Abstract
Nox5 belongs to the calcium-regulated subfamily of NADPH oxidases (Nox). Like other calcium-regulated Noxes, Nox5 has an EF hand-containing calcium-binding domain at its N-terminus, a transmembrane heme-containing region and a C-terminal dehydrogenase (DH) domain that binds FAD and NADPH. While Nox1-4 require regulatory subunits including p22phox, Nox5 activity does not depend on any subunits. We found that inactive point mutants and truncated forms of Nox5 (including the naturally expressed splice form Nox5S) inhibit full-length Nox5, consistent with formation of a dominant negative complex. Oligomerization of full-length Nox5 was demonstrated using co-immunoprecipitation of co-expressed, differentially tagged forms of Nox5 and occurred independently of calcium ion. Several approaches were used to show that the DH domain mediates oligomerization: Nox5 could be isolated as a multimer when the calcium-binding domain and/or the N-terminal polybasic region (PBR-N) were deleted, but deletion of the DH domain eliminated oligomerization. Further, a chimera containing the transmembrane domain of Ciona intestinalis voltage sensor-containing phosphatase (CiVSP) fused to the Nox5 DH domain formed a co-immunoprecipitating complex with, and functioned as a dominant inhibitor of, full-length Nox5. Radiation inactivation of Nox5 overexpressed in HEK293 cells or endogenously expressed in human aortic smooth muscle cells indicated molecular weights of about 350 kDa and 300 kDa, respectively, consistent with a tetramer as the functionally active unit. Thus, Nox5 forms a catalytically active oligomer in the membrane that is mediated by its dehydrogenase domain. As a result of oligomerization, the short, calcium-independent splice form Nox5S may function as an endogenous inhibitor of calcium-stimulated ROS generation by full-length Nox5.
The family of human reactive oxygen species (ROS)-generating NADPH-oxidases (Nox) consists of seven members that include Nox1, Nox2, Nox3, Nox4 and Nox5 plus the Dual oxidases Duox1 and Duox2 (1). Structural features and experimental data have been used to classify the Nox and Duox enzymes into two broad groups: those regulated by subunits including a membrane-associated subunit p22phox (Nox1-Nox4) and those that are regulated by calcium ion (Nox5 and Duox1 and 2) (2–4). In the former subgroup, Nox forms a tightly associated, mutually stabilizing heterodimer with p22phox. Nox1-3 enzymes require additional regulatory subunits such as p47phox, p67phox, p40phox, NOXO1, NOXA1, and the small GTPase Rac (5). The calcium-regulated subgroup possesses a calcium regulatory domain that contains single or multiple EF-hand calcium-binding motif(s). While Duox1 and Duox2 require the maturation factors DuoxA1 and DuoxA2, respectively, to facilitate their movement to the plasma membrane (6, 7), Nox5 apparently functions independently of subunits [(8) and vide infra].
Human Nox5 contains four EF-hand motifs in its calcium-binding domain, which is located at the N-terminus (9). Rapid activation of Nox5 by calcium ion has been observed in Nox5-overexpressing HEK293 cells (8, 9), human aortic smooth muscle cells (10), prostate cells (11) and vascular endothelial cells (12, 13). In addition to calcium, other co-activating mechanisms cooperate by lowering the concentration of calcium needed to trigger activation: these include phorbol 12-myristate 13-acetate-dependent phosphorylation of Nox5 (14), calmodulin (13, 15), and agonist-generated lipids including phosphatidic acid and arachidonate (Kawahara and Lambeth, manuscript submitted).
The physiological role of Nox5 has been extensively studied using Drosophila melanogaster where Nox5 (termed d-Nox) generates a ROS signal in smooth muscle to mediate hormone-induced smooth muscle contraction and egg laying (16). In human, Nox5 expression is abundant in the spleen, testis, and vascular tissue, cells of the gastrointestinal tract, reproductive systems, and is also seen in various cancers (17). Nox5 has been lost in rodent genomes (2), making it impossible to investigate its role in mouse knockout models. Results from in vitro studies in human cells indicate that Nox5-derived ROS enhances cell growth of prostate cancer cells (11), hairy cell leukemia cells (18), aortic smooth muscle cells (10), and microvascular endothelial cell (12). Recently, studies of human biopsy samples of coronary arteries demonstrated increased expression of Nox5 in coronary artery disease, using immunoblotting, immunofluorescence, and quantitative PCR (19).
The present studies were undertaken based on the observation (vide infra) that co-transfection of catalytically inactive point-mutated forms of several Nox isoforms along with the same wild-type isoform resulted in inhibition of wild-type enzyme activity, meeting the definition of a “dominant negative” genetic effect. There are at least two possible explanations for this type of phenomenon. One involves sequestering of a needed regulatory subunit or other component by the catalytically inactive enzyme, thereby removing a factor that is essential for the activity of the wild-type enzyme. A second class of explanations involves catalytically essential dimerization or higher-order oligomerization. According to the latter scenario, the inclusion of a catalytically inactive unit in the oligomer renders the whole inactive or partially active, for example, by creating a poorly active conformation of the whole. Because Nox5 does not require the participation of heterologous subunits, interpretation of data are more straightforward than for other Nox isoforms, and this isoform was therefore selected for an in-depth investigation of the possibility of homo-oligomerization. A variety of methods were used to demonstrate oligomerization, and to show that this interaction is essential for Nox5 function. In addition, several approaches were used to identify the region of Nox5 that mediates oligomerization.
MATERIALS AND METHODS
Materials
Human embryonic kidney (HEK) 293 cells and human aortic smooth muscle cells (HASMCs) were obtained from American Type Culture Collection (Manassas, VA) and Lonza (Walkersville, MD), respectively. Ionomycin, ferricytochrome c, superoxide dismutase (SOD), ethylenediaminetetraacetic acid (EDTA) and mouse monoclonal anti-β-actin antibody were purchased from Sigma (St. Louis, MO). Anti-human Nox5 antibody was made in rabbits immunized with synthetic peptides corresponding to residues 491-506 and 577-588 of human Nox5α. Anti-myc and -hexahistidine monoclonal antibodies, anti-GFP monoclonal antibody, and anti-GFP rabbit polyclonal antibody were purchased from Cell Signaling, Millipore, and Anaspec, respectively. Platelet-derived growth factor BB (PDGF-BB) was purchased from R&D Systems, Inc. Maltose dehydrogenase, alcohol dehydorogenase, and β-galactosidase were purchased from Sigma.
Cell culture
HEK293 cells were grown for 24 h in six-well plates and allowed to reach to 50% confluence in 2 ml DMEM with 10% (vol/vol) fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. HASMCs were maintained in smooth muscle growth medium (SmGM2: smooth muscle basal medium (SmBM) with addition of 5% FBS, 5 ng/mL insulin, 0.5 ng/mL EGF, 2 ng/mL FGF-B, and amphotericin B. Cells at passages 4 to 10 were used for the experiments.
Constructs
Expression vector encoding wild-type, untagged Nox5 (subcloned in PCMV-5A tag vector, Stratagene), C-terminally myc-tag Nox5 (PCMV-5A tag vector), N-terminally hexahistidine-tagged Nox5 (PCMV-5A tag vector), and N-terminally GFP-tagged Nox5 (pEGFP-C3 vector, BD bioscience) were previously described (20). Point mutations were introduced by PCR-mediated mutagenesis as described previously (8). Nox5-P567H was constructed by amplifying human Nox5α cDNA (GenBank accession No. AF353088) using primer set A (primer-1: GCATGGATCCACCATGAACACATCTGGAGA; P567H-primer 2, CAGAATGGAAGCAAAGTGGGTGATGCCGATGCC) and primer set B (P567H-primer 3, GGCATCGGCATCACCCACTTTGCTTCCATTCTG, primer-4 GCATAAGCTTCTAGAAATTCTCTTGGAAAAATCTGAAGCCGAAC). Nucleotides encoding the mutated amino acid are underlined in primers 2 and 3. Primers 1 and 4 also introduce BamHI (italics) and HindIII cleavage sites (underlined), respectively. Following a second round of PCR using primers 1 and 4 and digestion by each enzyme, the nucleotide fragments were subcloned into the pCMV-tag 5A (Stratagene, La Jolla, CA). Point mutations V276R, L277R, L277A, P278L, P438A, D656A and K658A of Nox5 were generated using a similar strategy. Truncated Nox5 (S59-F737) was constructed by amplifying the cDNA using primer set (S59-primer 5, GCATAAGCTTTCCTTCTTTGCAGAGCGATTCTTT, primer-6 GCATGGATCCCTAGAAATTCTCTTGGAAAAATCTGAAGCCGAAC). Primers 5 and 6 are designed to introduce BamHI (italics) and HindIII cleavage sites (underlined), respectively. The nucleotide PCR products were subcloned into the pEGFP-C3 (BD Bioscience, La Jolla, CA). Other truncations M173-F737, A202-F737, A202-K429, and A202-A407 were generated using a similar strategy.
