Abstract
Hydroxy-3-methyl-glutaryl-CoA reductase (HMGR) is the rate-controlling enzyme of cholesterol synthesis, and owing to its biological and pharmacological relevance, researchers have investigated several compounds capable of modulating its activity with the hope of developing new hypocholesterolemic drugs. In particular, polyphenol-rich extracts were extensively tested for their cholesterol-lowering effect as alternatives, or adjuvants, to the conventional statin therapies, but a full understanding of the mechanism of their action has yet to be reached. Our work reports on a detailed kinetic and equilibrium study on the modulation of HMGR by the most-abundant catechin in green tea, epigallocatechin-3-gallate (EGCG). Using a concerted approach involving spectrophotometric, optical biosensor, and chromatographic analyses, molecular docking, and site-directed mutagenesis on the cofactor site of HMGR, we have demonstrated that EGCG potently inhibits the in vitro activity of HMGR (Ki in the nanomolar range) by competitively binding to the cofactor site of the reductase. Finally, we evaluated the effect of combined EGCG-statin administration.
Keywords: cholesterol, reduced nicotinamide adenine dinucleotide phosphate, epigallocatechin-3-gallate, inhibition
Atherosclerotic cardiovascular diseases are frequent in individuals with abnormally high levels of blood cholesterol (1–3). Early stages of cholesterol biosynthesis in humans are rate-regulated by 3-hydroxy-3-methyl-glutaryl-CoA reductase (HMGR) (4, 5), a two-domain endoplasmic reticulum-bound enzyme involved in the mevalonate pathway (6). Because of this central role, HMGR is the target of several hypocholesterolemic drugs, of which statins are the most extensively studied (7–9) and among the most widely prescribed drugs worldwide, despite some recurrent side-effects (10–14). Recently, in efforts to identify nonconventional treatments for hypercholesterolemia, such as novel drugs based on a natural product, tea catechins (mainly gallate ester derivatives) have been tested successfully both in vitro and in vivo as cholesterol-lowering agents (15–25). Globally, these findings indicate that catechins exert concerted, multiple-level action involving the upregulation of the LDL receptor (15, 24), the reduction of cholesterol absorption (16, 26, 27), and the modulation of both synthetic (15, 28, 29) and metabolic pathways (24). Owing to the complexity of action and the short half-life of HMGR (30), the direct effect on HMGR activity is very difficult to recognize and isolate, and the literature supporting a physiologically significant inhibition of HMGR by catechins is limited (22). In this work, in order to clarify the direct interplay between epigallocatechin-3-gallate (EGCG) and HMGR, supported by experimental evidence on analogous enzymatic systems that point to the presence of a high-affinity binding site for EGCG on reduced nicotinamide adenine dinucleotide phosphate (NADPH) (31–37), we sought to identify site-directed binding of EGCG to HMGR by discussing the results of a predictive structural bioinformatic approach. In particular, on the basis of the information derived from computational analysis, we performed both enzymatic and binding studies, the results of which show collectively and unequivocally that EGCG strongly inhibits HMGR activity, and in all likelihood has a profound effect on the synthetic pathway of cholesterol.
MATERIALS AND METHODS
Materials
The plasmid expressing the catalytic subunit of human HMGR was kindly provided by Professor J. Deisenhofer (Howard Hughes Institute and Department of Biochemistry, University of Texas Southwestern Medical Center). Samples of liver biopsies from healthy patients were kindly supplied by Dr. L. Trozzi and Professor A. Benedetti (Department of Gastroenterology, Polytechnic University of Marche; Ancona, Italy). Carboxymethyl-dextran (CMD) cuvettes, 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC), N-hydroxysuccinimide (NHS), and ethanolamine were obtained from Farfield Group (Cheshire, UK), whereas Na2HPO4, CH3COONa, KCl, NaCl, Tween-20, EDTA, DTT, DMSO, EGCG, HMG-CoA, NADPH, nicotinamide adenine dinucleotide phosphate (NADP+), glucose-6-phosphate dehydrogenase, glucose-6-phosphate, GSH, glutathione-S transferase affinity columns, Tris, sucrose, phenylmethylsulfonyl fluoride, tosyl phenylalanyl chloromethyl ketone, pravastatin, and the primers for mutagenesis studies were all obtained from Sigma-Aldrich (Milan, Italy). All chemicals were of the highest grade available. The Cary 1E UV-vis spectrophotometer was obtained from Varian (Palo Alto, CA). The IAsys plus biosensor came from Thermo Fisher Scientific (Milan, Italy). The HPLC system Gold equipped with a UV-vis detector, and HPLC column heater were obtained from Beckman Coulter S.p.A. (Milan, Italy). The Luna C18 column (5 μm particle size, 250 × 4.6 mm, equipped with a 5 mm guard column) was purchased from Phenomenex Italia (Bologna, Italy).
Induction and expression of recombinant HMGR
Expression and purification of the human HMGR catalytic subunit were carried out as described previously (38). Briefly, the pGEX-cs plasmid was transformed into BL21 Escherichia coli strains containing the pLysC plasmid according to a standard protocol (39). Bacteria were grown at 37°C in 100 ml Luria-Bertani medium containing ampicillin at 60 µg ml−1 and chloramphenicol at 30 µg ml−1 to the A600 of 0.6. Overproduction of recombinant protein was induced by adding isopropyl β-d-thiogalactopyranoside (IPTG) to the final concentration of 0.4 mM. Next, cell growth was prolonged for an additional 3 h essentially as described by Parks et al. (40). A control culture was grown under the same conditions in IPTG-free medium. Fusion protein was purified by GST-glutathione affinity chromatography at 4°C, and its purity was finally checked by SDS-PAGE.
Preparation of liver microsomes
Human liver microsomes were prepared as previously reported (41). Liver tissue samples (0.5 g) were added to 4.5 ml of cold homogenization buffer (50 mM Tris-HCl buffer, 0.3 M sucrose, 10 mM EDTA, 10 mM DTT, and 50 mM NaCl at pH 7.4, in the presence of 1 mM PMSF and 1 mM TPCK) and homogenized using a bench-top Ultra-Turrax TP 18/10 homogenizer (Janke and Kunkel; Staufen, Germany). The homogenate was centrifuged at 20,000 g for 15 min at 4°C. The supernatant was collected and centrifuged at 100,000 g for 60 min at 4°C. The microsomal pellet was finally resuspended in the activity buffer. Total protein concentration was determined according to the method of Lowry et al. (42).
Bioinformatic analysis
To identify the most probable binding site for EGCG on HMGR, preliminary molecular docking analyses were performed on a Pentium4/Linux Red Hat-based platform using Autodock 4.0 and InsightII (release 2005) software. The X-ray crystal structure of human HMGR [pdb entry: 3CCT (43)] was retrieved from the Protein Data Bank (44). Hydrogen atoms were added to the protein prior to any analysis. Autodock, a software performing a Lamarckian genetic algorithm to explore the binding possibilities of a ligand in a binding pocket (45), was used with a grid of 48, 48, and 48 points (in the x, y, and z directions) around the HMG-CoA binding site, and with a grid of 54, 50, and 52 points around the NADPH binding site, with a grid spacing of 0.375 Å, a root-mean-square (rms) tolerance of 0.8 Å, and a maximum of 2,500,000 energy evaluations. Other parameters were set to default values (46). Next, InsightII was used to refine the ligand/receptor models through an energy minimization procedure, using the Discover module of the software with a consistent valence force field and a conjugate gradients algorithm to an rms derivative of 0.001 kcal/mol. Finally, ligand-receptor binding affinities were calculated using the experimentally validated (47) scoring functions of the Ludi algorithm, which takes into account five contributions: total number of hydrogen bonds, perturbed ionic interactions, lyophilic interactions, frozen internal degrees of freedom of the ligand, and loss of translational and rotational entropy of the ligand. Binding affinities were expressed throughout as predicted equilibrium dissociation constants (KD,p) derived from the Ludi Score, according to the standard relation (equation 1):
| (Eq. 1) |
and Energy Estimate 3 as scoring function (48). The output from InsightII, all modeling studies, and images were rendered with PyMOL (Python Molecular Graphics - 2006; DeLano Scientific LLC, San Carlos, CA). PyMOL was also used to calculate the length of theoretical hydrogen bonds, measured between the hydrogen and its putative binding partner.
