Abstract
Genetically engineered mice have been employed to understand the role of lipases in dietary fat digestion with the expectation that the results can be extrapolated to humans. However, little is known about the properties of mouse pancreatic triglyceride lipase (mPTL) and pancreatic lipase-related protein-2 (mPLRP2). In this study, both lipases were expressed in Pichia Pastoris GS115, purified to near homogeneity, and their properties were characterized. Mouse PTL displayed the kinetics typical of PTL from other species. Like mPTL, mPLRP2 exhibited strong activity against various triglycerides. In contrast to mPTL, mPLRP2 was not inhibited by increasing bile salt concentration. Colipase stimulated mPLRP2 activity 2- to 4-fold. Additionally, mPTL absolutely required colipase for absorption to a lipid interface, whereas mPLRP2 absorbed fully without colipase. mPLRP2 had full activity in the presence of BSA, whereas BSA completely inhibited mPTL unless colipase was present. All of these properties of mPLRP2 differ from the properties of human PLRP2 (hPLRP2). Furthermore, mPLRP2 appears capable of compensating for mPTL deficiency. These findings suggest that the molecular mechanisms of dietary fat digestion may be different in humans and mice. Thus, extrapolation of dietary fat digestion in mice to humans should be done with care.
Keywords: triglyceride, colipase, interaction affinity, binding, PTL deficiency
Dietary fats are predominantly triacylglycerols (TAG), which consist of three fatty acids linked to glycerol. Dietary TAGs must be hydrolyzed by lipases to release acyl chains and monoacylglycerol for efficient absorption. The digestion of dietary TAGs commences in the stomach, where gastric lipase partially releases the fatty acids from TAG (1). The emulsion particles move from the stomach and mix with digestive enzymes from the pancreas in the duodenum where digestion proceeds. The pancreas synthesizes and secretes multiple lipases, including the well-characterized colipase-dependent pancreatic triglyceride lipase (PTL), carboxyl ester lipase (CEL), and the recently described pancreatic lipase-related protein-2 (PLRP2) (2–5). All have the ability to hydrolyze TAGs. Under normal circumstances, the assimilation of dietary fat is extremely efficient, with >95% of dietary TAGs hydrolyzed and absorbed by the intestine (6). Steatorrhea is a major symptom in pancreatic insufficiency or exocrine pancreas failure, because gastric lipase cannot compensate for the loss of pancreatic lipases. Thus, pancreas-derived lipases are central to the efficient digestion of dietary TAG. Despite the critical function of pancreatic lipases in digestion, the physiological role of each lipase remains uncertain. A series of in vitro and in vivo studies of human and mouse pancreatic lipases have provided conflicting data about the roles of these lipases and have raised questions about the suitability of mouse models to understand the function of lipases in humans.
For many years, it has been assumed that PTL is the predominant triglyceride lipase in the digestion of dietary TAGs. Curiously, PTL is inhibited by normal constituents of the duodenum, such as bile acids, phospholipids, or dietary proteins. Colipase, another pancreatic protein, forms a complex with PTL and reverses the inhibition of PTL in the duodenum (7). The colipase-PTL complex likely accounts for roughly 60% of dietary TAG hydrolysis in human adults (1). The importance of colipase, and by extension, of PTL in dietary fat digestion was supported by a mouse model of colipase deficiency, in which adult mice had significant steatorrhea on a Western diet (2). Subsequently, another study reported a mouse model of PTL deficiency (6). Unexpectedly, although PTL deficiency delayed dietary fat absorption early in the digestive process, the overall TAG absorption efficiency was normal in PTL-deficient mice, even when they were fed a high-fat diet.
There may be several possible explanations for the differences in these mouse models. PTL may actually have a limited role in dietary fat digestion, or other pancreatic lipases may compensate for the loss of PTL. In either case, colipase must interact with other lipases. CEL was initially considered a candidate for the residual lipolytic activity in the intestinal lumen of PTL-deficient mice. Characterization of PTL−/−/CEL−/− mice eliminated CEL as a major compensatory lipase for PTL deficiency because PTL−/−/CEL−/− mice exhibited normal fat assimilation when fed a normal fat diet, and fat absorption only decreased modestly when fed a high-fat diet (8). Fat malabsorption in PTL−/−/CEL−/− mice was much less severe than in colipase-deficient mice (2). Furthermore, CEL does not require colipase for activity in vitro.
Mouse PLRP2 (mPLRP2) is another candidate to compensate for PTL deficiency. PLRP2-deficient mice experience fat malabsorption as suckling pups, a time when PTL expression is absent, indicating that mPLRP2 has TAG lipase activity in vivo (2, 5, 9). Additionally, mPLRP2 is the only source of colipase-dependent lipase activity in the neonate mouse pancreas (9). These two observations also offer a clear explanation for the steatorrhea in colipase-deficient mouse pups prior to the expression of PTL (2). Thus, colipase-stimulated mPLRP2 activity is a major contributor to TAG digestion in mouse pups. Although adult PLRP2-deficient mice had normal fat absorption, mPLRP2 may have an increased role when mouse PTL (mPTL) is absent.
The major difficulty with the hypothesis that mPLRP2 compensates for mouse PTL is the lack of kinetic characterization of mPLRP2. This characterization is critical for interpreting any results in mice because the kinetics of PLRP2 varies considerably among species. For instance, human PLRP2 (hPLRP2) has almost no activity in the presence of bile salt micelles and is not reactivated by colipase (10–12). The same is true for horse PLRP2 (13). It has been suggested that the major physiological role of hPLRP2 is as a galactolipase (10, 12, 14, 15). In contrast, rat PLRP2 has activity in the presence of bile salt micelles even in the absence of colipase. Colipase stimulates rat PLRP2 activity against short-, medium-, and long-chain TAGs (16). The kinetics of mPLRP2 has not been determined previously, and it is unknown whether purified mPLRP2 has activity in the presence of bile salt micelles or whether its activity is stimulated by colipase. In the present study, we expressed and purified recombinant mPTL and mPLRP2 and characterized their kinetic properties to provide additional insight into the role of PLRP2 in the mouse and to help determine whether the mouse mimics the process of dietary fat digestion in humans.