The cDNA of the voltage sensor containing phosphatase from Ciona intestinalis (CiVSP, GenBank accession No. AB183035) was kindly provided by Dr. Yasushi Okamura (Osaka Univeristy, Osaka, Japan). The N-terminus of the CiVSP (amino acids 1-239) containing the transmembrane region of the CiVSP protein was fused to the DH domain of Nox5 (amino acids 404-737 of Nox5α) to create the chimera referred to as “chimera CiVSP-N/Nox5-DH-C”. This chimera was made with either an N-terminal GFP tag or a C-terminal myc tag. These cDNAs were created by sequential PCR reactions followed by digestion and ligation into the appropriate vector. The CiVSP-N/Nox5-DH-C-myc was constructed by amplifying the cDNA using primer sets: 1. CiVSP with BamH1 and Kozak sense, 5′ TTTTGGATCCGCCACCATGGAGGGATTCGACGGTTCAG-3′; 2. CiVSP end at 239/Nox5a begin at 404 sense 5′-GAATATTTTATTCCCATCAACAAGAGAAGGCCATCGGACTG-3′ with the CiVSP sequence underlined; 3. Anti-sense of primer 2, 5′-CAGTCCGATGGCCTTCTCTTGTTGATGGGAATAAAATATTC-3′; 4. Nox5 delete stop HindIII anti-sense 5′-TTTTAAGCTTGAAATTCTCTTGGAAAAATCTGAAGCCG-3′. PCR reaction A used primers 1 and 3 with CiVSP cDNA and primers 2 and 4 with Nox5 cDNA. PCR reaction B used primers 1 and 4 with the two PCR products of PCR reaction A. The final chimera product was digested with BamH1 and HindIII and ligated into pCMV tag 5A (Stratagene), which includes a C-terminal myc tag. To make CiVSP-N/Nox5-DH-C, the primers were: 5. CiVSP Not1 sense, 5′-TTTTGCGGCCGCATGGAGGGATTCGACGGTTCAG-3′; 2 and 3 Same as above; 6. Nox5 stop HindIII anti-sense 5′-TTTTAAGCTTCTAGAAATTCTCTTGGAAAAATCTGAAGCCG-3′. PCR reactions A and B were performed as above. This chimera product was digested with Not1 and HindIII and then ligated with the GFP-containing PCR product with a C-terminal Not1 site and an N-terminal BamH1 site into pCMV tag 5A. The resulting cDNA is referred to as GFP-CiVSP-N/Nox5-DH-C. To construct GFP-CiVSP, a CiVSP sense primer 7 with an N-terminal HindIII restriction enzyme site 5′-TTTTAAGCTTATGGAGGGATTCGACGGTTCAG -3′ and an anti-sense primer 8 with a C-terminal BamH1 site, 5′-TTTTGGATCCCTAAATGTCTTCAGCATCTGAAAACG-3′ was used to PCR full-length CiVSP. This product was digested with BamH1 and HindIII and ligated into pEGFP-C3, which encodes an N-terminal GFP tag. The CiVSP-myc construct was made with primer 1 listed above and an anti-sense primer 9 with a HindIII site and deletion of the stop codon, 5′-TTTTAAGCTTAATGTCTTCAGCATCTGAAAACG-3′. All sequences were confirmed by commercial DNA sequencing (Beckman Coulter Genomics).
Immunoprecipitation
HEK293 cells were plated in 10 cm dishes and allowed to grow for 24 hours. Cells were then transfected with indicated amount of each cDNA construct using Fugene-6 transfection reagents (Roche) according to the manufacturer’s instructions. Cells were grown for 48 hours and harvested in Hank’s balanced salt solution (Gibco). Cell pellets were suspended in lysis buffer (20 mM Tris-HCl pH 7.5, 1% Triton X-100, 150 mM NaCl) supplemented with EDTA-free protease inhibitor cocktail (Complete Mini, Roche), 1 mM phenylmethylsulfonyl fluoride (PMSF) and 100 μM diisopropylfluorophosphate (DFP). Lysate mixtures were incubated on ice for 30 minutes and centrifuged. The supernatant containing proteins was incubated with primary antibody over night with rotation at 4°C. A 50% protein-G Sepharose mixture was added to the lysates and rotated at 4°C for two hours. The protein-G Sepharose pellet containing immunoprecipitated proteins was washed three times with the Tris-HCl buffer described above. Laemmli sample buffer (Bio-Rad) containing 5% 2-mercaptoethanol was added to the immunoprecipitated proteins. Samples were resolved by SDS-PAGE followed by Western blotting.
Measurement of ROS Generation in intact Cells
ROS was measured using luminol luminescence as previously described (8). HEK293 cells were grown and transfected as described above. At 48 h after transfection, cells were washed twice with Hanks’ balanced salt solution (HBSS) containing 1 mM calcium and 1 mM magnesium and harvested by centrifuging at 500 × g for 5 min. Cells (2 × 105) in HBSS with calcium and magnesium were mixed with 200 μM luminol plus 0.32 units of horseradish peroxidase in a 200-μl total volume in each well of a 96-well plate. Luminescence was quantified using a FluoStar™ luminometer (BMG Labtech), recording data every 30 seconds for 1 h. Cells were treated with DMSO vehicle alone or with 1 μM ionomycin in DMSO at 10 min after beginning data collection. The maximum luminescence was achieved within 1 min and is the value reported.
Measurement of ROS Generation in Permeabilized Cells
Superoxide in permeabilized HEK293 cells was measured using SOD-inhibitable cytochrome c reduction as previously described (20). After pretreatment with 5 μM thapsigargin to deplete intracellular calcium stores, cells were washed with calcium- and magnesium-free HBSS, and were resuspended in phosphate buffer saline (PBS, pH 7.2) containing protease inhibitor mixture (Complete EDTA-free, Roche). To permeabilize cells, reduced streptolysin-O (final concentration, 20 U/ml) was added to 107 cells in 10 ml and incubated for 10 min at 37°C. Cells were chilled to 4°C, centrifuged for 5 min at 1250 × g, and washed three times with PBS (pH 7.2). To measure superoxide, cells (~5 × 105 cell equivalents) were suspended in calcium calibration solution [30 mM MOPS buffer (pH 7.2) containing 100 mM KCl, 1 mM CaEGTA and 9 mM EGTA; Molecular Probes, Eugene, OR], which results in 17 μM free calcium ion concentration. Calcium-free buffer was the same, except that it lacked added calcium and contained 10 mM EGTA. After 5 min at room temperature, 1 μl NADPH solution was added to give a final concentration of 100 μM, with or without SOD (100 U/ml), and the rate of absorbance increase at 550 nm was measured for 10 min using a Synergy HT spectrophotometer (Bio-Tek Instruments, Winooski, VT). Permeabilized cells were then lysed with 20 mM HEPES buffer containing 1% Triton X-100, and the protein was quantified using the BCA protein assay reagent kit (Pierce, Rockford, IL). The rate of superoxide production was expressed as SOD-inhibitable cytochrome c reduction per min per mg protein.
In some experiments, H2O2 generation by permeabilized HEK293 cells and hASMCs for radiation inactivation analysis was assessed by the fluorescence increase at 620 nm (excitation wavelength of 540 nm) due to Amplex Red oxidation in the presence of HRP, as described (21). Permeabilized cells (~ 5 × 105 cell equivalents) were kept frozen on dry ice during irradiation. For activity measurements, cells were thawed and suspended as above in calcium calibration solution at 17μM free calcium ion or in calcium-free buffer. After 5 min at room temperature, hydrogen peroxide was measured by Amplex UltraRed with 200 μl of a reaction buffer [30 mM MOPS, (PH 7.2) containing 0.12M NaCl, 3 mM KCl, 100 μM Amplex Red, 200 mU/ml HRP, 20 μM FAD, 36 μM NADPH]. The rate of fluorescence increase at 620 nm was measured at 25C for 10 min using a Synergy HT spectrofluorimeter (Bio-Tek Instruments). The rate of calcium-dependent H2O2 production was expressed as fluorescence increase at 620 nm per 106 cells minus that obtained in the calcium-free buffer.
Immunoblotting
Whole cell extract was obtained from cultured cells lysed in Laemmli sample buffer containing protease inhibitor mixture (Complete), 100 μM diisopropyl fluorophosphate, and 25 mM tris(2-carboxyethyl)phosphine (TCEP). Protein extracted from 8 × 104 cell equivalents was resolved by 10% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) filters. After blocking with bovine serum albumin (3%), proteins were probed using their respective primary antibodies and HRP-conjugated secondary antibodies against rabbit and mouse IgG (Bio-Rad Laboratories). Primary antibodies were used at dilutions of 1:1000 (anti-Nox5, -hexahistidine, -myc, -GFP antibodies), and 1:20,000 (anti-β–actin antibody). Visualization was carried out using an enhanced chemiluminescent substrate kit (Pierce).
Radiation-inactivation
Protein samples (permeabilized cells or commercially obtained standard enzymes) in 1.5 ml microfuge tubes were flushed with liquid N2 for 30 min and stored in a freezer at −80 C and transported packed in dry ice pellets. Duplicate samples were maintained on dry ice during irradiation (~19,700 Rads/min), and were removed at various times and stored at −80 C until enzyme assay, performed as above. Irradiation was performed at the Centers for Disease Control, Atlanta, GA, using a Gammacell 220 Excel Irradiator Cobalt 60 radiation source (MDS Nordion, Ottawa, Canada). Radiation doses delivered by the Gammacell were calculated for each irradiation date using accurate decay charts for the cobalt 60 sealed radiation source: Dose = (Rads/min)* (minutes of irradiation). Enzymatic activities of enzymes from each irradiated sample were determined. Nox5 activity was measured as described above. Activities of standard enzymes maltose dehydrogenase (MDH), alcohol dehydorogenase (ADH), and β-galactosidase were measured as described previously (22). MDH was measured using pig heart cytoplasmic malate dehydrogenase, assayed in a reaction mixture containing 30 mM Hepes, 50 mM KCl, 0.5 mM EDTA and 0.2 mg/ml bovine serum albumin, 0.1 mM NADH, pH 7.5, and sample was added immediately before incubation. Activity was measured as the linear decrease in absorption at 340 nm after the addition of freshly made 1 mM-oxaloacetate. ADH was measured using yeast alcohol dehydrogenase, assayed in a reaction mixture containing 75 mM sodium pyrophosphate, 10 mM glycine, 75 mM semicarbazide. 1 mM EDTA, 1 mM NAD+ and 0.04% (v/v) ethanol, (pH 8.9). Activity was measured by the production of NADH, monitored at 340 nm. β-galactosidase was measured using Escherichia coli β-galactosidase. The assay was performed in a reaction mixture containing 100 mM sodium phosphate, 10 mM KCl, 1 mM MgSO4, 40 mM 2-mercaptoethanol and 2.5 mM o-nitrophenyl 8-D-galactoside, pH 7.0. Activity was measured at 420 nm as the production of o-nitrophenol.