Assay of HMG-CoA reductase activity
HMGR catalyses the four-electron reduction of HMG-CoA to CoA and mevalonate. The spectrophotometric assay of HMGR activity was derived from previously published methods (7), performing enzyme rate measurements with respect to both HMG-CoA and NADPH. Briefly, HMGR (4 nM) was incubated at 37°C with increasing concentrations of EGCG in 100 mM phosphate buffer containing 1 mM EDTA, 10 mM DTT, and 2% DMSO at pH 6.8 (activity buffer).
HMGR enzymatic activity was continuously monitored after addition of both free and EGCG-preincubated enzyme to assay mixtures containing NADPH and HMG-CoA. To clarify the mechanism of the interaction, the apparent dissociation constant (Kd,app) of the preformed HMGR-EGCG complex on the NADPH binding site was determined by measuring the decrease in catalytic activity upon addition of increasing levels of EGCG at different cofactor concentrations in the range 30–270 μM.
After 20 min preincubation (more-prolonged preincubation periods did not affect HMGR activity further; Fig. 3F), residual activities were measured at 340 nm by continuously monitoring the disappearance of NADPH after the initiation of the reaction. The residual activity (ai) was expressed as the ratio of the initial velocities of the product formation in the presence (v0,i) and in the absence (v0) of a given EGCG concentration [Ii] (equation 2):
| (Eq. 2) |
The experimental dataset was constituted by a set of residual activities (ai) measured at increasing EGCG concentrations [Ii]. Kd,app values were calculated for each cofactor concentration using the standard equation (49) (equation 3):
| (Eq. 3) |
where [Et] was the added enzyme concentration. Kd,app is related to Kd according to the following equation (equation 4):
| (Eq. 4) |
For each cofactor concentration (m = 4), the residual activity was measured at increasing EGCG concentrations (n = 10), and the resulting n×2×m matrix was globally analyzed by Marquardt-Levenberg nonlinear least squares fitting procedure (50) using equations 3 and 4 as fitting functions.
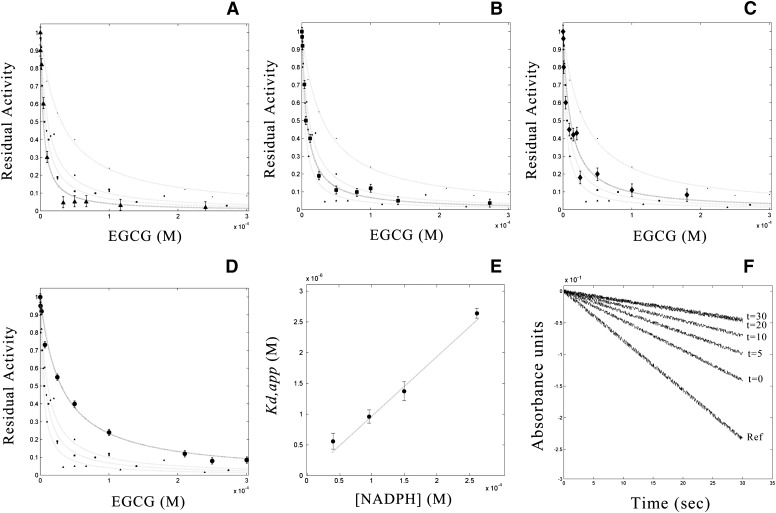
Fig.3.
Inhibition of human HMG-CoA activity by EGCG. HMG-CoA reductase residual activities versus tea catechin concentration measured at four different NADPH concentrations: the curves and related standard deviations are global fits of the data to equations 3 and 4 and yield the derivation of the apparent equilibrium dissociation constant (Kd,app) at different cofactor concentrations. Moreover, Marquardt-Levenberg nonlinear least squares fitting procedure is used to obtain the equilibrium dissociation constant Kd of the complex HMG-CoA/EGCG. ▴: 30 µM NADPH(A); ■: 90 µM NADPH(B); ¨: 150 µM NADPH(C); •: 270 µM NADPH(D). E: Linear fit to equation 4 of Kd,app values calculated for the EGCG-HMGR interaction versus cofactor concentrations. F: Time courses for the inhibition of HMGR activity by EGCG at different preincubation periods.
Chromatographic assay of HMGR activity
HMGR residual activity assays were performed upon 20 min preincubation of the enzyme (0.4 µM) with increasing levels of EGCG (0–6.54 mM). The preformed EGCG-HMGR complex was added to 1.55 µM HMG-CoA and 2.68 mM NADPH dissolved in the activity buffer, and stored for 60 min at 37°C. The resulting mixture (10 µl) was separated with a Phenomenex Luna C18 reverse-phase (RP)-HPLC column thermostatted at 26 ± 0.1°C (51). Data were analyzed according to equation 3.
Biosensor binding studies
The CMD surface was rinsed and equilibrated at 37°C with PBS-T [10 mM Na2HPO4, 2.7 mM KCl, 138 mM NaCl, 0.05% (v/v) Tween-20, pH 7.4] in the vibrostirred sensing chamber. Prior to activation, the surface was washed with detergent-free PBS, pH 7.4, to avoid a possible ‘mask’ effect of carboxyl groups. Next, the CMD surface was standardly activated with an equimolar EDC/NHS mixture (52). Then, HMG-CoA reductase (0.2 mg/ml) dissolved in the immobilization buffer [10 mM CH3COONa, pH 5.5, chosen on the basis of the isoelectric point (pI) of the enzyme (pI = 6.8)] was added and incubated for 10 min. Nonspecifically-bound ligand was removed by PBS wash, whereas nonreacted carboxylic groups were deactivated with 1 M ethanolamine, pH 8.5. Finally, EGCG was added at increasing concentrations. Raw data were analyzed with Fast Fit software (Fison Applied Sensor Technology; Affinity Sensors); the software makes it possible to use both mono-exponential and bi-exponential models (53) to fit experimental data. Kinetic raw data were globally fitted (54, 55) using a standard monophasic time course equation (equation 5):
| (Eq. 5) |
where kass and kdiss are the association and dissociation rate constants, respectively, Rt is the response at time t, R0 is the initial response, and Req is the maximal response at equilibrium for a given EGCG concentration (equation 6):
| (Eq. 6) |
and Rmax is the extent of binding at asymptotically high concentrations of [L].
Production of G807D mutant
The G at position 2470 on the sequence of HMGR (56) was replaced with an A to change the amino acid residue from Gly to Asp. The site-directed mutagenesis was carried out by the Quick Change Kit (Stratagene) using pGEX-cs plasmid as template (40) and the mutagenic oligos HMG-F11M (5′-GGAACGGTGGGTGATGGGACCAACCTAC-3′) and HMG-R22M (5′-GTAGGTTGGTCCCATCACCCACCGTTCC-3′). The mutation was confirmed by DNA sequencing according to the dideoxy chain termination reaction (39).