MATERIALS AND METHODS
DNA manipulations
Mouse cDNA clones for PTL (clone ID 30316303, Genebank ID BC061061) and PLRP2 (clone ID 6431510, Genebank ID BC094923) were purchased from Open Biosystems (Huntsville, AL). cDNAs encoding mature mPTL, mPLRP2, human PTL (hPTL), and hPLRP2 protein were amplified by PCR and subcloned into Pichia pastoris protein expression vectors pHILS1 and pPICZαA, respectively. In these plasmid constructs, the yeast PHO1 signal peptide and α-factor mating signal peptide were used to replace mouse PTL and PLRP2 native secretion signal peptides, respectively. The sequence of the cDNA was verified by dideoxynucleotide sequence analysis.
Expression and purification of recombinant proteins
Recombinant proteins were produced in Pichia pastoris GS115. Plasmid DNAs, pHILSI/mPTL and pPICZαA/mPLRP2, were linearized by Bgl II and Sac I, respectively. The linearized plasmid DNAs were transformed into the competent yeast strain GS115 by electroporation, and positive colonies were selected according to the method described previously (17). Media from the methanol-induced cultures were analyzed by immunoblot and lipase activity assay as described previously (18, 19).
Large-scale expression of recombinant proteins was accomplished according to the methods described, with minor modifications (17). For each transformation, the transformant with the highest secreted protein level was chosen to inoculate 10 ml buffered minimal glycerol-complex medium (pH 6.0). The overnight culture was then used to inoculate three liters of the same medium and grown until the optical density at 600 nm was between 6.0 and 10.0. The harvested yeasts were resuspended in 1,000 ml of buffered minimal methanol complex medium (pH 6.0). The cultures were allowed to grow at 28°C for 30 (mPLRP2) or 48 h (mPTL) with daily 1% methanol supplementation.
The clarified culture supernatant was further concentrated down to ∼50 ml at 4°C over a Pellicon XL Biomax 30 membrane (Millipore, Bedford, MA). The sample was dialyzed at 4°C overnight against double-distilled H2O containing 2 mM benzamidine, and then adjusted to contain 10 mM MES at pH 6.0 for mPTL and at pH 5.5 for mPLRP2. The sample was applied to a Mono S FPLC column (Pharmacia) equilibrated with 10 mM MES at the corresponding pH. Chromatography was controlled with an AktaExplorer system (Amersham Biosciences, Piscataway, NJ). The bound recombinant protein was eluted from the column using a linear concentration gradient from 0 to 200 mM NaCl within 60 min at a rate of 1 ml/min. Fractions containing recombinant protein were identified by A280, lipase activity assay, and further gel staining. The purest fractions were pooled and concentrated over an Amicon Ultra-15 30K MWCO centrifuge filter (Millipore), and the buffer was exchanged to 20 mM Tris-Cl (pH 8.0). The protein concentration was determined by UV light spectrophotometry at A280 using an extinction coefficient of E1cm1% = 1.24 for mPTL and E1cm1% = 1.33 for mPLRP2.
Determination of protein integrity
The purified proteins were analyzed by gel staining to confirm their homogeneity and integrity. Briefly, 6 µg of each isolated protein was resolved by 10% SDS-PAGE and stained with GelCode Blue Stain Reagent (Pierce, Rockford, IL). The gel staining was scanned by Molecular Imager VersaDoc MP 4000 System (Bio-Rad, Hercules, CA).
Protein extraction and Western blot
To quantify the relative protein amount of PTL and PLRP2 in the mouse pancreas, seven (four males and three females) 11-wk- old Balb/C mice were euthanized. Pancreas tissues were quickly excised and snap-frozen in liquid nitrogen and then stored at −80°C. Pancreatic protein extraction was carried out as described previously (9). Each whole pancreatic tissue (average wet weight ∼150 mg) was thawed and sonicated 6× 10 s in 1.5 ml of ice-cold protein lysis buffer [50 mM Tris-HCl, 0.5% Na-deoxycholate, 0.1% SDS, 150 mM NaCl, 1 mM EDTA (pH 7.5)] containing EDTA free complete protease inhibitor cocktail (Roche Diagnostics). The homogenate was centrifuged at 18,000 g for 10 min at 4°C. The resulting supernatant was further diluted 10 times with lysis buffer and mixed with 2× protein loading buffer (Bio-Rad) and boiled for 5 min. Then 10 µl of each sample was analyzed on a 10% SDS-PAGE, along with molecular weight markers and varying amounts of purified recombinant mPTL and mPLRP2 proteins. The resolved proteins were transferred onto nitrocellulose membranes (Bio-Rad). After incubation with blocking buffer (LI-COR, Lincoln, NE) for 1 h, the membrane was incubated with a rabbit polyclonal antibody against hPTL at 1:3000 dilution in blocking buffer for 1 h, and then with IRDye 680 goat anti-rabbit IgG (LI-COR) at 1:10,000 dilution for 30 min (19). The immunoblots were scanned by ODYSSEY INFRARED IMAGER, and band density was quantified by the rectangle method (LI-COR).
