoic acid and isomerisation in paediatric oncology--is changing shape the key to success? Biochem Pharmacol. 2005;69:1299–1306. doi: 10.1016/j.bcp.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Allenby G, Bocquel MT, Saunders M, Kazmer S, Speck J, Rosenberger M, Lovey A, Kastner P, Grippo JF, Chambon P, et al. Retinoic acid receptors and retinoid X receptors: interactions with endogenous retinoic acids. Proc Natl Acad Sci USA. 1993;90:30–34. doi: 10.1073/pnas.90.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Juchau MR. Glutathione S-transferases act as isomerases in isomerization of 13-cis-retinoic acid to all-trans-retinoic acid in vitro. Biochem J. 1997;327:721–726. doi: 10.1042/bj3270721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsukada M, Schroder M, Roos TC, Chandraratna RA, Reichert U, Merk HF, Orfanos CE, Zouboulis CC. 13-cis retinoic acid exerts its specific activity on human sebocytes through selective intracellular isomerization to all-trans retinoic acid and binding to retinoid acid receptors. J Invest Dermatol. 2000;115:321–327. doi: 10.1046/j.1523-1747.2000.00066.x. [DOI] [PubMed] [Google Scholar]
- 10.O'Reilly KC, Trent S, Bailey SJ, Lane MA. 13-cis-Retinoic acid alters intracellular serotonin, increases 5-HT1A receptor, and serotonin reuptake transporter levels in vitro. Exp Biol Med. 2007;232:1195–1203. doi: 10.3181/0703-RM-83. [DOI] [PubMed] [Google Scholar]
- 11.Gamble MV, Shang E, Zott RP, Mertz JR, Wolgemuth DJ, Blaner WS. Biochemical properties, tissue expression, and gene structure of a short chain dehydrogenase/ reductase able to catalyze cis-retinol oxidation. J Lipid Res. 1999;40:2279–2292. [PubMed] [Google Scholar]
- 12.Gollapalli DR, Rando RR. The specific binding of retinoic acid to RPE65 and approaches to the treatment of macular degeneration. Proc Natl Acad Sci USA. 2004;101:10030–10035. doi: 10.1073/pnas.0401936101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCaffery PJ, Adams J, Maden M, Rosa-Molinar E. Too much of a good thing: retinoic acid as an endogenous regulator of neural differentiation and exogenous teratogen. Eur J Neurosci. 2003;18:457–472. doi: 10.1046/j.1460-9568.2003.02765.x. [DOI] [PubMed] [Google Scholar]
- 14.Kraft JC, Juchau MR. Xenopus laevis: a model system for the study of embryonic retinoid metabolism. III. Isomerization and metabolism of all-trans-retinoic acid and 9-cis-retinoic acid and their dysmorphogenic effects in embryos during neurulation. Drug Metab Dispos. 1995;23:1058–1071. [PubMed] [Google Scholar]
- 15.Herrmann K. Teratogenic effects of retinoic acid and related substances on the early development of the zebrafish (Brachydanio rerio) as assessed by a novel scoring system. Toxicol In Vitro. 1995;9:267–283. doi: 10.1016/0887-2333(95)00012-w. [DOI] [PubMed] [Google Scholar]
- 16.Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–3094. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- 17.Okuno M, Kojima S, Matsushima-Nishiwaki R, Tsurumi H, Muto Y, Friedman SL, Moriwaki H. Retinoids in cancer chemoprevention. Curr Cancer Drug Targets. 2004;4:285–298. doi: 10.2174/1568009043333023. [DOI] [PubMed] [Google Scholar]
- 18.Reynolds CP, Matthay KK, Villablanca JG, Maurer BJ. Retinoid therapy of high-risk neuroblastoma. Cancer Lett. 2003;197:185–192. doi: 10.1016/s0304-3835(03)00108-3. [DOI] [PubMed] [Google Scholar]
- 19.Bremner JD, McCaffery P. The neurobiology of retinoic acid in affective disorders. Prog Neuro-Psychopharmacol Biol Psychiatry. 2008;32:315–331. doi: 10.1016/j.pnpbp.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blaner WS. Cellular metabolism and actions of 13-cis-retinoic acid. J Am Acad Dermatol. 2001;45:S129–135. doi: 10.1067/mjd.2001.113714. [DOI] [PubMed] [Google Scholar]