EGCG/pravastatin combined effect on HMG-CoA reductase activity
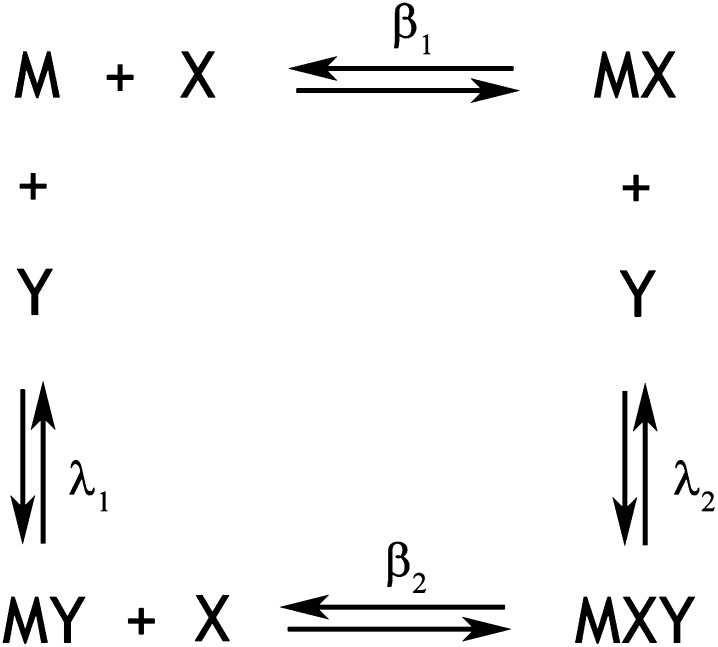
The combined effect of pravastatin and EGCG on the enzymatic activity of HMGR was evaluated, monitoring the effect of increasing levels of the statin in the range 0.6–54 µM both in the presence and in the absence of a fixed concentration of the catechin ([EGCG] = 66 µM). The data on the effect of EGCG on pravastatin binding to HMGR were analyzed according to a classic linkage relation: in detail, we considered a macromolecule M (namely HMGR) having two heterotropically associated binding sites, targeted by pravastatin (X) and EGCG (Y), respectively. The macromolecule M can exist in four states: M, MX, MY, and MXY, only M being productive (Fig. 1). Therefore, the inhibition of HMGR by pravastatin in the presence of EGCG is described by a general phenomenological equation (57) (equation 7):
| (Eq. 7) |
where β1, λ1, and β2 are the stepwise equilibrium association constants for the equilibria described in Fig. 1.
Fig.1.
Schematic representation of the mechanism of synergistic inhibition of HMGR (M) by EGCG (Y) and pravastatin (X). The phenomenological description of the multiple equilibria consists of the formation of two complexes (MX and MY) upon binding of the statin at the substrate site and of EGCG at the cofactor site, both with the ability of binding to the other ligand in a ternary complex (MXY).
RESULTS
Bioinformatic analysis
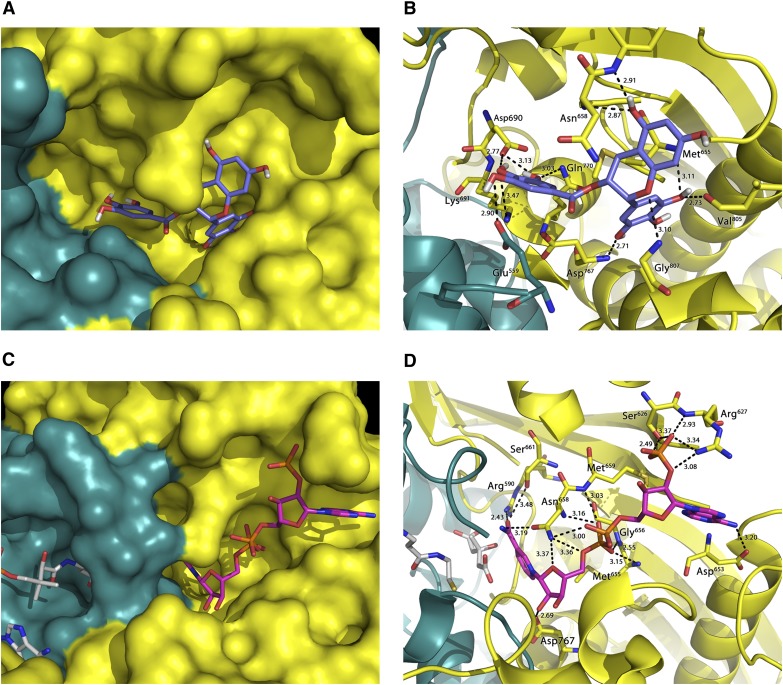
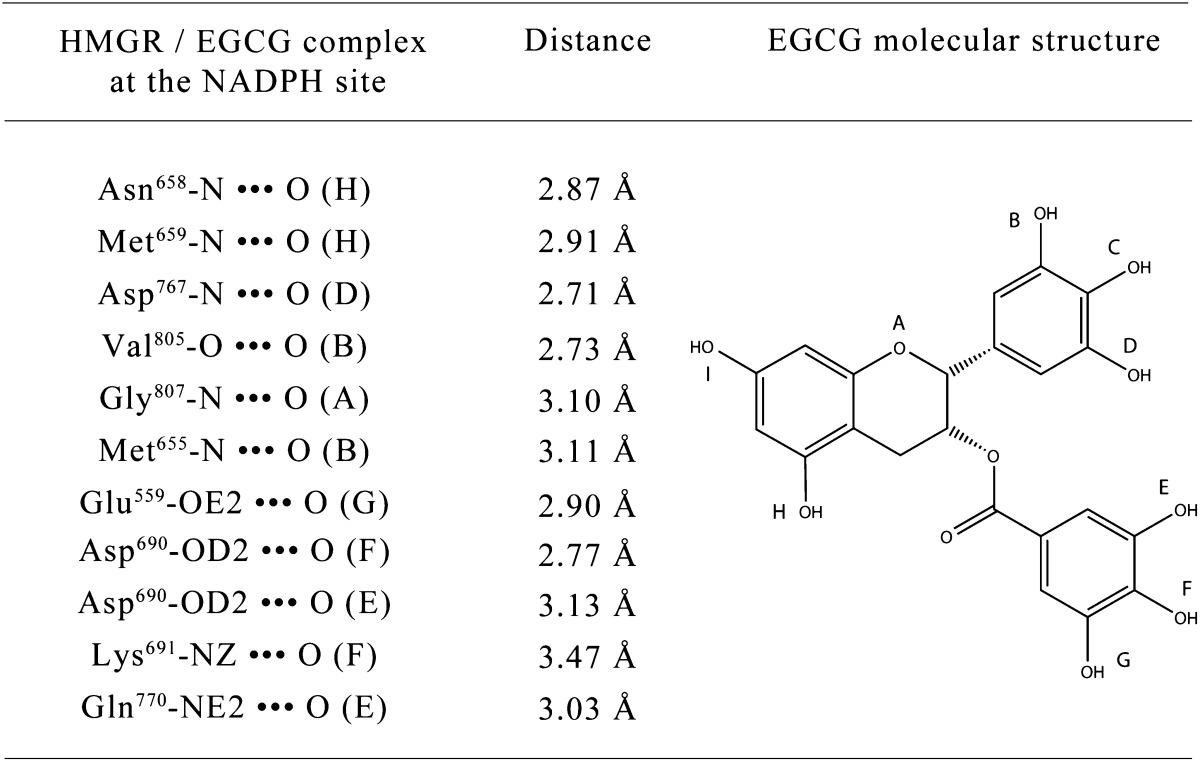
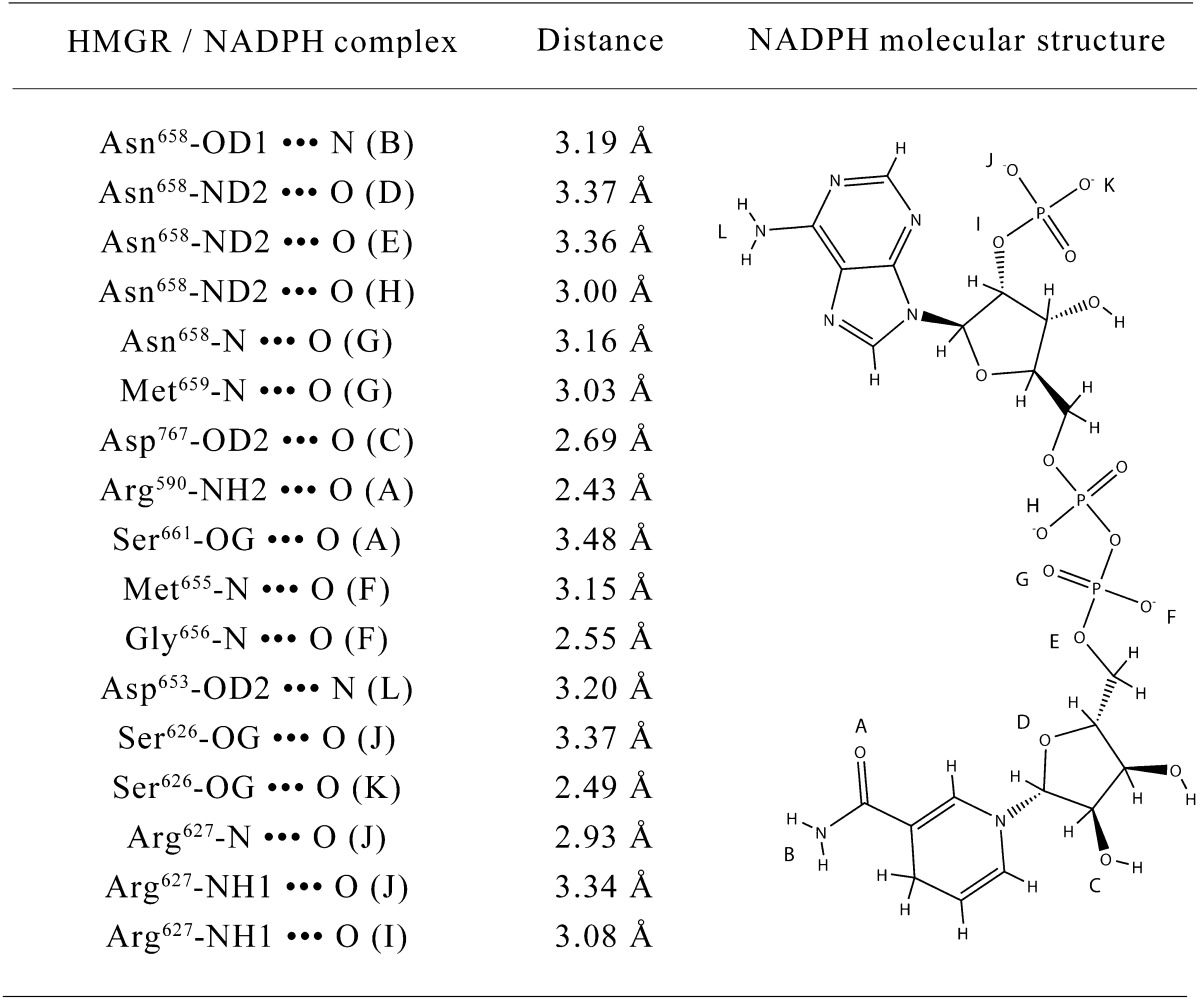
Docking studies of EGCG and NADPH on the X-ray crystal structure of the human HMGR disclosed new insights regarding both the interaction strength and the binding geometry of the complex. The cofactor binding site was calculated to be the most likely to accommodate an EGCG molecule (Fig. 2), with a dominant nonelectrostatic energy contribution. In fact, as derived from the Ludi Score (equation 1), EGCG showed a high affinity for the cofactor binding site (KD,p = 71 nM), and thus is a strong candidate for NADPH competitor. In detail, the predicted EGCG-HMGR complex was characterized by the formation of 11 theoretical H bonds involving Glu559, Asp690, Lys691, Gln770, Val805, Gly807, and Met659, Met655, Asn658, Asp767 (the latter being common to the NADPH/enzyme complex) (see Fig. 2, Table 1, and Table 2) and by a significant VdW energy contribution of −11.30 kcal/mol. Irrespective of the higher number of theoretical H bonds (n = 17), comparative analyses of NADPH docking to its own site revealed a lower affinity (KD,p = 220 nM). This difference can be essentially