Lipase activity assay
Triglyceride lipase activity was determined at 25°C using the pH-stat method as previously described (20–22). Unless otherwise stated, the reaction was carried out for 5 min in the presence or absence of a 5-fold molar excess of recombinant human colipase at the indicated sodium taurodeoxycholate (NaTDC) concentration. As taurocholate is one of the major bile salt species in duodenal juice from mice, we also measured the lipase activity of mPTL and mPLRP2 against triolein at varying sodium taurocholate (NaTC) concentrations. To determine the effects of intestinal luminal proteins on mPTL and mPLRP2 lipase activity, assays were performed in 12 mM NaTC using triolein as substrate in the presence of 3.75% of BSA with or without colipase (23).
Galactolipase activity was determined at 25°C using the pH-stat method. An amount of 5 mg of digalactosyldiacylglyceride was emulsified in 15 ml of the standard assay buffer containing 4 mM NaTDC. Then 40 µg of mPLRP2 or 3 µg hPLRP2 was assayed. Phospholipase activity was measure in the pH-stat using 200 mg of egg yolk phosphaditylcholine emulsified in assay buffer containing 1 mM Tris-Cl (pH 8.0), 20 mM calcium, and 13.3 mM deoxycholate. Then 20 µg of PLRP2 was assayed. Activity was determined from the rate over 5 min for each substrate. Colipase did not affect activity the assays, and it was not included.
To evaluate whether mouse or human PLRP2 substantively compensates for lipase activity when the corresponding PTL is absent, we assayed the total lipase activity of 2.5 µg PTL and 1.0 µg PLRP2 in the presence of 0.5 µg colipase (the molar ratio of colipase:PTL = 1:1). We then measured the total lipase activity of 2.5 µg PTL and 1.0 µg PLRP2 alone under the same conditions. The relative lipase activity in each assay was expressed as a percentage of the activity when both PTL and PLRP2 were included in the assay.
Colipase titration
The interactions of mPTL and mPLRP2 with colipase were assayed by adapting the method described previously (24). The assays were performed by measuring lipase activity over a range of colipase concentration (0-53 nM) with a constant concentration of lipase (2.6 nM) and excess substrate (0.2 ml of trioctanoin) in 4 mM NaTDC assay buffer in a total volume of 15 ml.
Interfacial absorption assays
The interfacial absorption of mPTL and mPLRP2 was assayed by mixing the enzyme with trioctanoin emulsion and then measuring the lipase activity remaining in the water phase after the separation of the oil phase. The ability of colipase to anchor lipases to trioctanoin was done as described for intralipid (20). An amount of 0.5 ml of trioctanoin was emulsified in 14.5 ml of binding buffer [50 mM Tris-HCl (pH 8.0), 2.0 mM Ca2Cl, 150 mM NaCl] in the presence of 4 mM NaTDC. Aliquots (0.5 ml) of the emulsion were dispensed in 1.5 ml microcentrifuge tubes, and then the desired amount of colipase and either 6 µg of mPTL or mPLRP2 were added. The mixture was incubated at room temperature with shaking for 2 min, transferred to an Amicon Micron-100 Ultrafiltration tube (Millipore), subjected to centrifugation at 800 g for 10–30 min. The lipase activity of 50 µl clear filtrate was assayed against tributyrin in 0.5 mM NaTDC in the absence of colipase. The assay included controls with lipase alone or lipase plus colipase. Lipase absorption ability was calculated as follows: Bound (%) = 100 − the filtrate lipase activity / the filtrate lipase activity in control.
RESULTS
Expression and purification of recombinant mPTL and mPLRP2
The recombinant mPTL and mPLRP2 were produced in Pichia Pastoris GS115 in large scale after 30 or 48 h methanol induction, and then purified through one-step Mono S column purification system. The final yield was ∼8-10 mg for mPTL, and ∼1 mg for mPLRP2. The homogeneity of purified recombinant proteins was ascertained by 10% SDS-PAGE and was followed by gel staining (Fig. 1). Protein purity was 91% for mPTL and 98% for mPLRP2. The major band for mPTL was around 50 kDa. The major bands for both preparations and the faster migrating minor bands in the PTL sample reacted with PTL antisera, indicating a minor amount of proteolytic degradation in the PTL preparation (data not shown). mPLRP2 contained two bands around 53 kDa, which represents differences in glycosylation (16). The identity of purified protein was verified by robust lipase activity against various TAGs as described below.
Fig. 1.
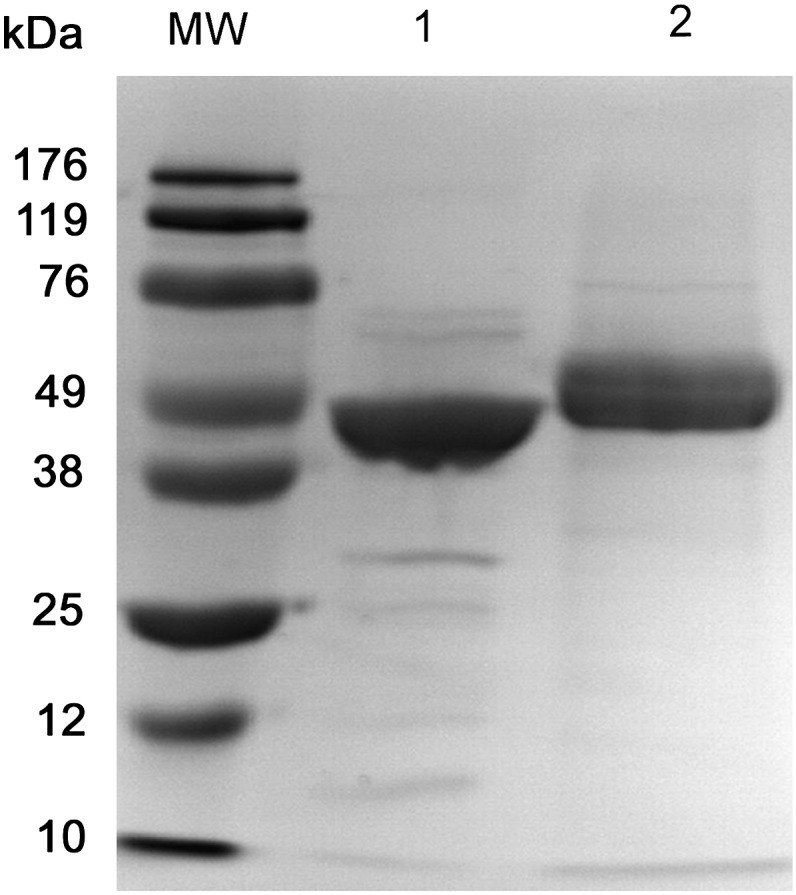
SDS-PAGE of purified recombinant mPTL and mPLRP2. An amount of 6 µg of each protein was dissolved in 2× protein sample buffer and separated by 10% SDS-PAGE. The gel was stained with GelCode Blue reagent. The positions of the molecular marker are given on the left of the figure. Lane 1: mPTL. Lane 2: mPLRP2. MW, molecular weight marker.