The functional sizes of MDH, ADH, and β-galactosidase were previously reported as 70 kDa, 146 kDa, and 464 kDa, respectively. As described in detail elsewhere (23, 24), the relationship between increasing dose of radiation and the remaining enzyme activity is expressed by the equation: log (AD/AO) = -kD, where AD is the activity after a given dose of radiation, AO is the starting activity, D is radiation dose and k is a constant that is directly proportional to the molecular size of the enzyme. The functional sizes of enzyme standards and of Nox5 were estimated using two different methods: 1) the empirically determined equation of Beauregard and Potier (25) log Mr = 5.89 – log D37 – 0.0028*T, where D37 is the dose in MRad at which 37% of activity remains, and T is the temperature in degrees C at which the samples were irradiated (−78.5C on dry ice). 2) Slopes of log % activity vs. radiation dose were normalized to the slope of the β-galactosidase standard (26), relative slopes of standards were plotted against literature values for functional molecular weight, and a best fit line was determined using linear regression using GraphPad Prism software (GraphPad Software, San Diego, CA). The molecular weight of Nox5 was estimated by interpolation on the best fit line.
Statistical Analysis
Data are presented as ROS-generating activity, read as relative luminescence units (for luminol), relative fluorescence units (for Amplex Red), or absorbance at 550 nm (for cytochrome c reduction) and are expressed as means ± SD. Means were calculated from at least three independent transfection experiments, and each assay was performed in triplicate for each transfection. Statistical significance was determined by t test by the GraphPad Prism software.
RESULTS
Inactive mutants of Nox5 inhibit wild-type Nox5 activity
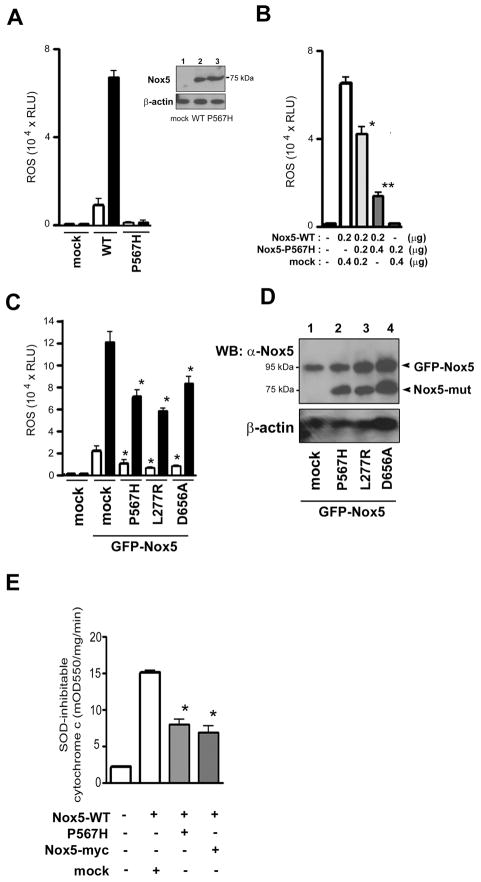
Proline-415 of Nox2, when mutated to a histidine, expresses in normal amounts but cannot bind NADPH and is therefore inactive (27–29). The homologous position in Nox5α is Proline-567, which we mutated to generate a catalytically inactive form of Nox5. Human Nox5α protein expressed in HEK293 cells generates ROS in response to the calcium ionophore ionomycin (Figure 1A, WT, filled bar). As predicted, Nox5-P567H was expressed to the same levels as wild-type Nox5, but was completely inactive, both in basal and ionomycin-stimulated activity (Figure 1A, P567H). Unexpectedly, co-expression of Nox5-P567H along with wild-type Nox5 resulted in a dose-dependent inhibition of wild-type activity (Figure 1B), thus conforming to the genetic definition of a dominant negative effect. In addition, dominant negative effects were also seen between the same isoenzyme forms using Nox1, Nox2 and Nox4 combined with their homologous NADPH-binding site mutations (T. Kawahara and D. Lambeth, unpublished data). We used the Nox5 isoenzyme to investigate the mechanism of dominant negative inhibition, because Nox5 does not require any known binding or regulatory partner proteins, thereby simplifying interpretation of data.
Figure 1. Inhibition of wild-type Nox5 by inactive mutants of Nox5.
(A) ROS generation was measured in HEK293 cells transfected with vector alone (mock), wild-type Nox5 (WT), or Nox5 (P567H). ROS production was measured by luminol-based luminescence as in Materials and Methods and expressed as relative luminescence units (RLU) per 1 × 105 cells. Cells were treated with DMSO (open bars) or 1 μM ionomycin (filled bars) luminescence was recorded. Data are means ± SD (n = 3). The inset shows an immunoblot of the protein samples using anti-Nox5 antibody or β-actin as a loading control Lane 1 = mock, Lane 2 = Nox5 (WT), Lane 3 = Nox5 (P567H). Similar results were obtained in two independent experiments. (B) ROS production from HEK293 cells co-transfected with the indicated DNA amounts of empty vector (mock), wild-type Nox5 (Nox5-WT), or Nox5 (P567H) was measured by luminol luminescence, expressed as RLU per 1 × 105 cells. Cells were activated with 1 μM ionomycin. Data are the mean ± SD (n = 4); *, P < 0.05, **, P < 0.005, compared with Nox5 WT + mock. (C) ROS generation (luminol luminescence) by co-expression of GFP-tagged wild-type Nox5 with Nox5 mutations P567H, L277R, or D656A. Equal amounts (0.2 μg each) of DNA encoding GFP-Nox5 and inactive mutations were used. Cells were treated with DMSO (open bars) or 1 μM ionomycin (filled bars). Values are means ± SD (n = 6); P < 0.05, compared with GFP-Nox5 + mock. (D) Protein expression of GFP-wild-type Nox5 and non-tagged mutant forms of Nox5 was determined with immunoblots using anti-Nox5 antibody. Protein loading was assessed with immunoblots using antibody to β-actin. Similar results were obtained in two independent experiments. (E) HEK293 cells transfected with 0.2 μg of wild-type Nox5 and 0.4 μg of either Nox5 (P567H), Nox5-myc or empty vector were permeabilized as in Materials and Methods, and used to measure SOD-inhibitable cytochrome c reduction activated by 17 μM free calcium ion. Data are the mean ± SD (n = 4); *, P < 0.05, compared with Nox5 WT + mock.
An N-terminally GFP-tagged Nox5 construct and native Nox5 previously had been shown to have equivalent activity (20). We used the larger molecular weight of the GFP-tagged Nox5 to assess protein co-expression of wild-type Nox5 versus mutant Nox5. Like its non-tagged version, GFP-tagged wild-type Nox5 showed high ionomycin-stimulated activity, which was inhibited by co-expression of Nox5-P567H (Figure 1C); the mutant Nox5 did not decrease the expression of GFP-Nox5 (Figure 1D), indicating that the activity effect was not due to suppressed protein expression. To investigate the generality of this effect, we generated additional point-mutated forms of Nox5, selecting residues that were evolutionally conserved in all EF-hand-containing Noxes from plants to vertebrates (2). These residues are V276 and L277, both located in the B-loop which joins the second and third transmembrane helix; P438, located in a region between the 6th transmembrane region and predicted FAD binding site, and D656 and K658, located in the dehydrogenase domain between the second and third predicted NADPH-binding subregions. All these point mutants were expressed at normal levels; mutants L277R and D656A were almost completely inactive (supplemental Figure S1). We coexpressed the two inactive mutants with wild-type, GFP-tagged Nox5 and assayed basal and ionomycin-stimulated activity. Like Nox5-P567H, Nox5-L277R and Nox5-D656A inhibited the ROS activity of coexpressed (GFP-tagged) WT Nox5, but did not decrease the expression of GFP-Nox5 (Figure 1, C and D).
It was unclear from initial experiments using intact cells whether the inhibition of enzyme activity itself caused the dominant negative responses, or whether they might represent indirect effects such as suppressing intracellular calcium ion fluxes or altering the localization of Nox5 at the plasma membrane. To investigate the issue, we measured NADPH-dependent superoxide generation in streptolysin O-permeabilized Nox5-expressing HEK293 cells. The pore size in permeabilized cells is large enough to permit entry of SOD, cytochrome c, and calcium ions through the cell membrane into the cytoplasm; consequently, superoxide production is measured independently of localization and intracellular calcium ion concentration can be maintained at 17 μM (20). As in intact cells (supplemental Figure S2), co-expression of Nox5-P567H suppressed wild-type Nox5 activity in permeabilized cells (Figure 1E). Similarly, a C-terminal expression tag on Nox5 abolishes both basal and calcium-stimulated Nox5 activity (20). As with the point-mutated forms of Nox5, an inactive myc-tagged Nox5 significantly inhibited the activity of GFP-tagged Nox5 in both intact (supplemental Figure S2) and permeabilized (Figure 1E) cells. Thus, these results suggest that dominant negative forms of Nox5 affect primarily the activity of the Nox5 enzyme itself and that indirect mechanisms are not responsible for suppression of activity. Among other mechanisms, dominant negative effects can result from formation of dimers or higher-order oligomers that result in an inactive overall conformation of both the active and inactive partner proteins.