attributed to the lower entropic contribution due to conformational degrees of freedom; in fact, the docking of NADPH into its own site forced the ligand and the binding pocket into more-restricted conformations with respect to EGCG (1 and 4 conformations per lowest energy cluster, respectively), resulting in a lower value of conformational entropy and an unfavorable contribution to the total free energy of binding (7).
Fig.2.
Molecular docking of EGCG in the NADPH binding site of HMG-CoA reductase (A: surface representation; B: cartoon and stick representation with h-bonds formed) and the same representation of the complex NADPH/reductase (C, D). Reductase chain A and chain B are colored in yellow and light blue, respectively.
TABLE 1.
H-bonds involved in the HMG-CoA reductase / EGCG complex at NADPH site
TABLE 2.
H-bonds involved in the complex between HMG-CoA reductase and NADPH in its specifi c binding site
Additionally, EGCG was docked into the substrate binding site (predicted KD,p = 2.01 µM), with a resulting 30-fold lower affinity with respect to the NADPH binding site, and a VdW contribution of −8.76 kcal/mol (electrostatic energies were comparable), supporting the hypothesis of EGCG essentially as an NADPH site-directed ligand.
Effect of EGCG on HMGR activity
A preincubation period (20 min) was required to allow the establishment of equilibrium between the enzyme, the inhibitor, and the enzyme-inhibitor complex (Fig. 3F), in particular at EGCG concentrations lower than 1 µM (for equal treatment time, a 20 min preincubation was performed throughout). The initial velocity of HMGR was manifestly dependent on EGCG, as unequivocally shown in the residual activity plot; in particular, in the presence of EGCG, we obtained hyperbolic inhibition isotherms, whose steepness decreased at increasing cofactor concentrations (Fig. 3). At saturating NADPH, the apparent dissociation constants (Kd,app) of the complex HMG-CoA/EGCG increased linearly with cofactor concentrations, unambiguously suggesting the competitive binding of tea catechin on the NADPH binding site (Fig. 3) (49). The experimentally measured equilibrium dissociation constant for the EGCG/HMG-CoA complex at the cofactor site (Kd = 77.0 ± 12.0 nM) was in excellent agreement with the computationally predicted value. Comparative analysis showed that the inhibition of HMGR activity measured without preincubation was significantly lower, in agreement with the time required for the establishment of the equilibrium between HMGR and EGCG (Fig. 3F).
No relevant differences in Kd were observed when experiments were repeated at different HMG-CoA concentrations.