Bile salts and colipase effects
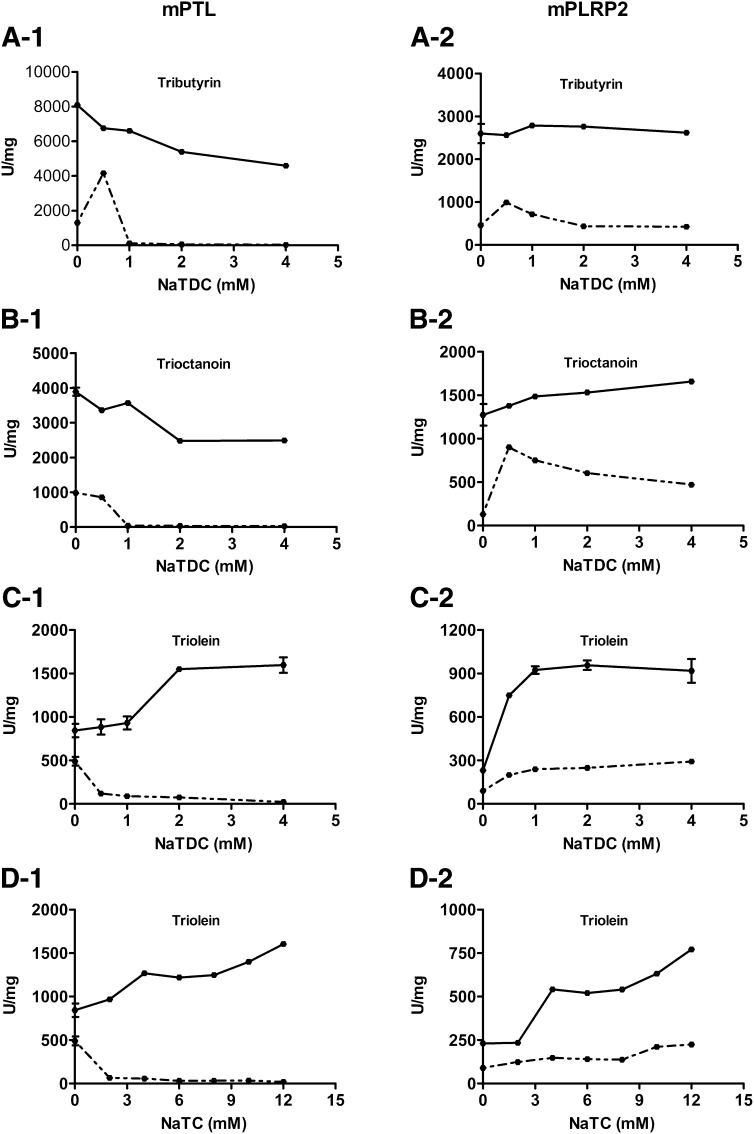
We first characterized the effects of bile salts and colipase on the catalytic activity of mPTL and mPLRP2 against various TAGs. In the absence of colipase, mPTL activity was stimulated by low NaTDC concentration, and it was completely inhibited by micellar concentrations of bile salt (>1.0 mM NaTDC) (Fig. 2). In contrast, mPLRP2 had activity against all three TAG substrates over NaTDC concentrations from 0 to 4 mM. Colipase increased lipase activities of both mPTL and mPLRP2 at all concentrations of bile salts tested, but the increase was greater for mPTL. As taurocholate is the major bile salt species in rodent bile, we also measured the lipase activity of mPTL and mPLRP2 against triolein in varying concentrations of NaTC. Although both lipases exhibited lower activity when assayed with NaTC, a similar pattern was observed with NaTDC (Fig. 2D). For each substrate and bile salt, the ratio of mPTL- and mPLRP2-specific activity in the presence of colipase remained relatively constant, ranging between 2 and 4. The activity of mPLRP2 exceeded that of hPLRP2 for three different triglyceride substrates (Table 1) (10). In particular, the activity of hPLRP2 was quite low against the long-chain substrate triolein in the presence of bile salt micelles. In contrast, hPLRP2 had better activity against phospholipids and galactolipids than did mPLRP2, although the mouse enzyme can hydrolyze both substrates (Table 1).
Fig. 2.
Effects of bile salts and colipase on the activities of mPTL and mPLRP2 on various TAGs. Lipolytic activities were measured using the standard 5 min pH-stat assay with 2 µg mPTL or mPLRP2 with emulsified tributyrin (A), trioctanoin (B), and triolein (C) at varying concentrations of NaTDC or with triolein at varying concentrations of NaTC (D), in the absence (dotted lines) or presence (solid lines) of a 5 molar excess of colipase. Each data point (mean ± SD) is the result of triplicate experiments.