- 21.O'Reilly K, Bailey SJ, Lane MA. Retinoid-mediated regulation of mood: possible cellular mechanisms. Exp Biol Med. 2008;233:251–258. doi: 10.3181/0706-MR-158. [DOI] [PubMed] [Google Scholar]
- 22.Lane MA, Bailey SJ. Role of retinoid signalling in the adult brain. Prog Neurobiol. 2005;75:275–293. doi: 10.1016/j.pneurobio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Krezel W, Kastner P, Chambon P. Differential expression of retinoid receptors in the adult mouse central nervous system. Neuroscience. 1999;89:1291–1300. doi: 10.1016/s0306-4522(98)00342-x. [DOI] [PubMed] [Google Scholar]
- 24.Maden M. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat Rev Neurosci. 2007;8:755–765. doi: 10.1038/nrn2212. [DOI] [PubMed] [Google Scholar]
- 25.Tang GW, Russell RM. 13-cis-retinoic acid is an endogenous compound in human serum. J Lipid Res. 1990;31:175–182. [PubMed] [Google Scholar]
- 26.Arnhold T, Tzimas G, Wittfoht W, Plonait S, Nau H. Identification of 9-cis-retinoic acid, 9,13-di-cis-retinoic acid, and 14-hydroxy-4,14-retro-retinol in human plasma after liver consumption. Life Sci. 1996;59:169–177. doi: 10.1016/0024-3205(96)00408-0. [DOI] [PubMed] [Google Scholar]
- 27.Bhat PV, Jetten AM. Metabolism of all-trans-retinol and all-trans-retinoic acid in rabbit tracheal epithelial cells in culture. Biochim Biophys Acta. 1987;922:18–27. doi: 10.1016/0005-2760(87)90240-2. [DOI] [PubMed] [Google Scholar]
- 28.Lansink M, van Bennekum AM, Blaner WS, Kooistra T. Differences in metabolism and isomerization of all-trans-retinoic acid and 9-cis-retinoic acid between human endothelial cells and hepatocytes. Eur J Biochem. 1997;247:596–604. doi: 10.1111/j.1432-1033.1997.00596.x. [DOI] [PubMed] [Google Scholar]
- 29.Redmond TM, Poliakov E, Kuo S, Chander P, Gentleman S. RPE65, visual cycle retinol isomerase, is not inherently 11-cis specific: Support for a carbocation mechanism of retinol isomerization. J Biol Chem. 2009;285:1919–1927. doi: 10.1074/jbc.M109.027458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redmond TM, Poliakov E, Yu S, Tsai JY, Lu Z, Gentleman S. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc Natl Acad Sci USA. 2005;102:13658–13663. doi: 10.1073/pnas.0504167102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi Y, Moiseyev G, Chen Y, Ma JX. Identification of conserved histidines and glutamic acid as key residues for isomerohydrolase activity of RPE65, an enzyme of the visual cycle in the retinal pigment epithelium. FEBS Lett. 2005;579:5414–5418. doi: 10.1016/j.febslet.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi Y, Moiseyev G, Ablonczy Z, Chen Y, Crouch RK, Ma JX. Identification of a novel palmitylation site essential for membrane association and isomerohydrolase activity of RPE65. J Biol Chem. 2009;284:3211–3218. doi: 10.1074/jbc.M807248200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolaeva O, Takahashi Y, Moiseyev G, Ma JX. Purified RPE65 shows isomerohydrolase activity after reassociation with a phospholipid membrane. FEBS J. 2009;276:3020–3030. doi: 10.1111/j.1742-4658.2009.07021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moiseyev G, Crouch RK, Goletz P, Oatis J, Jr, Redmond TM, Ma JX. Retinyl esters are the substrate for isomerohydrolase. Biochemistry. 2003;42:2229–2238. doi: 10.1021/bi026911y. [DOI] [PubMed] [Google Scholar]
- 35.Moiseyev G, Takahashi Y, Chen Y, Gentleman S, Redmond TM, Crouch RK, Ma JX. RPE65 is an iron(II)-dependent isomerohydrolase in the retinoid visual cycle. J Biol Chem. 2006;281:2835–2840. doi: 10.1074/jbc.M508903200. [DOI] [PubMed] [Google Scholar]