Michaelis-Menten parameters
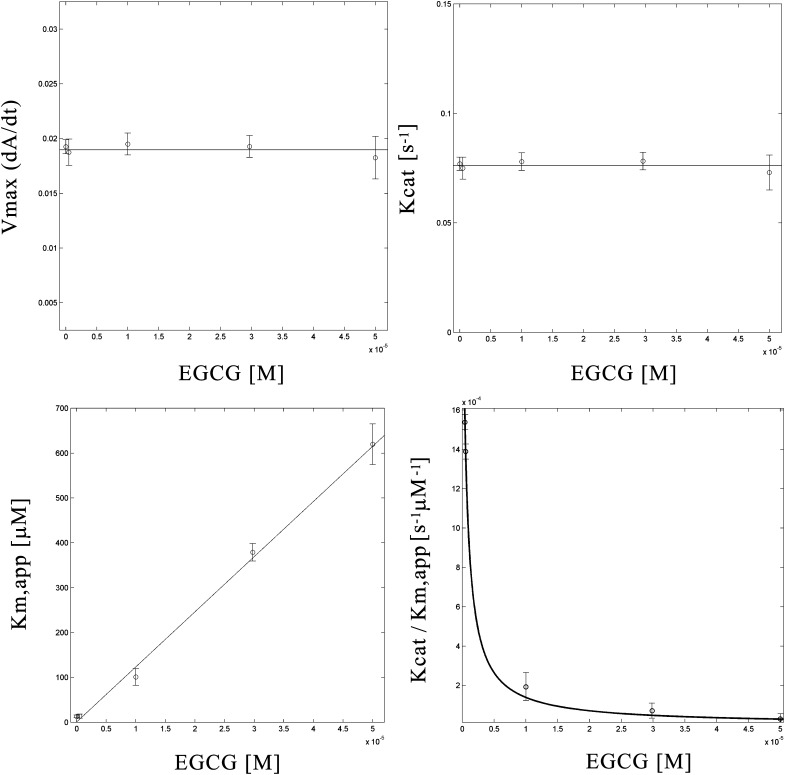
The Michaelis constant () was experimentally determined by plotting the initial rate (V0) against increasing NADPH concentrations. The kinetic parameters, Vmax, and the catalytic constant Kcat for each inhibitor concentration were derived from raw data analysis. As shown in Fig. 4 ( increased with EGCG, whereas Vmax remained unaffected), EGCG acted as a competitive inhibitor of NADPH. Additionally, calculated values of Kcat/ provided a measure of the catalytic efficiency of the enzyme, revealing the reduced efficiency of HMGR at increasing EGCG concentrations.
Fig.4.
Enzyme kinetic constants for the inhibition of HMGR by EGCG. The dependences of Vmax, Km, Kcat, and Kcat/Km from EGCG concentrations were fitted to the proper equations for a competitive inhibition model.
Biosensor binding studies
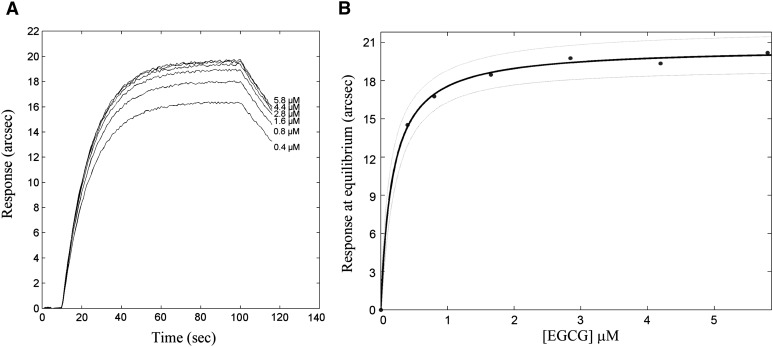
A more-detailed evaluation of the binding kinetics of EGCG to the enzyme was performed on an IAsys plus biosensor system. The sensing surface containing anchored HMGR was optimized on the basis of different experiments on the variations of enzyme concentration and immobilization buffer composition. Under the experimental conditions described in the Materials and Methods section, readout of 800 arcsec was obtained, corresponding to a partial Langmuir monolayer for a 90 kDa protein (1.30 ng mm−2); this “conveniently-low” HMGR surface density minimized possible hindering effects during recognition events. Then, EGCG was added at increasing concentrations in the range 0.4–6 µM, and association kinetics were followed up to equilibrium. Dissociation steps were performed by addition of fresh PBS buffer (Fig. 5A), whereas baseline recovery was achieved by multiple PBS-T washes at pH 7.4, because detergents decreased the stability of the complex. The longevity of the bio-layer was also tested; surface stability and enzyme activity were retained after at least 50 association/dissociation events. This experimental approach confirmed the high-affinity interaction between soluble EGCG and blocked HMG-CoA reductase (KD = 88.8 ± 31.4 nM), both association (kass = 29,500 ± 2,900 M−1s−1) and dissociation (kdiss = [2.6 ± 0.9]·10−3s−1) phases significantly contributing to the stabilization of the complex. Monophasic time courses were always observed upon the addition of soluble EGCG, both in the presence and in the absence of saturating HMG-CoA, whereas presaturation of the cofactor site with 1 mM NADPH prior to EGCG addition substantially annihilated any response.
Fig.5.
Overlay of association and dissociation kinetics of soluble EGCG to CMD-bound HMG-CoA reductase (A); dependence of the extent of binding on ligand concentration. Nonlinear fit (solid line) and 95% confidence-bound (dashed lines) are reported. Each experimental point was the average of five replicates (B).
To assess the validity of the monophasic model in fitting each time course, a standard F-test procedure was used; the biphasic model was statistically nonsignificant at 95% confidence (58). Time courses measured at several ligand concentrations were globally analyzed using equations 5 and 6, which share common parameters (kass, kdiss, and Rmax).
The binding response at equilibrium (extent of binding) was calculated for each time course with a fully comparable equilibrium dissociation constant ( = 98.9 ± 23.8 nM). The hyperbolic nature of the saturation plot (Fig. 5B) demonstrated the noncooperative binding of EGCG to human HMG-CoA reductase.
Chromatographic assay of HMGR activity
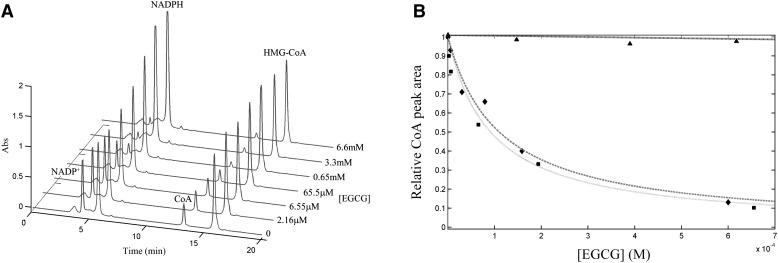
According to the RP-HPLC assay of HMGR activity in both bacterial and human liver microsomes, we observed an evident decrease in both HMG-CoA/NADPH consumption and mevalonate/NADP+ production rates upon 20 min preincubation of HMGR with increasing EGCG (Fig. 6). In detail, at 2.68 mM NADPH, we obtained comparable values of Kd,app for the bacterial-expressed and liver HMGR (91.2 ± 23.4 µM and 109.9 ± 35.3 µM, respectively), in strong agreement with the linear relation between Kd,app and NADPH concentration derived from the spectrophotometric inhibition assay.
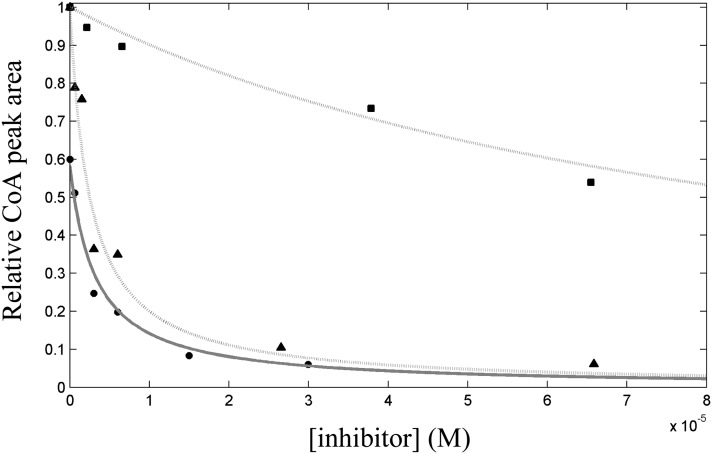
Fig.6.
Chromatographic analysis of HMGR modulation by EGCG. The superimposition of elution profiles of the species involved in the equilibrium (CoA:mevalonate stoichiometric ratio = 1:1) at different EGCG concentrations is presented. Peak area corresponding to CoA produced in the enzyme-free assay was each time subtracted from corresponding peaks in each sample (A). Residual activity plots of both bacterial expressed (■) and microsomal liver HMGR (¨), and HMGR mutant (▴) measured monitoring CoA/mevalonate production (B). Raw data were fitted to equation 3.