Formation of a homo-oligomer of Nox5
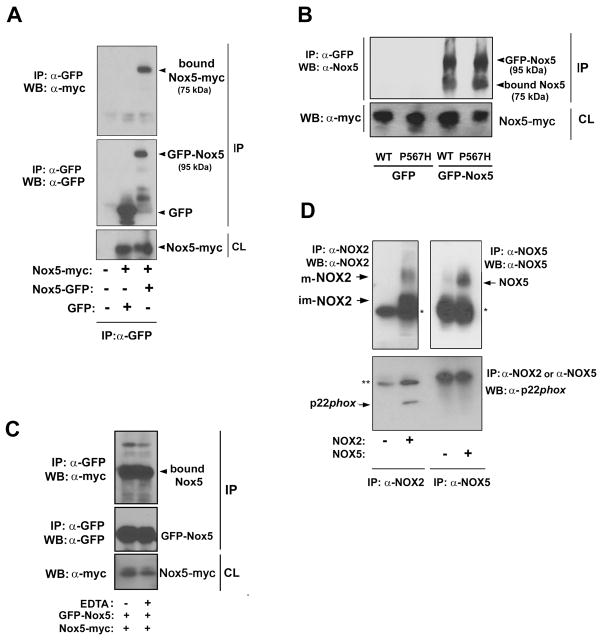
Many examples exist of membrane-associated proteins whose function requires in-membrane oligomerization. These include tetramer formation by voltage-gated potassium channels (30), transient receptor potential cation channels (31) (32), (33), and ligand-dependent dimerization of epidermal growth factor receptor (34). Based on the dominant inhibitory effects described above, we hypothesized that Nox5 requires homo-oligomerization for activity. To test this possibility, myc-tagged Nox5 and GFP-tagged Nox5 were co-expressed in HEK293 cells. Immunoprecipitation using anti-GFP antibody resulted in pull-down not only of GFP-tagged Nox5 but also myc-tagged Nox5 (Figure 2A), demonstrating association of the two proteins. Co-expression of GFP alone plus Nox5-myc followed by immunoprecipitation with anti-GFP antibody failed to pull down Nox5-myc. Similarly, un-tagged versions of both wild type Nox5 and the dominant inhibitory Nox5-P567H also co-immunoprecipitated with GFP-tagged Nox5 using anti-GFP antibody (Figure 2B). The two other untagged dominant inhibitory Nox5 mutants shown in Figure 1C (V277R and D656A) also co-immunoprecipitated with GFP-Nox5 (data not shown). Thus, Nox5 protein forms a homo-oligomer, providing a likely explanation for the inhibitory effects of inactive forms of Nox5.
Figure 2. Co-immunoprecipitation of Nox5 with itself.
(A) Equal amounts (5 μg each to 10-cm-dish) of expression vectors encoding myc-tagged Nox5 and GFP-tagged Nox5 were cotransfected in HEK293 cells. The cell lysate was immunoprecipitated with anti-GFP antibody, proteins were resolved by SDS-PAGE and were immunoblotted with an anti-myc antibody (top panel) or an anti-GFP antibody (middle panel). Nox5-myc expression in the cell lysate (CL) was demonstrated with immunoblots using anti-myc antibody (bottom panel). (B) Myc-tagged Wild-type Nox5 or the myc-tagged Nox5 (P567H) were co-expressed along with GFP-tagged Nox5 or GFP alone in HEK293 cells. The cell lysates were immunoprecipitated with anti-GFP antibody, and proteins were resolved by SDS-PAGE and immunoblotted with an anti-Nox5 antibody (upper panel). Expression of myc-Nox5 in the cell lysate (CL) was demonstrated with immunoblots using anti-myc antibody (lower panel). (C) Myc-tagged Nox5 and GFP-tagged Nox5 were co-expressed and immunoprecipitated with anti-GFP antibody in the presence or absence of 1 mM EDTA (EDTA + and −, respectively). Proteins were resolved by SDS-PAGE and immunoblotted with an anti-myc antibody (top panel) and an anti-GFP antibody (middle panel). Nox5-myc expression in the cell lysate (CL) was demonstrated with immunoblots using anti-myc antibody (bottom panel). (D) Equal amounts of cDNA encoding Nox2 or Nox5 were transfected in HEK 293 cells. Cell lysates were immunoprecipitated with anti-Nox2 antibody or anti-Nox5 antibody, proteins were resolved by SDS-PAGE and immunoblotted with anti-Nox2 antibody (top and left panel), anti-GFP antibody (top and right panel), and anti-p22phox antibody (bottom panel). * is the IgG heavy chain. ** is the IgG light chain. Abbreviations: im-Nox2, immature Nox2; m-Nox2, mature Nox2.
Because Nox5 is activated by calcium, we tested the hypothesis that calcium might regulate oligomerization, resulting in an active quaternary structure of the enzyme. Co-immunoprecipitation analyses demonstrated that the GFP-tag Nox5 co-precipitated with myc-tag Nox5 equally well in the presence and absence of 1 mM EDTA (Figure 2C). Thus, Nox5 oligomerization was not affected by calcium ion.
Absence of p22phox subunit in the Nox5 complex
Nox1, Nox2, Nox3 and Nox4 belong to the p22phox-dependent subgroup of Nox enzymes wherein the Nox catalytic subunit forms a hetero-complex with p22phox, affecting stability of the catalytic unit as well as providing a docking site for regulatory subunits (35) (36) (8). Nox5 expression and activity are independent of p22phox (8), but one study reported that overexpressed p22phox bound to Nox5 (12). To rule out that p22phox might be a partner in the Nox5 oligomer, a side-by-side experiment was carried out using Nox2 versus Nox5 to test whether each isoform could immunoprecipitate with p22phox. p22phox was readily detected during co-immunoprecipitation with Nox2 (Figure 2D). However Nox5 did not pull down any detectable p22phox protein. This is consistent with previous observations on the lack of effects of p22phox depletion on Nox5 activity, and shows that Nox5 fails to form a detectable hetero-complex with p22phox. Therefore, oligomerization of Nox5 does not involve participation of p22phox.
Role of the Nox5 DH domain in oligomer formation
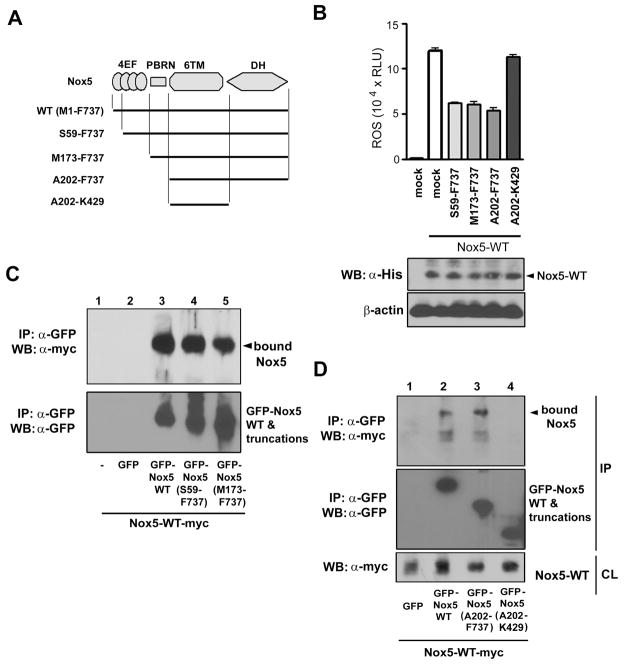
Nox5 protein can be divided into four functional domains (Figure 3A): the four EF-hand containing motifs, the N-terminal polybasic region (PBRN) that regulates subcellular localization of Nox5 through its interaction with phospholipids (20), the transmembrane, heme-containing domain composed of six α-helices, and the FAD/NADPH-binding dehydrogenase (DH) domain (37). To identify the domain that mediates Nox5 oligomerization, four N-terminally GFP-tagged deletion mutants were prepared: 1) Nox5 (S59-F737) in which only the first EF-hand motif was deleted; 2) Nox5 (M173-F737) in which the entire EF-hand domain containing all four EF-hand motifs was deleted; 3) Nox5 (A202-F737) in which the EF-hand domain and PBR-N were deleted; and 4) Nox5 (A202-K429) in which the EF-hands, the PBRN domains, and the DH domain were all deleted (i.e., only the transmembrane domain remains). All four truncated forms were inactive (both basal and ionomycin-stimulated activity) when expressed in cells (data not shown). Co-expression of the three deletion mutants containing the DH domain with wild type Nox5 resulted in inhibition of ionomycin-stimulated Nox5 activity without changing the expression of full-length wild-type Nox5 protein (Figure 3B). In contrast, removal of the DH domain eliminated the ability of Nox5 (A202-K429) to inhibit wild-type activity. These data suggests that the EF-hand and PBRN regions are not crucial for oligomerization of Nox5 but that the DH domain is a candidate for mediating oligomerization.
Figure 3. Effects of truncated forms of Nox5 on ROS generation by and binding to full-length Nox5.
(A) Diagram of truncated forms of Nox5. Abbreviations: 4EF, four EF-hand-containing calcium-binding domain; PBRN, N-terminal polybasic region; 6TM, six transmembrane region; DH, dehydrogenase domain. All truncated forms encoded GFP at their N-termini. (B) Expression vectors encoding the indicated truncated Nox5 forms (0.4 μg) were cotransfected with N-terminally hexahistidine-tagged full-length Nox5α (Nox5-WT, 0.25 μg) in HEK293 cells. ROS production (luminol luminescence) was measured as in Materials and Methods. The lower panels showed protein expression of Nox5-WT using anti-hexahistidine-tag antibody (α-His), and protein loading was assessed using antibody to β-actin. (C and D) Expression vectors encoding the myc-tagged full-length Nox5 and the indicated GFP-tagged truncations of Nox5 were cotransfected into HEK293 cells. Cell lysates were immunoprecipitated with anti-GFP antibody, and the proteins were resolved by SDS-PAGE and immunoblotted with an anti-myc antibody (top panel in C and D) and an anti-GFP antibody (bottom panel in C and middle panel of D). Nox5-myc expression was evaluated in the cell lysate (CL) with immunoblots using anti-myc antibody (bottom panel of D). The data are representative of three separate experiments.