- 36.Rupp B, Wullimann MF, Reichert H. The zebrafish brain: a neuroanatomical comparison with the goldfish. Anat Embryol. 1996;194:187–203. doi: 10.1007/BF00195012. [DOI] [PubMed] [Google Scholar]
- 37.Castro A, Becerra M, Manso MJ, Anadon R. Calretinin immunoreactivity in the brain of the zebrafish, Danio rerio: distribution and comparison with some neuropeptides and neurotransmitter-synthesizing enzymes. II. Midbrain, hindbrain, and rostral spinal cord. J Comp Neurol. 2006;494:792–814. doi: 10.1002/cne.20843. [DOI] [PubMed] [Google Scholar]
- 38.Mueller T, Wullimann MF. Atlas of Early Zebrafish Brain Development. Elsevier; Denvers, MA: 2005. Atlas of Cellular Markers in Zebrafish Neurogenesis; pp. 29–135. [Google Scholar]
- 39.Duester G. Involvement of alcohol dehydrogenase, short-chain dehydrogenase/reductase, aldehyde dehydrogenase, and cytochrome P450 in the control of retinoid signaling by activation of retinoic acid synthesis. Biochemistry. 1996;35:12221–12227. doi: 10.1021/bi961176+. [DOI] [PubMed] [Google Scholar]
- 40.Reimers MJ, Hahn ME, Tanguay RL. Two zebrafish alcohol dehydrogenases share common ancestry with mammalian class I, II, IV, and V alcohol dehydrogenase genes but have distinct functional characteristics. J Biol Chem. 2004;279:38303–38312. doi: 10.1074/jbc.M401165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shou S, Scott V, Reed C, Hitzemann R, Stadler HS. Transcriptome analysis of the murine forelimb and hindlimb autopod. Dev Dyn. 2005;234:74–89. doi: 10.1002/dvdy.20514. [DOI] [PubMed] [Google Scholar]
- 42.Gough WH, VanOoteghem S, Sint T, Kedishvili NY. cDNA cloning and characterization of a new human microsomal NAD+-dependent dehydrogenase that oxidizes all-trans-retinol and 3alpha-hydroxysteroids. J Biol Chem. 1998;273:19778–19785. doi: 10.1074/jbc.273.31.19778. [DOI] [PubMed] [Google Scholar]
- 43.Driessen CA, Winkens HJ, Kuhlmann ED, Janssen AP, van Vugt AH, Deutman AF, Janssen JJ. The visual cycle retinol dehydrogenase: possible involvement in the 9-cis retinoic acid biosynthetic pathway. FEBS Lett. 1998;428:135–140. doi: 10.1016/s0014-5793(98)00473-6. [DOI] [PubMed] [Google Scholar]
- 44.Gagnon I, Duester G, Bhat PV. Kinetic analysis of mouse retinal dehydrogenase type-2 (RALDH2) for retinal substrates. Biochim Biophys Acta. 2002;1596:156–162. doi: 10.1016/s0167-4838(02)00213-3. [DOI] [PubMed] [Google Scholar]
- 45.Crandall J, Sakai Y, Zhang J, Koul O, Mineur Y, Crusio WE, McCaffery P. 13-cis-retinoic acid suppresses hippocampal cell division and hippocampal-dependent learning in mice. Proc Natl Acad Sci USA. 2004;101:5111–5116. doi: 10.1073/pnas.0306336101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakai Y, Crandall JE, Brodsky J, McCaffery P. 13-cis Retinoic acid (accutane) suppresses hippocampal cell survival in mice. Ann N Y Acad Sci. 2004;1021:436–440. doi: 10.1196/annals.1308.059. [DOI] [PubMed] [Google Scholar]
- 47.O'Reilly KC, Shumake J, Gonzalez-Lima F, Lane MA, Bailey SJ. Chronic administration of 13-cis-retinoic acid increases depression-related behavior in mice. Neuropsychopharmacology. 2006;31:1919–1927. doi: 10.1038/sj.npp.1300998. [DOI] [PubMed] [Google Scholar]
- 48.Ishikawa J, Sutoh C, Ishikawa A, Kagechika H, Hirano H, Nakamura S. 13-cis-retinoic acid alters the cellular morphology of slice-cultured serotonergic neurons in the rat. Eur J Neurosci. 2008;27:2363–2372. doi: 10.1111/j.1460-9568.2008.06191.x. [DOI] [PubMed] [Google Scholar]