Mutagenesis of Gly-807 in the cofactor site of HMGR
To further ascertain the EGCG selective binding at the cofactor site HMGR, we constructed a point mutation of Gly807. Gly807 was shown not to influence catalysis, because the G807D mutant retained the in vitro activity toward HMG-CoA with respect to wild-type HMGR. Conversely, the substitution was critical in diminishing (and nearly abolishing) the capacity of the mutant to bind EGCG, with a Kd,app at least 400-fold higher than wild-type HMGR (Fig. 6B).
Combined effect of EGCG/pravastatin on HMG-CoA reductase activity
We explored the effect exerted on HMGR activity by increasing levels of the statin in both the presence and absence of a fixed concentration of the catechin. Analysis of raw data according to equation 7 showed that the affinity of pravastatin for HMGR shifted from β1 (108 M−1) to β2 (0.47·108 M−1) upon binding of EGCG at the cofactor site, eliciting a mild negative allosteric heterotropic effect (β2/β1 = 0.47) in the binding of the two inhibitors to HMGR. The inhibition observed upon coadministration of EGCG and pravastatin was higher than the effects exerted by each single inhibitor, but (slightly) lower than the total expected theoretical effect (Fig. 7).
Fig.7.
Residual activity plot of HMGR in the presence of EGCG (■), pravastatin (▴), and pravastatin and EGCG (•). Raw data for the HMGR inhibition by pravastatin in the presence of EGCG were fitted to equation 7.
DISCUSSION
Gallate catechins were demonstrated to modulate the expression and activity of cholesterogenic enzymes, but to date, both in vivo and in vitro studies hardly linked the hypocholesterolemic effect of catechin to the direct modulation of HMGR.
The present work showed for the first time that EGCG (commonly the most-abundant tea catechin) potently and reversibly inhibits HMG-CoA reductase from different sources to a comparable extent. The integrated experimental approaches used to derive both equilibrium and kinetic parameters (Table 3) concordantly proved EGCG to be a competitive ligand of HMGR at the cofactor site (KD in the nanomolar range), emphasizing the significant inference of both association and dissociation events in the stabilization of the complex. In detail, from a thermodynamic point of view, the mono-exponential nature of the binding kinetics, the significant values of both rate and equilibrium constants, and a plateau value for the Rmax at “saturating” EGCG values (Fig. 5B) collectively provided solid evidences for the presence of a highly specific binding site for EGCG on HMGR. On the other hand, the biosensor competitive binding assay clearly demonstrated that EGCG selectively overlaps with the NADPH binding site: in this assay, NADPH presaturated enzyme completely lost the ability to bind to EGCG, whereas this ability was effectively fully retained upon presaturation with the HMG-CoA (see supplementary material). Again, the spectrophotometric assay of HMGR activity provided further confirmation of exclusive competition between EGCG and NADPH for the same site, because the residual activity increased with NADPH; conversely, increases in HMG-CoA concentration did not significantly affect HMGR residual activity. Finally, the increase in apparent equilibrium dissociation constants with NADPH (Fig. 3E), and the equilibrium shift between HMGR-bound and soluble EGCG upon buffer wash (see supplementary material) clearly demonstrated the reversibility of the interaction.
TABLE 3.
Experimentally measured and predicted kinetic and affinity constants for EGCG at the cofactor site are listed. Biosensor-derived KD values were calculated directly from kdiss/kass and from fit to the plot of the maximum responses at equilibrium against EGCG concentration (*)
| EGCG |
|||||
|---|---|---|---|---|---|
| Biosensor assay |
NADPH | ||||
| Docking analysis(KD,p, nM) | Activity assay (Kd, nM) | kass (M−1 s−1) | kdiss (s−1) | KD (nM) | Docking analysis(KD,p, nM) |
| 70.96 | 77.0 ± 12.0 | 29500 ± 2900 | (2.6 ± 0.9)·10 | 88.8 ± 31.4 | 219.19 |
| 98.9 ± 43.8* | |||||
Site-directed mutagenesis ultimately substantiated these results; in fact, the replacement of glycine at position 807 of the cofactor site by an aspartate residue was crucial (Fig. 2B) in almost annihilating the binding affinity of HMGR for EGCG.
On the strength of these results, the physiological relevance of HMGR inhibition by EGCG can be asserted by considering the collective data available on the in vivo levels of NADPH ([NADPH] + [NADP+] = 20–370 μg/g wet liver weight) (59) and EGCG ([EGCG] = 0.3–7.5 μM in the blood of normal green tea consumers) (60), and their respective binding affinities for NADPH binding site (Km,NADPH = 21 μM (7), and KD,EGCG = 77 nM).
Hence, on the basis of these parameters, we can reasonably assert that inhibition of the enzyme occurs under physiological conditions, even irrespective of the possible binding of EGCG to other enzymes having NADPH/NADP+ as cofactor, but not sharing the same binding affinity. HMGR binding affinity for EGCG is up to 10,000-fold higher than the other NADPH enzymes previously studied, and owing to the concentration of EGCG in blood/liver upon oral administration (60), HMGR is likely to be inhibited by the catechin, even in the presence of the other “EGCG-sequestering agents.”
Nevertheless, the wide spectrum of EGCG molecular targets opens other scenarios on the modulation of HMGR activity, because EGCG is likely also to indirectly inhibit HMGR by activating AMP-activated protein kinase (61, 62), which in turn is responsible for the phosphorylation and inhibition of several enzymes, including HMGR (63).
Similarly, according to our results, the high specificity of EGCG for the NADPH site of HMGR is particularly interesting from a pharmacological point of view, inasmuch as the EGCG-statin synergistic inhibition of HMGR provides consistent information in the perspective of the coadministration of catechin-rich food and beverages during statin treatment of hypercholesterolemic states. Collectively, the results presented here confirm the role of tea catechins, EGCG in particular, as precursors for the development of novel drugs for the treatment of hypercholesterolemic disorders.
Supplementary Material
Acknowledgments
The authors would like to thank Professor J. Deisenhofer for providing the expression plasmid of the human HMGR, and Dr. L. Trozzi and Professor A. Benedetti for providing biopsies from human liver. The authors would like to thank Dr. Sheila Beatty for her assistance in manuscript revision.
Footnotes
Abbreviations:
- CMD
- carboxymethyl-dextran
- EDC
- 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide
- EGCG
- epigallocatechin-3-gallate
- HMGR
- hydroxy-3-methyl-glutaryl-CoA reductase
- IPTG
- isopropyl β-d-thiogalactopyranoside
- NADP+
- nicotinamide adenine dinucleotide phosphate
- NADPH
- reduced nicotinamide adenine dinucleotide phosphate
- NHS
- N-hydroxysuccinimide
- rms
- root-mean-square
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three figures and a reference.