The deletion mutants were further tested for their ability to form a complex with full-length Nox5. The GFP-tagged truncations Nox5 (S59-F737), Nox5 (M173-F737), and Nox5 (A202-F737), and Nox5 (A202-K429) were co-expressed with myc-tagged full length Nox5 and immunoprecipitated using anti-GFP antibody. All of the truncation mutants containing the DH domain formed a complex with full-length Nox5 (Figures 3C and D). In contrast, Nox5 (A202-KF737) which lacked the DH domain failed to complex with full-length Nox5 (Figure 3D). All of the truncation mutants were expressed approximately equally, and did not affect the expression of full-length Nox5 (bottom panel, Figure 3D). Thus, the DH domain must be present to form an oligomer of Nox5, and the ability of truncation mutants to function as dominant inhibitors paralleled their ability to bind to full-length Nox5.
Essential role of the DH domain in the interaction of Nox5
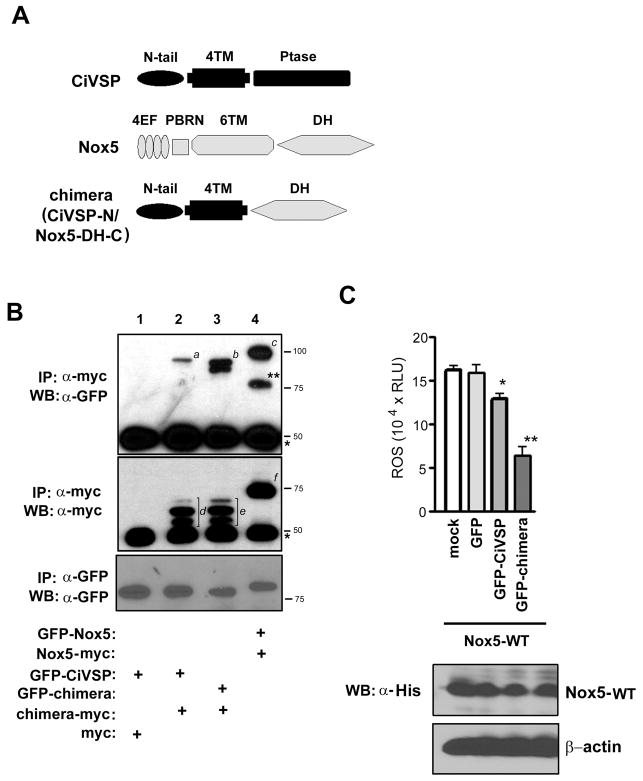
To test whether the DH domain is sufficient to form an oligomer, chimeric vectors were designed in which the Nox5 DH domain was linked at the N-terminus to the transmembrane domain of Ciona intestinalis voltage-sensor containing phosphatase (CiVSP), as shown in Figure 4A. In the chimera, the catalytic phosphatase domain of CiVSP was replaced by the DH domain of Nox5, now joined to the transmembrane region of CiVSP, which is comprised of four predicted transmembrane α-helices. The expressed CiVSP protein exists as a monomer (38), so any oligomerization must be due to the Nox5 DH domain. Two different CiVSP/Nox5 chimera were generated, one with a C-terminal myc tag, the other with a N-terminal GFP tag. Immunoprecipitation with anti-myc anitibody was then used to test whether the GFP-tagged material was co-immunoprecipitated. As shown in Figure 4B (lane 3), GFP-tagged chimera containing Nox5-DH co-precipitated with the myc-tagged chimera. The GFP-tagged full-length CiVSP (GFP-CiVSP) bound only slightly to the myc-tagged chimera (Figure 4B, lane 2). The doublet (b in Fig. 4B) or triplet (d and e) bands seen in the CiVSP-N/Nox5-DH-C chimeras can plausibly be explained by the introduction or exposure of proteolysis sites in the chimeric protein. Interaction between two full-length Nox5 proteins was shown as positive control (GFP-Nox5 and Nox5-myc, Figure 4B, lane 4). Consistent with previous reports, CiVSP tagged with GFP did not interact with CiVSP tagged with myc (data not shown). Immunoprecipitation with anti-GFP antibody was performed to show expression of GFP-tagged proteins (a bottom panel in Figure 4B, GFP-CiVSP (lanes 1 and 2), GFP-chimera (lane 3), GFP-Nox5 (lane 4). Therefore, in a membrane-anchored form that uses a heterologous non-interacting transmembrane domain, Nox5 DH domain self-associates to form an oligomer.
Figure 4. Oligomerization is mediated by the Nox5 dehydrogenase domain.
(A) Schematic diagram of full-length of CiVSP (CiVSP), full-length Nox5 (Nox5) and a chimera comprised of the transmembrane (TM) portion of CiVSP fused to the Nox5 DH domain (chimera, CiVSP-N/Nox5-DH-C). Abbreviations are: N-tail, N-terminal cytoplasmic tail region of CiVSP; 4TM, the four-helix transmembrane region of CiVSP; Ptase, the phosphatase domain of CiVSP. Other abbreviations are as in the legend to Figure 3A. (B) Equal amounts (5 μg) of the indicated vector were cotransfected into HEK293 cells. Lysates were immunoprecipitated using an anti-myc monoclonal antibody, resolved by SDS-PAGE and immunoblotted with an anti-GFP antibody (top panel) and an anti-myc antibody (middle panel). * is the IgG heavy chain. Italic letters (a–f) refer to immunostaining protein bands [a, GFP-CiVSP; b, GFP-chimera (CiVSP-N/Nox5-DH-C); c, GFP-Nox5; d and e, chimera-myc (CiVSP-N/Nox5-DH-C); f, Nox5-myc]. A protein band appeared around 77 kDa in lane 4 (**) is the degradative product of GFP-Nox5 protein. To evaluate the expression of GFP-tagged proteins in the cell lysate, GFP-fused proteins were immunoprecipitated using anti-GFP monoclonal antibody, resolved by SDS-PAGE and immunoblotted using anti-GFP antibody (bottom panel). Similar results were obtained in four independent experiments. (C) ROS production in HEK293 cells co-transfected with empty vector (mock), full-length, N-terminally hexahistidine-tagged Nox5 (Nox5-WT), and full-length, N-terminally GFP-tagged CiVSP (GFP-CiVSP) or the chimera consisting of the N-terminally GFP-tagged CiVSP TM domain linked to the Nox5 DH domain [GFP-chimera (CiVSP-N/Nox5-DH-C)] was measured using luminol luminescence. Equal expression of full-length hexahistidine-tagged Nox5 (Nox5-WT) in cell lysates (CL) was evaluated using immunoblotting with an anti-hexahistidine-tag antibody (α-His, bottom), and protein loading was assessed with immunoblots using antibody to β-actin. Data are the mean ± SD (n = 4); *, P < 0.05, **, P < 0.01, compared with Nox5 WT + GFP. Similar results were obtained in two independent experiments.
We also tested whether the CiVSP-Nox5 chimera functioned as a dominant negative by inhibiting the activity of wild type Nox5. Cells co-expressing wild type Nox5 plus the chimera showed about 50% decreased ROS generation (Figure 4C), whereas those expressing CiVSP plus wild-type Nox5 showed only about a 10–15% decrease in activity. This weak inhibition might be due to an effect of the phospholipid phosphatase activity of holo-CiVSP (39), since phosphatidiylinocytol (4,5)-biphosphate regulates Nox5 ROS generation by regulating its localizaion to the plasma membrane (20). Alternatively, there might be weak binding between GFP-CiVSP and Nox5 DH. Whatever the explanation, the effect is small compared with inhibition by the chimera containing the Nox5 DH domain. Expression of full-length Nox5 was not affected by the co-expressed full-length CiVSP or by chimeric protein (Figure 4C, lower panel). Therefore, the DH domain mediates both oligomerization of Nox5 and its ability to function as a dominant negative inhibitor of wild-type Nox5.
Estimated size of the Nox5 functional unit in intact membranes
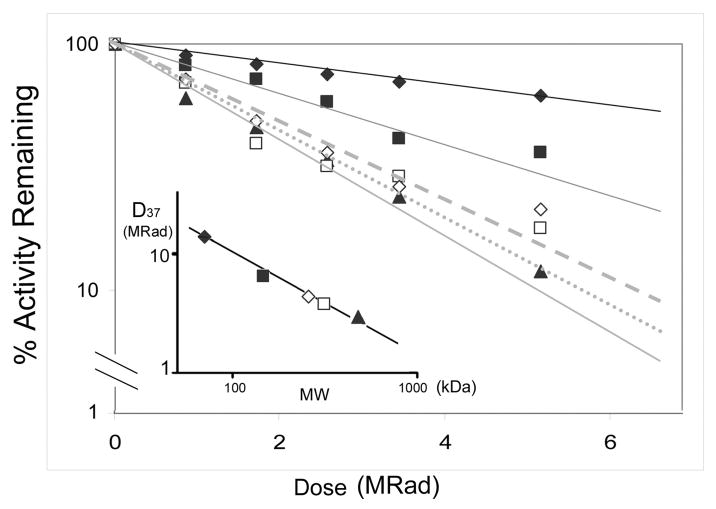
Radiation inactivation has been used to calculate the molecular size of the active enzymatic unit, which may be a monomer or a multimer, of both soluble and membrane-associated and soluble enzymes (24, 26). The theoretical basis for the method has been extensively discussed (23) (40) (41), and is based on the concept that high energy radiation will produce dose-dependent random inactivating modifications in enzyme amino acid residues in proportion to the size of the active unit. Thus, if a monomer is active within a larger oligomer, the monomer molecular size will be reported, whereas if the multimer is needed for activity, then the multimer size will be reported. The method has the advantage that enzyme size can be characterized in situ (e.g., membranes or whole cells), without the necessity to perturb the system, for example by adding detergents. Proteins whose size and oligomerization state are well known were used as controls: these are MDH (70 kDa, dimer), ADH (146 kDa, dimer), and β-galactosidase (464 kDa, tetramer). These frozen standard enzymes were exposed to radiation for various times over the course of 6 hours, and their activities were later determined as in Experimental Procedures. Rates of inactivation are shown in Figure 5, which demonstrates that larger enzymes are inactivated more rapidly than smaller ones. Figure 5, (inset) illustrates this relationship in a standard curve relating functional molecular size to radiation dose. For determination of Nox5 size, frozen, permeabilized HEK293 cells expressing Nox5α were exposed to radiation, and calcium-dependent ROS generation was measured. Figure 5 shows that Nox5 (monomer Mr = 84 kDa) was inactivated much more rapidly than MDH (70 kDa), and comparison with the standards revealed a molecular size of ~350 kDa. Separate radiation inactivation experiments and different activity assays (cytochrome c reduction and Amplex red fluorescence) to measure Nox5 activity revealed approximately 30% variability in the size estimates obtained, but all methods yielded size estimates above 300 kDa. These data are most consistent with a tetramer (n = 4.2), assuming that no other protein participates as a catalytically essential component of the complex.