TABLE 1.
Activity of mouse and human PLRP2 against various lipids
| Lipase Activity (U/mg) |
||
|---|---|---|
| Lipid | mPLRP2 | hPLRP2 |
| Tributyrin | 2623 ± 14 | 200 ± 9.0 |
| Trioctanoin | 1659 ± 18 | 190 ± 3.0 |
| Triolein | 918 ± 143 | 25 ± 0.4 |
| Digalactosyldiacylglycerol | 10.5 ± 0.8 | 100 ± 7.0 |
| Phosphatidylcholine | 5.9 ± 0.4 | 20 ± 6.4 |
Activities were measured at pH 8.0 using the pH-stat method as described in Materials and Methods. A 5-fold molar excess of colipase was included in the assays of the triglycerides. The assays were done at least three times. Data is mean ± SD.
Interaction with colipase
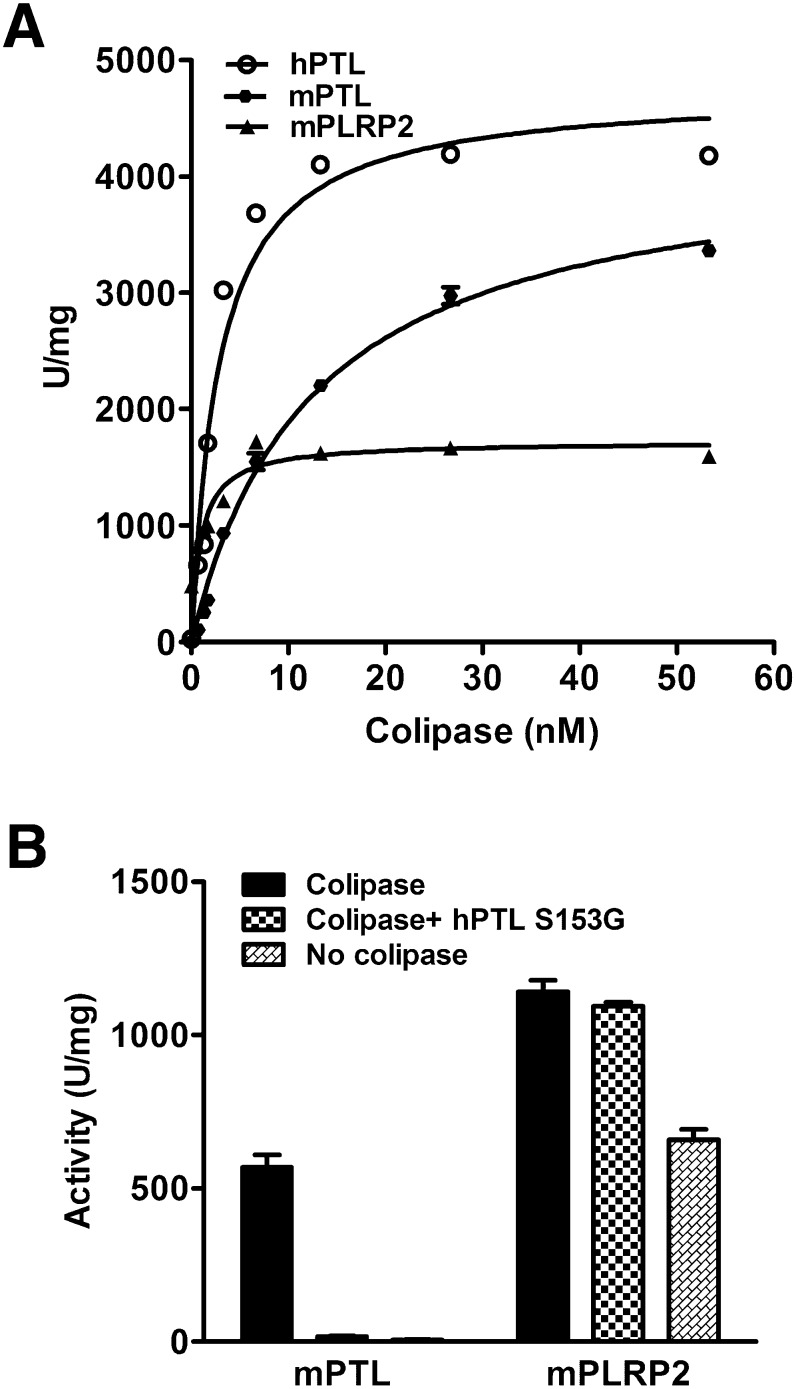
To better characterize the differences in colipase interaction between mPTL and mPLRP2, we measured the ability of colipase to activate these two lipases over a range of colipase concentrations in the presence of excess trioctanoin and a constant amount of lipase. From these data, we determined the concentration of colipase that restored half-maximal activity to the lipases (Kd) and the concentration that restored maximal activity (Bmax). The apparent Kd and Bmax were determined by nonlinear regression. Given the large excess of substrate in the assays, the apparent Kd most likely reflected the interaction between colipase and lipase rather than the interaction between colipase and the substrate. The colipase titration curves for mPTL and mPLRP2 are presented in Fig. 3A. As expected from the previous experiment, the Bmax of mPTL was higher than that of mPLRP2 (4244 versus 1717 U/mg). Unexpectedly, the apparent Kd for mPTL was 6-fold higher than that determined for mPLRP2 (12.5 versus 2.0 nM). The same values for hPTL were 4740 and 2.85 nM. Interestingly, the plateau for hPTL and mPLRP2 began at lower colipase concentrations than that for mPTL.
Fig. 3.