REFERENCES
- 1.Berry J. D., Liu K., Folsom A. R., Lewis C. E., Carr J. J., Polak J. F., Shea S., Sidney S., O'Leary D. H., Chan C., et al. 2009. Prevalence and progression of subclinical atherosclerosis in younger adults with low short-term but high lifetime estimated risk for cardiovascular disease: the coronary artery risk development in young adults study and multi-ethnic study of atherosclerosis. Circulation. 119: 382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steinberger J., Daniels S. R., Eckel R. H., Hayman L., Lustig R. H., McCrindle B., Mietus-Snyder M. L. 2009. Progress and challenges in metabolic syndrome in children and adolescents: a scientific statement from the American Heart Association Atherosclerosis, Hypertension, and Obesity in the Young Committee of the Council on Cardiovascular Disease in the Young; Council on Cardiovascular Nursing; and Council on Nutrition, Physical Activity, and Metabolism. Circulation. 119: 628–647. [DOI] [PubMed] [Google Scholar]
- 3.Winston G. J., Barr R. G., Carrasquillo O., Bertoni A. G., Shea S. 2009. Gender and racial-ethnic differences in cardiovascular disease risk factor treatment and control among persons with diabetes in the Multi-Ethnic Study of Atherosclerosis (MESA). Diabetes Care. 32: 1467–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown M. S., Goldstein J. L. 1980. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J. Lipid Res. 21: 505–517. [PubMed] [Google Scholar]
- 5.Edwards P. A., Lemongello D., Kane J., Shechter I., Fogelman A. M. 1980. Properties of purified rat hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase and regulation of enzyme activity. J. Biol. Chem. 255: 3715–3725. [PubMed] [Google Scholar]
- 6.Goldstein J. L., Brown M. S. 1990. Regulation of the mevalonate pathway. Nature. 343: 425–430. [DOI] [PubMed] [Google Scholar]
- 7.Carbonell T., Freire E. 2005. Binding thermodynamics of statins to HMG-CoA reductase. Biochemistry. 44: 11741–11748. [DOI] [PubMed] [Google Scholar]
- 8.Corsini A., Maggi F. M., Catapano A. L. 1995. Pharmacology of competitive inhibitors of HMG-CoA reductase. Pharmacol. Res. 31: 9–27. [DOI] [PubMed] [Google Scholar]
- 9.Istvan E. S., Deisenhofer J. 2001. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 292: 1160–1164. [DOI] [PubMed] [Google Scholar]
- 10.Ertas F. S., Ertas N. M., Gulec S., Atmaca Y., Tanju S., Sener C., Erol C. 2006. Unrecognized side effect of statin treatment: unilateral blepharoptosis. Ophthal. Plast. Reconstr. Surg. 22: 222–224. [DOI] [PubMed] [Google Scholar]
- 11.Kashani A., Sallam T., Bheemreddy S., Mann D. L., Wang Y., Foody J. M. 2008. Review of side-effect profile of combination ezetimibe and statin therapy in randomized clinical trials. Am. J. Cardiol. 101: 1606–1613. [DOI] [PubMed] [Google Scholar]
- 12.Kent D. M. 2005. Improved perioperative outcomes from carotid endarterectomy: yet another statin side effect? Stroke. 36: 2058–2059. [DOI] [PubMed] [Google Scholar]
- 13.Mukhtar R. Y., Reckless J. P. 2005. Statin-induced myositis: a commonly encountered or rare side effect? Curr. Opin. Lipidol. 16: 640–647. [DOI] [PubMed] [Google Scholar]
- 14.Ornato J. P. 2003. Should you worry about the side effects of statins? Statin dose, health, and other drugs affect side-effect risk. Health News. 9: 1–2. [PubMed] [Google Scholar]
- 15.Bursill C. A., Abbey M., Roach P. D. 2007. A green tea extract lowers plasma cholesterol by inhibiting cholesterol synthesis and upregulating the LDL receptor in the cholesterol-fed rabbit. Atherosclerosis. 193: 86–93. [DOI] [PubMed] [Google Scholar]
- 16.Yang T. T., Koo M. W. 2000. Chinese green tea lowers cholesterol level through an increase in fecal lipid excretion. Life Sci. 66: 411–423. [DOI] [PubMed] [Google Scholar]
- 17.Chen Z. Y., Jiao R., Ma K. Y. 2008. Cholesterol-lowering nutraceuticals and functional foods. J. Agric. Food Chem. 56: 8761–8773. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda I., Hamamoto R., Uzu K., Imaizumi K., Nagao K., Yanagita T., Suzuki Y., Kobayashi M., Kakuda T. 2005. Dietary gallate esters of tea catechins reduce deposition of visceral fat, hepatic triacylglycerol, and activities of hepatic enzymes related to fatty acid synthesis in rats. Biosci. Biotechnol. Biochem. 69: 1049–1053. [DOI] [PubMed] [Google Scholar]
- 19.Ito Y., Ichikawa T., Iwai T., Saegusa Y., Ikezawa T., Goso Y., Ishihara K. 2008. Effects of tea catechins on the gastrointestinal mucosa in rats. J. Agric. Food Chem. 56: 12122–12126. [DOI] [PubMed] [Google Scholar]
- 20.Ito Y., Ichikawa T., Morohoshi Y., Nakamura T., Saegusa Y., Ishihara K. 2008. Effect of tea catechins on body fat accumulation in rats fed a normal diet. Biomed. Res. 29: 27–32. [DOI] [PubMed] [Google Scholar]
- 21.Bose M., Lambert J. D., Ju J., Reuhl K. R., Shapses S. A., Yang C. S. 2008. The major green tea polyphenol, (-)-epigallocatechin-3-gallate, inhibits obesity, metabolic syndrome, and fatty liver disease in high-fat-fed mice. J. Nutr. 138: 1677–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh D. K., Banerjee S., Porter T. D. 2009. Green and black tea extracts inhibit HMG-CoA reductase and activate AMP kinase to decrease cholesterol synthesis in hepatoma cells. J. Nutr. Biochem. 20: 816–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Tian W. 2001. Green tea epigallocatechin gallate: a natural inhibitor of fatty-acid synthase. Biochem. Biophys. Res. Commun. 288: 1200–1206. [DOI] [PubMed] [Google Scholar]
- 24.Bursill C. A., Roach P. D. 2006. Modulation of cholesterol metabolism by the green tea polyphenol (-)-epigallocatechin gallate in cultured human liver (HepG2) cells. J. Agric. Food Chem. 54: 1621–1626. [DOI] [PubMed] [Google Scholar]
- 25.Franiak-Pietryga I., Koter-Michalak M., Broncel M., Duchnowicz P., Chojnowska-Jezierska J. 2009. Anti-inflammatory and hypolipemic effects in vitro of simvastatin comparing to epicatechin in patients with type-2 hypercholesterolemia. Food Chem. Toxicol. 47: 393–397. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda H., Chisaka T., Kubomura Y., Yamahara J., Sawada T., Fujimura H., Kimura H. 1986. Effects of crude drugs on experimental hypercholesterolemia. I. Tea and its active principles. J. Ethnopharmacol. 17: 213–224. [DOI] [PubMed] [Google Scholar]
- 27.Muramatsu K., Fukuyo M., Hara Y. 1986. Effect of green tea catechins on plasma cholesterol level in cholesterol-fed rats. J. Nutr. Sci. Vitaminol. (Tokyo). 32: 613–622. [DOI] [PubMed] [Google Scholar]
- 28.Abe I., Seki T., Umehara K., Miyase T., Noguchi H., Sakakibara J., Ono T. 2000. Green tea polyphenols: novel and potent inhibitors of squalene epoxidase. Biochem. Biophys. Res. Commun. 268: 767–771. [DOI] [PubMed] [Google Scholar]
- 29.Seiki S., Frishman W. H. 2009. Pharmacologic inhibition of squalene synthase and other downstream enzymes of the cholesterol synthesis pathway: a new therapeutic approach to treatment of hypercholesterolemia. Cardiol. Rev. 17: 70–76. [DOI] [PubMed] [Google Scholar]
- 30.Taylor F. R. 1992. Correlation among oxysterol potencies in the regulation of the degradation of 3-hydroxy-3-methylglutaryl CoA reductase, the repression of 3-hydroxy-3-methylglutaryl CoA synthase and affinities for the oxysterol receptor. Biochem. Biophys. Res. Commun. 186: 182–189. [DOI] [PubMed] [Google Scholar]