Figure 5. Radiation inactivation of overexpressed and endogenously expressed Nox5.
Frozen samples of permeabilized HEK 293 cells transfected with Nox5 (open squares) or of human vascular smooth muscle cells containing endogenous Nox5 (open diamonds) were irradiated for the indicated times as described in Materials and Methods. Frozen standard proteins of known functional molecular size were irradiated at the same time: these are maltose dehydrogenase (70 kDa, filled diamonds), alcohol dehydrogenase (146 kDa, filled squares), and β-galactosidase (464 kDa, filled triangles). Enzymatic activity was assayed in the thawed samples as detailed in Materials and Methods, and is expressed as the % activity remaining after a given dose of irradiation. The inset shows a standard curve of molecular weight (log MW) versus log D37 that is the dose in MRad at which 37% of activity remains, which allow calculation of the functional molecular weight of Nox5 in HEK cells (open square) as 350 kDa, and that of Nox5 in vascular smooth muscle cells as 300 kDa, corresponding to calculated oligomer sizes of n = 4.3 and n = 3.7, respectively. The result was representative of four independent experiments.
To insure that the multimeric functional size did not result from use of an overexpression system, we used a cell system that expresses Nox5 endogenously. Primary cultured proliferating human aortic smooth muscle cells (hASMC) express both Nox5α (83.8 kDa monomer size) and Nox5β (82 kDa monomer size) as major forms along with minor amounts of Nox5γ (86 kDa) and Nox5δ (85 kDa) (10). It was previously shown that hASMC produces ROS in response to increased calcium and that the source of the calcium ion-dependent ROS generation was Nox5, based on inhibition by siRNA (10). We confirmed that hASMCs grown in the presence of PDGF produced ROS that was depenent upon calcium, and that siRNA of Nox5, but not a control siRNA, significantly suppressed expression and activity (supplemental Figures S3–S4). Streptolysin-O permeabilized hASMCs were frozen and irradiated along with standard proteins, and calcium-dependent activity was measured (Figure 5, diamonds). The inactivation rate was similar to that seen in the HEK 293 cells, clearly indicating an oligomer as the minimal functional unit, and most consistent with a tetramer. These results also reinforce conclusions based on functional analysis and co-immunoprecipitation experiments that show that active Nox5 functions as an oligomer.
Discussion
The key finding of this study is that Nox5 forms an oligomer, most likely a tetramer, and that oligomerization is needed for full activity. Furthermore, the oligomerization was mediated by the dehydrogenase domain. Oligomerization was demonstrated by the following approaches. First, we showed that inactive mutants of Nox5 inhibited calcium-activated ROS generation by wild-type Nox5. While this sort of dominant negative effect does not itself prove oligomerization, this is one of several possible explanations. Second, Nox5 immunoprecipitated with itself, demonstrated by immunoprecipitating epitope-tagged Nox5 with either wild-type Nox5 or a different epitope-tagged version of Nox5. Third, deletion of the DH domain eliminated oligomer formation and also eliminated the inhibitory effect on Nox5 activity (i.e., the ability of dominant negative forms of Nox5 to inhibit depended on the occurrence of oligomer formation). Fourth, the DH domain of Nox5 anchored to a heterologous transmembrane domain (from CiVSP) mediates oligomerization whereas the CiVSP transmembrane domain alone is monomeric. In addition, the CiVSP-linked Nox5 DH-domain inhibited wild-type Nox5 activity, providing convincing evidence that the DH domain is a key molecular determinant mediating Nox5 oligomerization. Fifth, radiation inactivation analysis in whole permeabilized cells revealed that the functional Nox5 size of both overexpressed and endogenous Nox5 was most consistent with a tetramer, and was much larger than the monomer molecular weight. Thus, Nox5 forms a functionally important oligomer within the membrane, and this oligomer is critically important for calcium-activated ROS generation by Nox5.
The calculation of a tetrameric size for the active form of Nox5 assumes that no additional proteins are present in a catalytically essential complex with Nox5. Indeed, Nox5 was selected for the present study because it is not known to require any additional subunits such as regulatory subunits or p22phox. However, in this context, two additional proteins, c-Abl and calmodulin, are reported to stimulate Nox5 activity in response to H2O2 and elevated calcium, respectively, and to form complexes with Nox5 under certain activation conditions (42) (15). In the case of c-Abl, H2O2 stimulates a calcium flux and tyrosine phosphorylation of c-Abl, and results in its translocation to a membrane compartment containing Nox5. In addition, after cells are treated with H2O2, immunoprecipitation of c-Abl results in co-immunoprecipitation of Nox5. However, both the irradiation inactivation and immunoprecipitation experiments described above were carried out without H2O2 stimulation of the cells, so significant complex formation with c-Abl would not have occurred under our conditions. Likewise, ionomycin was used only to assay activity, and calcium-free conditions were used for both co-immunoprecipitation experiments and radiation inactivation experiments, so calcium-induced complex formation with calmodulin is not expected to be an issue under our conditions. Thus, in addition to the phox-like regulatory proteins, calmodulin and c-Abl can be ruled out as members of the Nox5 complex demonstrated here.
One possible explanation for dominant inhibitory effects is that the inactive Nox5 binds to signaling proteins such as calmodulin or c-Abl, described above, or protein kinase C, which decreases the concentration needed to activate Nox5. However, in the case of ionomycin activation, high calcium (around 1 mM) fully activates Nox5, and other effectors do not cause further activation (15, 42). In the present study, we observed inhibition of Nox5 activity by co-expression of an inactive Nox5 mutant in the ionomycin-treated HEK293 cells, as well as in cells permeabilized in the presence of a maximally activating concentration of free calcium ion. These results demonstrated clearly that the functional effect of the dominant negative forms of Nox5 is not due to binding of the mutant Nox5 to either of these activator proteins.
The molecular details by which oligomerization of Nox5 occurs are not known, but comparison with known oligomeric homologs suggests some possibilities. Our data point to the dehydrogenase domain as the primary interaction site mediating oligomerization. We are not aware of homologous flavoprotein dehydrogenases that have been proven to form oligomers in solution. However, cytochrome b5 reductase from Physarium forms a dimer in the crystal structure (43). It is not yet clear whether this represents an artifact of crystal packing or a true dimer structure. Nevertheless, the structure provides a reasonable starting point for proposing specific amino acid residues that function in oligomerization. While mutagenesis studies are underway to identify such key residues, we have not yet succeeded in generating an oligomer interface mutant that exists as a monomer. It is therefore not yet possible to test whether the monomer retains some activity. Nevertheless, radiation inactivation experiments imply that the functional unit as it exists in native cells and membranes is an oligomer. In addition, several EF-hand proteins, such as S100 proteins (44), calpain (45) (46), and polcalins (47), exist as dimers and this self-interaction may facilitate regulatory coupling and cooperativity, e.g. upon calcium binding. Often, dimerization in these cases is mediated by non-canonical EF-hand regions that occur as un-paired odd-numbered motifs (45), and such regions are absent in the Nox5 calcium-binding domain. The recently reported (48) crystal structure of the EF-hand domain of a plant NADPH-oxidase, OsRbohB, showed a dimer structure, but it is not clear whether the dimer occurs naturally, or is a crystallization artifact. In the case of human Nox5, the present study shows that oligomers of Nox5 form in the absence of the EF-hand domain and that the dehydrogenase domain is critical for mediating oligomerization. We cannot rule out the possibility that in an assembled Nox5 oligomer, the calcium-binding domains might also associate in a functionally important manner.
Dimerization and higher order oligomerization can perform a variety of functions. First, such oligomerization may result in stabilization of the protein, extending its lifetime in the cell. The heterodimer between Nox2 and p22phox, for example, results in stabilization of both proteins so that in the absence of either protein, its partner is degraded more rapidly (49). In addition, oligomerization frequently has regulatory consequences, such as cooperativity seen for oxygen binding to hemoglobin, and substrate binding to many regulated enzymes. We are not aware of any published data that suggest cooperative behavior for regulators or substrates for Nox5, but this possibility cannot be ruled out, as reported experiments were not designed to reveal such kinetic behavior. Finally, inactive forms of Nox may occur naturally and may be involved in adjusting the activity of the active forms of Nox5. Five splice variants of human Nox5 ---- Nox5α, β, γ, δ and Nox5S ---- have been described. Nox5α, β, γ, and δ all include a calcium-binding domain containing all four EF-hand motifs, but with insertions or extensions of different length; other regions including PBRN, transmembrane domain, and DH domain are present in these variants. Nox5S, however, is a variant in which the calcium-binding domain is absent; this form is identical to the truncation mutant M173-F737 in which the calcium-binding domain has been deleted. Nox5S is expressed in human Barrett’s adenocarcinoma cell lines where it promotes cell proliferation (50, 51), and in human microvascular endothelial cells (HMEC) where it participates in thrombin signaling (12). Nox5S appears to produce ROS under some circumstances by as-yet unknown activation mechanisms, but because it lacks the calcium-binding domain, cannot respond directly to calcium. The present study suggests that Nox5S can also function as an endogenous suppressor of calcium-activated forms of Nox5 (i.e., those that retain a calcium-binding domain), since Nox5(M173-F737) bound to Nox5α and inhibited its ionomycin-induced ROS generation (Figure 3). HMEC cells expressed NOX5β (an active long form) as well as significant amount of Nox5S, suggesting that the activity of NOX5β in HMEC cells might be regulated by the protein level of Nox5S.