The colipase dependence of mPTL, hPTL, and mPLRP2. A: The assays were done using the pH-stat method in the presence of 4 mM NaTDC. Each assay included 2.6 nM of each lipase, 180 mM trioctanoin, and the indicated concentrations of colipase. B: Competition between S153G and mPTL or mPLRP2 for colipase. The assays were done in the pH-stat with trioctanoin and 4 mM NaTDC. An amount of 10 µg mPTL or 10 µg mPLRP2 (13 nM lipase), 1 µg colipase (6.5 nM, molar ratio of colipase to lipase of 0.5), and 10 µg S153G PTL when indicated were mixed and added to the assay reagents. Each assay was performed in triplicate.
To determine if there was a difference in the ability of mPLRP2 and mPTL to interact with colipase, we investigated the ability of the mouse lipases to compete with S153G, an inactive hPTL, for colipase binding (25). This mutant interacts with lipids and colipase indistinguishably from wild-type hPTL (26). We measured activity against trioctanoin in 4 mM NaTDC using a lipase-to-colipase molar ratio of 1 to 0.5, a range where lipase activity varies linearly with colipase concentration. Consequently, lipase activity is directly proportional to the formation of a colipase-PTL complex. The mouse lipase and colipase were premixed with or without S153G and added to the reaction mixture together. In the absence of S153G, mPTL had lower activity than mPLRP2 as predicted by the titration curves, suggesting that mPTL has lower affinity for colipase than does mPLRP2 (Fig. 3B). mPLRP2 had significant activity without colipase, and colipase stimulated activity another 40% under these conditions. The inclusion of S153G completely eliminated mPTL activity, but it had no significant effect on mPLRP2 activity. The results suggest that the interaction of mPTL with colipase is weaker than the interaction of either hPTL or mPLRP2 with colipase.
Ability of colipase to anchor lipases to substrate
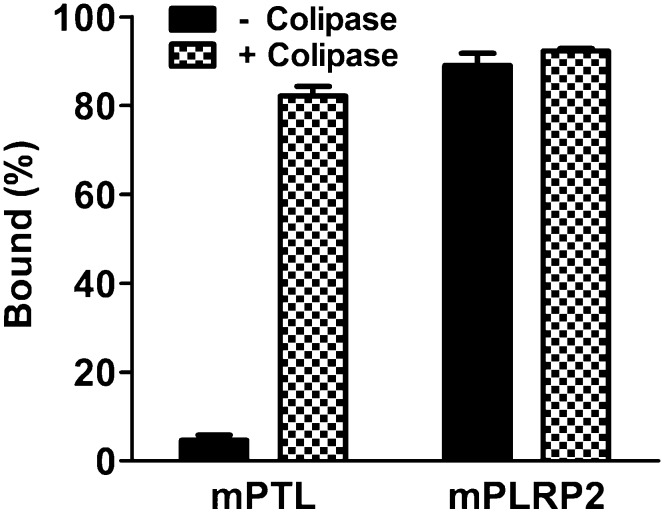
Current models of colipase function hypothesize that colipase restores activity to bile salt-inhibited lipase by anchoring lipase to the substrate. As the lipase activity of mPLRP2 and mPTL was at the same scale and the colipase affinity of mPLRP2 was greater than that of mPTL, we further measured the ability of colipase to anchor mPTL and mPLRP2 onto trioctanoin in 4 mM NaTDC. Like PTL from other species, mPTL lacked the ability to absorb to trioctanoin without colipase (Fig. 4). The addition of 5-fold molar excess of colipase anchored ∼85% of mPTL absorbed onto trioctanoin. In contrast, 90% of mPLRP2 absorbed to trioctanoin, regardless of the presence or absence of colipase. The interfacial-specific activity was calculated from the binding data and the overall specific activity of mPLRP2 in the presence and absence of colipase. In the absence of colipase, the specific activity of mPLRP2 at the lipid interface was 560 U/mg. In the presence of colipase, the interfacial-specific activity increased to 1,850 U/mg. Thus, the main effect of colipase is to increase the specific activity of mPLRP2 at the interface, whereas the main effect of colipase is to increase interfacial absorption of mPTL.
Fig. 4.
Binding of mPTL and mPLRP2 to trioctanoin. Binding was determined with 6.0 µg mPTL or mPLRP2 and expressed as the percentage of total lipase activity bound to the lipid phase after centrifugation, as described in “Materials and Methods.” Filled bar, mPTL; hatched bar, mPLRP2.