- 49.Jin M, Li S, Moghrabi WN, Sun H, Travis GH. Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell. 2005;122:449–459. doi: 10.1016/j.cell.2005.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moiseyev G, Chen Y, Takahashi Y, Wu BX, Ma JX. RPE65 is the isomerohydrolase in the retinoid visual cycle. Proc Natl Acad Sci USA. 2005;102:12413–12418. doi: 10.1073/pnas.0503460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Lintig J, Vogt K. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving beta-carotene to retinal. J Biol Chem. 2000;275:11915–11920. doi: 10.1074/jbc.275.16.11915. [DOI] [PubMed] [Google Scholar]
- 52.Kiser PD, Golczak M, Lodowski DT, Chance MR, Palczewski K. Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proc Natl Acad Sci USA. 2009;106:17325–17330. doi: 10.1073/pnas.0906600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moiseyev G, Takahashi Y, Chen Y, Kim S, Ma JX. RPE65 from cone-dominant chicken is a more efficient isomerohydrolase compared with that from rod-dominant species. J Biol Chem. 2008;283:8110–8117. doi: 10.1074/jbc.M703654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maeda A, Maeda T, Imanishi Y, Golczak M, Moise AR, Palczewski K. Aberrant metabolites in mouse models of congenital blinding diseases: formation and storage of retinyl esters. Biochemistry. 2006;45:4210–4219. doi: 10.1021/bi052382x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bok D, Ruiz A, Yaron O, Jahng WJ, Ray A, Xue L, Rando RR. Purification and characterization of a transmembrane domain-deleted form of lecithin retinol acyltransferase. Biochemistry. 2003;42:6090–6098. doi: 10.1021/bi0342416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petrat F, Paluch S, Dogruoz E, Dorfler P, Kirsch M, Korth HG, Sustmann R, de Groot H. Reduction of Fe(III) ions complexed to physiological ligands by lipoyl dehydrogenase and other flavoenzymes in vitro: implications for an enzymatic reduction of Fe(III) ions of the labile iron pool. J Biol Chem. 2003;278:46403–46413. doi: 10.1074/jbc.M305291200. [DOI] [PubMed] [Google Scholar]
- 57.Okano T, Yoshizawa T, Fukada Y. Pinopsin is a chicken pineal photoreceptive molecule. Nature. 1994;372:94–97. doi: 10.1038/372094a0. [DOI] [PubMed] [Google Scholar]
- 58.Pasqualetti M, Bertolucci C, Ori M, Innocenti A, Magnone MC, De Grip WJ, Nardi I, Foa A. Identification of circadian brain photoreceptors mediating photic entrainment of behavioural rhythms in lizards. Eur J Neurosci. 2003;18:364–372. doi: 10.1046/j.1460-9568.2003.02770.x. [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Fernandez JM, Jimenez AJ, Gonzalez B, Pombal MA, Foster RG. An immunocytochemical study of encephalic photoreceptors in three species of lamprey. Cell Tissue Res. 1997;288:267–278. doi: 10.1007/s004410050812. [DOI] [PubMed] [Google Scholar]
- 60.Kojima D, Mano H, Fukada Y. Vertebrate ancient-long opsin: a green-sensitive photoreceptive molecule present in zebrafish deep brain and retinal horizontal cells. J Neurosci. 2000;20:2845–2851. doi: 10.1523/JNEUROSCI.20-08-02845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peirson SN, Halford S, Foster RG. The evolution of irradiance detection: melanopsin and the non-visual opsins. Philos Trans R Soc Lond B Biol Sci. 2009;364:2849–2865. doi: 10.1098/rstb.2009.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pepperberg DR, Brown PK, Lurie M, Dowling JE. Visual pigment and photoreceptor sensitivity in the isolated skate retina. J Gen Physiol. 1978;71:369–396. doi: 10.1085/jgp.71.4.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 66.Ma J, Zhang J, Othersen KL, Moiseyev G, Ablonczy Z, Redmond TM, Chen Y, Crouch RK. Expression, purification, and MALDI analysis of RPE65. Invest Ophthalmol Vis Sci. 2001;42 :1429–1435. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.