In addition, oligomerization could in theory function to concentrate and elevate local concentrations of reactive oxygen within a small area, increasing its effectiveness as a signal molecule, for example in the regulation of enzymes such as protein tyrosine phosphatases, transcription factors, and ion channels, as has been well documented previously (1, 52). This might be especially effective if the Nox enzyme is co-localized with its target protein in an organelle or membrane subregion. In this context, in vascular smooth muscle, Nox1 has been reported in caveolae, while Nox4 is localized to focal adhesions (53).
It will be important to address in future studies whether other isoforms of Nox also form oligomers, and if so, what are the functional consequences? Inactive mutated forms of Nox1, Nox2 and Nox4 also function as dominant negative inhibitors (Kawahara, unpublished data), but in these cases they might act by competing for p22phox and/or regulatory subunits. Because of this, it is not straightforward to distinguish this mechanism from one involving homo-oligomerization. Interestingly, cytochrome b558 (the complex formed from Nox2 plus p22phox), when detergent extracted and purified from human neutrophil membrane, was heterogeneous in size by sedimentation equilibrium analysis, but the smallest detectable species showed a molecular mass of ~350 kDa (54). If one assumes minimal binding by phospholipids or other bound proteins, then this size corresponds to an oligomer of n = 3–4 (the exact number depending on the contribution of glycosylation to the overall size of gp91phox). While it is not clear in this report whether the observed oligomerization occurs naturally or was induced upon detergent extraction of the protein from its native environment, the possibility should be entertained that oligomerization may be a general property of the Nox family of enzymes, and may impact enzymatic function and ROS signaling by the several mechanisms as discussed above.
Supplementary Material
Acknowledgments
We are grateful to Dr. Yasushi Okamura for the CiVSP cDNA clone.
ABBREVIATIONS
- DH
dehydrogenase
- CiVSP
Ciona intestinalis voltage sensor-containing phosphatase
- ROS
reactive oxygen species
- Nox
NADPH-oxidases
- SOD
superoxide dismutase
- TM
transmembrane
- EDTA
ethylenediaminetetraacetic acid
- HEK
Human embryonic kidney
- HASMC
human aortic smooth muscle cells
- MDH
maltose dehydrogenase
- ADH
alcohol dehydorogenase
- HRP
horse radish peroxidase
- HMEC
human microvascular endothelial cells
Footnotes
This work was supported by NIH R01 grants CA084138 and CA105116 and by EHS program project grant ES011163.
SUPPORTING INFORMATION AVAILABLE.
Supporting figures are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 2.Kawahara BT, Quinn MT, Lambeth JD. Molecular evolution of the reactive oxygen-generating NADPH oxidase (Nox/Duox) family of enzymes. BMC Evol Biol. 2007;7:109. doi: 10.1186/1471-2148-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambeth JD, Kawahara T, Diebold B. Regulation of Nox and Duox enzymatic activity and expression. Free radical biology & medicine. 2007;43:319–331. doi: 10.1016/j.freeradbiomed.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sumimoto H. Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. Febs J. 2008;275:3249–3277. doi: 10.1111/j.1742-4658.2008.06488.x. [DOI] [PubMed] [Google Scholar]
- 5.Kawahara T, Lambeth JD. Molecular evolution of Phox-related regulatory subunits for NADPH oxidase enzymes. BMC Evol Biol. 2007;7:178. doi: 10.1186/1471-2148-7-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grasberger H, Refetoff S. Identification of the maturation factor for dual oxidase. Evolution of an eukaryotic operon equivalent. The Journal of biological chemistry. 2006;281:18269–18272. doi: 10.1074/jbc.C600095200. [DOI] [PubMed] [Google Scholar]
- 7.Luxen S, Noack D, Frausto M, Davanture S, Torbett BE, Knaus UG. Heterodimerization controls localization of Duox-DuoxA NADPH oxidases in airway cells. J Cell Sci. 2009;122:1238–1247. doi: 10.1242/jcs.044123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawahara T, Ritsick D, Cheng G, Lambeth JD. Point mutations in the proline-rich region of p22phox are dominant inhibitors of Nox1- and Nox2-dependent reactive oxygen generation. The Journal of biological chemistry. 2005;280:31859–31869. doi: 10.1074/jbc.M501882200. [DOI] [PubMed] [Google Scholar]
- 9.Banfi B, Tirone F, Durussel I, Knisz J, Moskwa P, Molnar GZ, Krause KH, Cox JA. Mechanism of Ca2+ activation of the NADPH oxidase 5 (NOX5) The Journal of biological chemistry. 2004;279:18583–18591. doi: 10.1074/jbc.M310268200. [DOI] [PubMed] [Google Scholar]
- 10.Jay DB, Papaharalambus CA, Seidel-Rogol B, Dikalova AE, Lassegue B, Griendling KK. Nox5 mediates PDGF-induced proliferation in human aortic smooth muscle cells. Free radical biology & medicine. 2008;45:329–335. doi: 10.1016/j.freeradbiomed.2008.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brar SS, Corbin Z, Kennedy TP, Hemendinger R, Thornton L, Bommarius B, Arnold RS, Whorton AR, Sturrock AB, Huecksteadt TP, Quinn MT, Krenitsky K, Ardie KG, Lambeth JD, Hoidal JR. NOX5 NAD(P)H oxidase regulates growth and apoptosis in DU 145 prostate cancer cells. Am J Physiol Cell Physiol. 2003;285:C353–369. doi: 10.1152/ajpcell.00525.2002. [DOI] [PubMed] [Google Scholar]
- 12.BelAiba RS, Djordjevic T, Petry A, Diemer K, Bonello S, Banfi B, Hess J, Pogrebniak A, Bickel C, Gorlach A. NOX5 variants are functionally active in endothelial cells. Free radical biology & medicine. 2007;42:446–459. doi: 10.1016/j.freeradbiomed.2006.10.054. [DOI] [PubMed] [Google Scholar]
- 13.Montezano AC, Burger D, Paravicini TM, Chignalia AZ, Yusuf H, Almasri M, He Y, Callera GE, He G, Krause KH, Lambeth D, Quinn MT, Touyz RM. Nicotinamide adenine dinucleotide phosphate reduced oxidase 5 (Nox5) regulation by angiotensin II and endothelin-1 is mediated via calcium/calmodulin-dependent, rac-1-independent pathways in human endothelial cells. Circ Res. 2010;106:1363–1373. doi: 10.1161/CIRCRESAHA.109.216036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagnandan D, Church JE, Banfi B, Stuehr DJ, Marrero MB, Fulton DJ. Novel mechanism of activation of NADPH oxidase 5(NOX5): Calcium-sensitization via phosphorylation. The Journal of biological chemistry. 2006 doi: 10.1074/jbc.M608966200. [DOI] [PubMed] [Google Scholar]
- 15.Tirone F, Cox JA. NADPH oxidase 5 (NOX5) interacts with and is regulated by calmodulin. FEBS Lett. 2007;581:1202–1208. doi: 10.1016/j.febslet.2007.02.047. [DOI] [PubMed] [Google Scholar]
- 16.Ritsick DR, Edens WA, Finnerty V, Lambeth JD. Nox regulation of smooth muscle contraction. Free radical biology & medicine. 2007;43:31–38. doi: 10.1016/j.freeradbiomed.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng G, Cao Z, Xu X, van Meir EG, Lambeth JD. Homologs of gp91phox: cloning and tissue expression of Nox3, Nox4, and Nox5. Gene. 2001;269:131–140. doi: 10.1016/s0378-1119(01)00449-8. [DOI] [PubMed] [Google Scholar]
- 18.Kamiguti AS, Serrander L, Lin K, Harris RJ, Cawley JC, Allsup DJ, Slupsky JR, Krause KH, Zuzel M. Expression and activity of NOX5 in the circulating malignant B cells of hairy cell leukemia. J Immunol. 2005;175:8424–8430. doi: 10.4049/jimmunol.175.12.8424. [DOI] [PubMed] [Google Scholar]
- 19.Guzik TJ, Chen W, Gongora MC, Guzik B, Lob HE, Mangalat D, Hoch N, Dikalov S, Rudzinski P, Kapelak B, Sadowski J, Harrison DG. Calcium-dependent NOX5 nicotinamide adenine dinucleotide phosphate oxidase contributes to vascular oxidative stress in human coronary artery disease. J Am Coll Cardiol. 2008;52:1803–1809. doi: 10.1016/j.jacc.2008.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawahara T, Lambeth JD. Phosphatidylinositol (4,5)-bisphosphate Modulates Nox5 Localization via an N-Terminal Polybasic Region. Mol Biol Cell. 2008;19:4020–4031. doi: 10.1091/mbc.E07-12-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serrander L, Jaquet V, Bedard K, Plastre O, Hartley O, Arnaudeau S, Demaurex N, Schlegel W, Krause KH. NOX5 is expressed at the plasma membrane and generates superoxide in response to protein kinase C activation. Biochimie. 2007;89:1159–1167. doi: 10.1016/j.biochi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Nugent JH. Molecular-size standards for use in radiation-inactivation studies on proteins. The Biochemical journal. 1986;239:459–462. doi: 10.1042/bj2390459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verkman AS, Skorecki K, Ausiello DA. Radiation inactivation of oligomeric enzyme systems: theoretical considerations. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:150–154. doi: 10.1073/pnas.81.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beauregard G, Potier M. Radiation inactivation of membrane proteins: molecular weight estimates in situ and after Triton X-100 solubilization. Analytical biochemistry. 1984;140:403–408. doi: 10.1016/0003-2697(84)90185-4. [DOI] [PubMed] [Google Scholar]