Compensation of total lipase activity by mPTL or mPLRP2
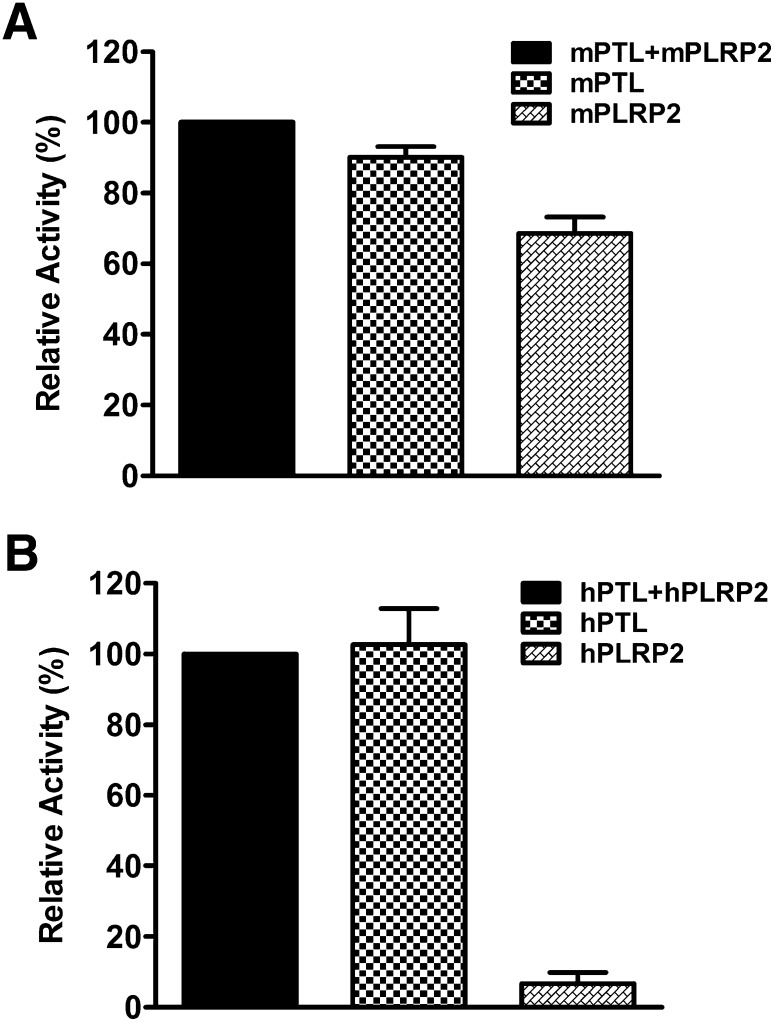
To determine if mPLRP2 can compensate for mPTL deficiency under physiological conditions, we set up assays against triolein containing physiological ratios of the two lipases and colipase. We first quantified the molar ratio of mPTL to mPLRP2 from adult mouse pancreas extracts by Western blot analysis (Fig. 5A). The molar ratio of mPTL to mPLRP2 was 2.8 ± 0.8 (Fig. 5B), which is similar to the molar ratio of PTL to PLRP2 protein in human pancreatic juice assayed by ELISA (27). To closely mimic the molar ratio of mPTL to mPLRP2 in vivo, we measured the total lipase activity of 2.5 µg mPTL and 1.0 µg mPLRP2 in the presence of limited colipase. We chose a 1:1 molar ratio of colipase to mPTL because that is the approximate ratio of the two proteins in pancreatic juice of various species, including humans (28). We also assayed the lipase activity of 2.5 µg mPTL and 1.0 µg mPLRP2 alone under the same conditions to mimic the situation when one of the lipases is absent. Under these simulated conditions, mPTL had 90% of the activity of the two lipases together (Fig. 6A). Importantly, mPLRP2 alone still had 70% of the total lipase activity for both lipases together (Fig. 6A). These findings indicate that under physiological conditions, mPTL and mPLRP2 can compensate for the absence of the other lipase. In contrast, the same experiment with human PTL and PLRP2 showed that hPLRP2 could not compensate for the absence of hPTL (Fig. 6B).
Fig. 5.
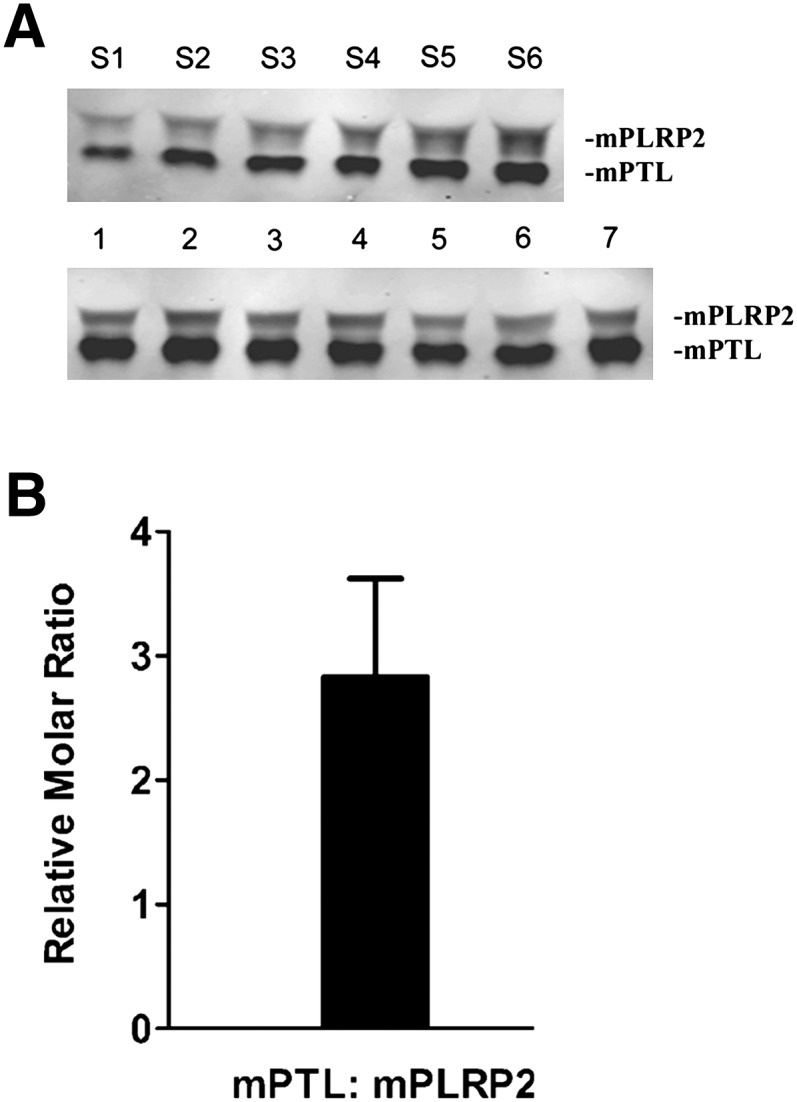
Immunoblot and quantification of mPTL and mPLRP2. A: Pancreas total protein extract samples were run on a 10% SDS-PAGE along with purified recombinant mPTL and mPLRP2, transferred onto nitrocellulose membrane, and blotted with rabbit anti-human PTL polyclonal antibody. Top band: mPLRP2; Lower band: mPTL. Lanes S1-S6, purified recombinant mPTL and mPLRP2 protein samples with varying amounts; Lanes 1-7, individual pancreas protein extraction sample from seven adult mice. B: Quantification of protein molar ratio of mPTL:mPLRP2.