- 31.Spina M., Cuccioloni M., Mozzicafreddo M., Montecchia F., Pucciarelli S., Eleuteri A. M., Fioretti E., Angeletti M. 2008. Mechanism of inhibition of wt-dihydrofolate reductase from E. coli by tea epigallocatechin-gallate. Proteins. 72: 240–251. [DOI] [PubMed] [Google Scholar]
- 32.Sharma S. K., Parasuraman P., Kumar G., Surolia N., Surolia A. 2007. Green tea catechins potentiate triclosan binding to enoyl-ACP reductase from Plasmodium falciparum (PfENR). J. Med. Chem. 50: 765–775. [DOI] [PubMed] [Google Scholar]
- 33.Navarro-Peran E., Cabezas-Herrera J., Garcia-Canovas F., Durrant M. C., Thorneley R. N., Rodriguez-Lopez J. N. 2005. The antifolate activity of tea catechins. Cancer Res. 65: 2059–2064. [DOI] [PubMed] [Google Scholar]
- 34.Navarro-Peran E., Cabezas-Herrera J., Hiner A. N., Sadunishvili T., Garcia-Canovas F., Rodriguez-Lopez J. N. 2005. Kinetics of the inhibition of bovine liver dihydrofolate reductase by tea catechins: origin of slow-binding inhibition and pH studies. Biochemistry. 44: 7512–7525. [DOI] [PubMed] [Google Scholar]
- 35.Du Y., Wu Y., Cao X., Cui W., Zhang H., Tian W., Ji M., Holmgren A., Zhong L. 2009. Inhibition of mammalian thioredoxin reductase by black tea and its constituents: implications for anticancer actions. Biochimie. 91: 434–444. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y., Zhang H., Holmgren A., Tian W., Zhong L. 2008. Inhibitory effect of green tea extract and (-)-epigallocatechin-3-gallate on mammalian thioredoxin reductase and HeLa cell viability. Oncol. Rep. 20: 1479–1487. [PubMed] [Google Scholar]
- 37.Banerjee T., Sharma S. K., Surolia N., Surolia A. 2008. Epigallocatechin gallate is a slow-tight binding inhibitor of enoyl-ACP reductase from Plasmodium falciparum. Biochem. Biophys. Res. Commun. 377: 1238–1242. [DOI] [PubMed] [Google Scholar]
- 38.Istvan E. S., Palnitkar M., Buchanan S. K., Deisenhofer J. 2000. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. EMBO J. 19: 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J., Russell D. W. 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 40.Parks A. L., Cook K. R., Belvin M., Dompe N. A., Fawcett R., Huppert K., Tan L. R., Winter C. G., Bogart K. P., Deal J. E., et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292. [DOI] [PubMed] [Google Scholar]
- 41.Angelin B., Einarsson K., Liljeqvist L., Nilsell K., Heller R. A. 1984. 3-Hydroxy-3-methylglutaryl coenzyme A reductase in human liver microsomes: active and inactive forms and cross-reactivity with antibody against rat liver enzyme. J. Lipid Res. 25: 1159–1166. [PubMed] [Google Scholar]
- 42.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193: 265–275. [PubMed] [Google Scholar]
- 43.Sarver R. W., Bills E., Bolton G., Bratton L. D., Caspers N. L., Dunbar J. B., Harris M. S., Hutchings R. H., Kennedy R. M., Larsen S. D., et al. 2008. Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Med. Chem. 51: 3804–3813. [DOI] [PubMed] [Google Scholar]
- 44.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. 2000. The Protein Data Bank. Nucleic Acids Res. 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., Olson A. J. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30: 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mozzicafreddo M., Cuccioloni M., Cecarini V., Eleuteri A. M., Angeletti M. 2009. Homology modeling and docking analysis of the interaction between polyphenols and mammalian 20S proteasomes. J. Chem. Inf. Model. 49: 401–409. [DOI] [PubMed] [Google Scholar]
- 47.Bohm H. J. 1992. The computer program LUDI: a new method for the de novo design of enzyme inhibitors. J. Comput. Aided Mol. Des. 6: 61–78. [DOI] [PubMed] [Google Scholar]
- 48.Bohm H. J. 1998. Prediction of binding constants of protein ligands: a fast method for the prioritization of hits obtained from de novo design or 3D database search programs. J. Comput. Aided Mol. Des. 12: 309–323. [DOI] [PubMed] [Google Scholar]
- 49.Bieth J. G. 1974. Some Kinetic Consequences of the Tight Binding of Protein-Proteinase-Inhibitors to Proteolytic Enzymes and Their Application to the Determination of Dissociation Constants. Springer-Verlag, Berlin. [Google Scholar]
- 50.Brooks S. P. 1992. A simple computer program with statistical tests for the analysis of enzyme kinetics. Biotechniques. 13: 906–911. [PubMed] [Google Scholar]
- 51.Mozzicafreddo M., Cuccioloni M., Eleuteri A. M., Angeletti M. 2010. Rapid RP-HPLC assay of HMG-CoA reductase activity. J. Lipid. Res. 51: 2460–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edwards P. R., Lowe P. A., Leatherbarrow R. J. 1997. Ligand loading at the surface of an optical biosensor and its effect upon the kinetics of protein-protein interactions. J. Mol. Recognit. 10: 128–134. [DOI] [PubMed] [Google Scholar]
- 53.Hall D. R., Winzor D. J. 1997. Use of a resonant mirror biosensor to characterize the interaction of carboxypeptidase A with an elicited monoclonal antibody. Anal. Biochem. 244: 152–160. [DOI] [PubMed] [Google Scholar]
- 54.De Crescenzo G., Grothe S., Zwaagstra J., Tsang M., O'Connor-McCourt M. D. 2001. Real-time monitoring of the interactions of transforming growth factor-beta (TGF-beta) isoforms with latency-associated protein and the ectodomains of the TGF-beta type II and III receptors reveals different kinetic models and stoichiometries of binding. J. Biol. Chem. 276: 29632–29643. [DOI] [PubMed] [Google Scholar]
- 55.Roden L. D., Myszka D. G. 1996. Global analysis of a macromolecular interaction measured on BIAcore. Biochem. Biophys. Res. Commun. 225: 1073–1077. [DOI] [PubMed] [Google Scholar]
- 56.Luskey K. L., Stevens B. 1985. Human 3-hydroxy-3-methylglutaryl coenzyme A reductase. Conserved domains responsible for catalytic activity and sterol-regulated degradation. J. Biol. Chem. 260: 10271–10277. [PubMed] [Google Scholar]
- 57.Wyman J., Gill S. J. 1990. Binding and Linkage—Functional Chemistry of Biological Macromolecules. University Science Books, Mill Valley, CA. [Google Scholar]
- 58.Bevington P. R., Robinson D. K. 1992. Data Reduction and Error Analysis for the Physical Sciences. McGraw-Hill Book Company, New York. [Google Scholar]
- 59.Greenbaum A. L., Clark J. B., McLean P. 1965. The estimation of the oxidized and reduced forms of the nicotinamide nucleotides. Biochem. J. 95: 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ullmann U., Haller J., Decourt J. P., Girault N., Girault J., Richard-Caudron A. S., Pineau B., Weber P. 2003. A single ascending dose study of epigallocatechin gallate in healthy volunteers. J. Int. Med. Res. 31: 88–101. [DOI] [PubMed] [Google Scholar]
- 61.Hwang J. T., Ha J., Park I. J., Lee S. K., Baik H. W., Kim Y. M., Park O. J. 2007. Apoptotic effect of EGCG in HT-29 colon cancer cells via AMPK signal pathway. Cancer Lett. 247: 115–121. [DOI] [PubMed] [Google Scholar]
- 62.Hwang J. T., Kwon D. Y., Yoon S. H. 2009. AMP-activated protein kinase: a potential target for the diseases prevention by natural occurring polyphenols. New Biotechnol. 26: 17–22. [DOI] [PubMed] [Google Scholar]
- 63.Hardie D. G. 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8: 774–785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.