- 25.Beauregard G, Potier M. Temperature dependence of the radiation inactivation of proteins. Analytical biochemistry. 1985;150:117–120. doi: 10.1016/0003-2697(85)90448-8. [DOI] [PubMed] [Google Scholar]
- 26.Pan RS, Chien LF, Wang MY, Tsai MY, Pan RL, Hsu BD. Functional size of photosynthetic electron transport chain determined by radiation inactivation. Plant physiology. 1987;85:158–163. doi: 10.1104/pp.85.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dinauer MC, Curnutte JT, Rosen H, Orkin SH. A missense mutation in the neutrophil cytochrome b heavy chain in cytochrome-positive X-linked chronic granulomatous disease. J Clin Invest. 1989;84:2012–2016. doi: 10.1172/JCI114393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roos D, Boer M, Kuribayashi F, Meischl C, Weening R, Segal A, Ahlin A, Nemet K, Hossle J, Bernatowska-Matuszkiewicz E, Middleton-Price H. Mutations in the X-linked and autosomal recessive forms of chronic Granulomatous disease. Blood. 1996;87:1663–1681. [PubMed] [Google Scholar]
- 29.Segal AW. The NADPH oxidase and chronic granulomatous disease. Molecular Medicine Today. 1996;2:129–135. doi: 10.1016/1357-4310(96)88723-5. [DOI] [PubMed] [Google Scholar]
- 30.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 31.Mio K, Ogura T, Hara Y, Mori Y, Sato C. The non-selective cationpermeable channel TRPC3 is a tetrahedron with a cap on the large cytoplasmic end. Biochem Biophys Res Commun. 2005;333:768–777. doi: 10.1016/j.bbrc.2005.05.181. [DOI] [PubMed] [Google Scholar]
- 32.Stewart AP, Egressy K, Lim A, Edwardson JM. AFM imaging reveals the tetrameric structure of the TRPM8 channel. Biochem Biophys Res Commun. 2010;394:383–386. doi: 10.1016/j.bbrc.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 33.Barrera NP, Shaifta Y, McFadzean I, Ward JP, Henderson RM, Edwardson JM. AFM imaging reveals the tetrameric structure of the TRPC1 channel. Biochem Biophys Res Commun. 2007;358:1086–1090. doi: 10.1016/j.bbrc.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 34.Hurwitz DR, Emanuel SL, Nathan MH, Sarver N, Ullrich A, Felder S, Lax I, Schlessinger J. EGF induces increased ligand binding affinity and dimerization of soluble epidermal growth factor (EGF) receptor extracellular domain. The Journal of biological chemistry. 1991;266:22035–22043. [PubMed] [Google Scholar]
- 35.Sumimoto H, Hata K, Mizuki K, Ito T, Kage Y, Sakaki Y, Fukumaki Y, Nakamura M, Takeshige K. Assembly and activation of the phagocyte NADPH oxidase: Specific Interaction of the N-terminal Src homology 3 domain of p47phox with p22phox is required for activation of the NADPH oxidase complex. Journal of Biological Chemistry. 1996;36:22152–22158. doi: 10.1074/jbc.271.36.22152. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y, Marchal CC, Casbon AJ, Stull N, von Lohneysen K, Knaus UG, Jesaitis AJ, McCormick S, Nauseef WM, Dinauer MC. Deletion mutagenesis of p22phox subunit of flavocytochrome b558: identification of regions critical for gp91phox maturation and NADPH oxidase activity. The Journal of biological chemistry. 2006;281:30336–30346. doi: 10.1074/jbc.M607191200. [DOI] [PubMed] [Google Scholar]
- 37.Vignais PV. The superoxide-generating NADPH oxidase: structural aspects and activation mechanism. Cell Mol Life Sci. 2002;59:1428–1459. doi: 10.1007/s00018-002-8520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohout SC, Ulbrich MH, Bell SC, Isacoff EY. Subunit organization and functional transitions in Ci-VSP. Nat Struct Mol Biol. 2008;15:106–108. doi: 10.1038/nsmb1320. [DOI] [PubMed] [Google Scholar]
- 39.Iwasaki H, Murata Y, Kim Y, Hossain MI, Worby CA, Dixon JE, McCormack T, Sasaki T, Okamura Y. A voltage-sensing phosphatase, Ci-VSP, which shares sequence identity with PTEN, dephosphorylates phosphatidylinositol 4,5-bisphosphate. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7970–7975. doi: 10.1073/pnas.0803936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kempner ES, Schlegel W. Size determination of enzymes by radiation inactivation. Analytical biochemistry. 1979;92:2–10. doi: 10.1016/0003-2697(79)90617-1. [DOI] [PubMed] [Google Scholar]
- 41.Kepner GR, Macey RI. Membrane enzyme systems. Molecular size determinations by radiation inactivation. Biochim Biophys Acta. 1968;163:188–203. doi: 10.1016/0005-2736(68)90097-7. [DOI] [PubMed] [Google Scholar]
- 42.El Jamali A, Valente AJ, Lechleiter JD, Gamez MJ, Pearson DW, Nauseef WM, Clark RA. Novel redox-dependent regulation of NOX5 by the tyrosine kinase c-Abl. Free radical biology & medicine. 2008;44:868–881. doi: 10.1016/j.freeradbiomed.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim S, Suga M, Ogasahara K, Ikegami T, Minami Y, Yubisui T, Tsukihara T. Structure of Physarum polycephalum cytochrome b5 reductase at 1.56 A resolution. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:274–279. doi: 10.1107/S1744309107010731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Potts BC, Smith J, Akke M, Macke TJ, Okazaki K, Hidaka H, Case DA, Chazin WJ. The structure of calcyclin reveals a novel homodimeric fold for S100 Ca(2+)-binding proteins. Nat Struct Biol. 1995;2:790–796. doi: 10.1038/nsb0995-790. [DOI] [PubMed] [Google Scholar]
- 45.Blanchard H, Grochulski P, Li Y, Arthur JS, Davies PL, Elce JS, Cygler M. Structure of a calpain Ca(2+)-binding domain reveals a novel EF-hand and Ca(2+)- induced conformational changes. Nat Struct Biol. 1997;4:532–538. doi: 10.1038/nsb0797-532. [DOI] [PubMed] [Google Scholar]
- 46.Lin GD, Chattopadhyay D, Maki M, Wang KK, Carson M, Jin L, Yuen PW, Takano E, Hatanaka M, DeLucas LJ, Narayana SV. Crystal structure of calcium bound domain VI of calpain at 1.9 A resolution and its role in enzyme assembly, regulation, and inhibitor binding. Nat Struct Biol. 1997;4:539–547. doi: 10.1038/nsb0797-539. [DOI] [PubMed] [Google Scholar]
- 47.Verdino P, Westritschnig K, Valenta R, Keller W. The cross-reactive calcium-binding pollen allergen, Phl p 7, reveals a novel dimer assembly. EMBO J. 2002;21:5007–5016. doi: 10.1093/emboj/cdf526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oda T, Hashimoto H, Kuwabara N, Akashi S, Hayashi K, Kojima C, Wong HL, Kawasaki T, Shimamoto K, Sato M, Shimizu T. Structure of the N-terminal regulatory domain of a plant NADPH oxidase and its functional implications. The Journal of biological chemistry. 2010;285:1435–1445. doi: 10.1074/jbc.M109.058909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu L, Zhen L, Dinauer MC. Biosynthesis of the phagocyte NADPH oxidase cytochrome b558. Role of heme incorporation and heterodimer formation in maturation and stability of gp91phox and p22phox subunits. The Journal of biological chemistry. 1997;272:27288–27294. doi: 10.1074/jbc.272.43.27288. [DOI] [PubMed] [Google Scholar]
- 50.Fu X, Beer DG, Behar J, Wands J, Lambeth D, Cao W. cAMP-response element-binding protein mediates acid-induced NADPH oxidase NOX5-S expression in Barrett esophageal adenocarcinoma cells. The Journal of biological chemistry. 2006;281:20368–20382. doi: 10.1074/jbc.M603353200. [DOI] [PubMed] [Google Scholar]
- 51.Si J, Fu X, Behar J, Wands J, Beer DG, Souza RF, Spechler SJ, Lambeth D, Cao W. NADPH oxidase NOX5-S mediates acid-induced cyclooxygenase-2 expression via activation of NF-kappaB in Barrett’s esophageal adenocarcinoma cells. The Journal of biological chemistry. 2007;282:16244–16255. doi: 10.1074/jbc.M700297200. [DOI] [PubMed] [Google Scholar]
- 52.Burdon R. Superoxide and hydrogen peroxide in relation to mammalian cell proliferation. Free Radical Biology and Medicine. 1995;18:775–794. doi: 10.1016/0891-5849(94)00198-s. [DOI] [PubMed] [Google Scholar]
- 53.Hilenski LL, Clempus RE, Quinn MT, Lambeth JD, Griendling KK. Distinct subcellular localizations of Nox1 and Nox4 in vascular smooth muscle cells. Arteriosclerosis, thrombosis, and vascular biology. 2004;24:677–683. doi: 10.1161/01.ATV.0000112024.13727.2c. [DOI] [PubMed] [Google Scholar]
- 54.Nugent JH, Gratzer W, Segal AW. Identification of the haem-binding subunit of cytochrome b-245. The Biochemical journal. 1989;264:921–924. doi: 10.1042/bj2640921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.