Fig. 6.
Mutual compensation of lipase activity by PTL and PLRP2. The assays were done by the standard pH-stat method using excess triolein as substrate with limited colipase. Each assay included either 2.5 µg PTL + 1.0 µg PLRP2, 2.5 µg PTL, or 1.0 µg PLRP2 in the presence of 0.5 µg colipase. The results were expressed as the percentage of total lipase activity of 2.5 µg PTL + 1.0 µg PLRP2. Each data point (mean ± SD) is the result of triplicate experiments. A: Mouse lipase. B: Human lipase.
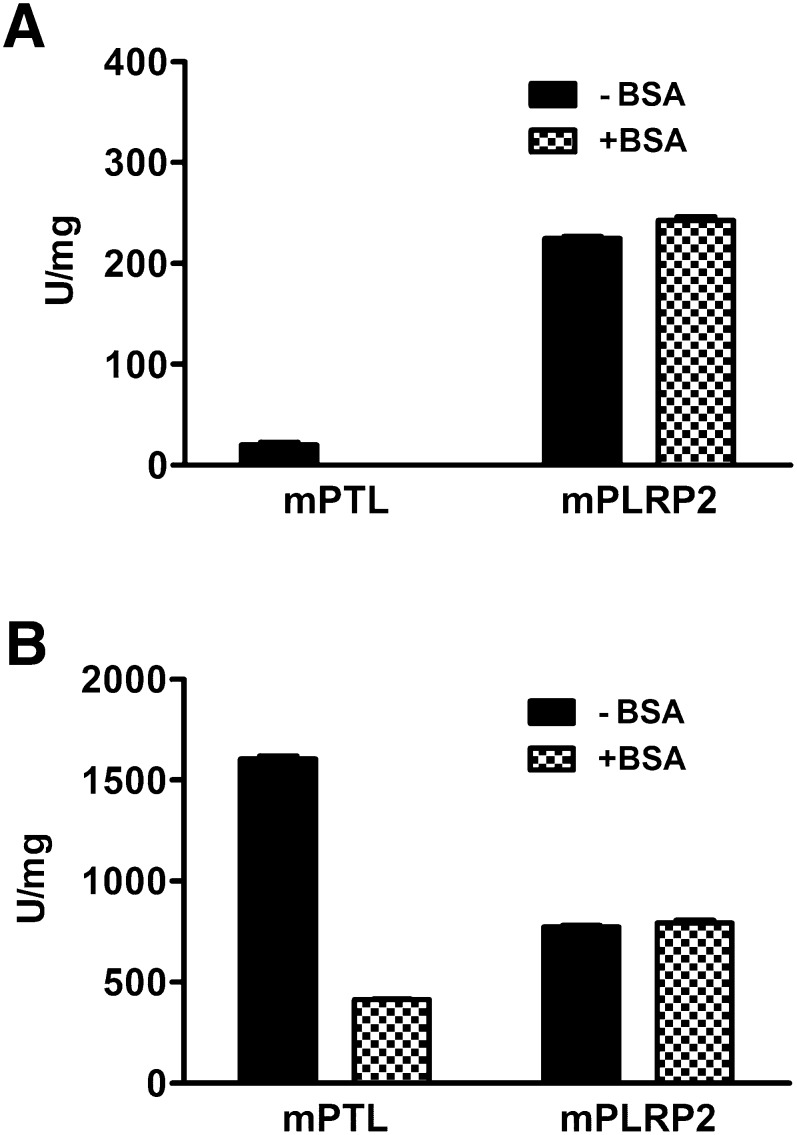
Effects of BSA on mPTL and mPLRP2 lipase activity
It's well known that denatured dietary proteins add further complexity to the lipid-water interface and present obstacles for PTL binding with interfacial substrates (29). To determine if mPLRP2 was similarly affected by protein, we tested the effect of BSA on the lipase activity of mPTL and mPLRP2 in 12 mM NaTC. In the absence of colipase, BSA completely inhibited mPTL, but it had no effect on mPLRP2 activity (Fig. 7A). In the presence of colipase, the lipase activity of mPTL was inhibited by BSA (1,600 compared with 400 U/mg), whereas the lipase activity of mPLRP2 remained unchanged (800 U/mg) (Fig. 7B). The addition of BSA reversed the ratio of specific activity of mPTL to mPLRP2 from 2:1 to 1:2.
Fig. 7.
Effects of BSA on mPTL and mPLRP2 lipase activity. Lipolytic activities were measured in the standard 5 min pH-stat assay with 2 µg mPTL or mPLRP2 on emulsified triolein in 12 mM NaTC with or without 3.75% BSA in the absence (A) or presence (B) of a 5 molar excess of colipase. Each data point (mean ± SD) is the result of triplicate experiments.
DISCUSSION
Dietary fat assimilation has been an interesting research field for years. Strategies have been explored to improve dietary fat digestion and absorption for proper development in neonates, and to modestly reduce fat digestion and absorption in treating obesity in adults. Genetically modified mice are common, useful models to examine the function of lipases in dietary fat digestion. Extrapolating results in mice to humans is always problematic, particularly when clear examples of human deficiency are not readily available to compare with the mouse model. In the absence of information regarding human deficiencies, in vitro biochemical functional studies comparing human to mouse lipases can provide a solid foundation to interpret data obtained from mouse models and to better appreciate the physiologic implications in humans. Despite the use of mouse models to study the function of lipases in digestion, little is known about the biochemical properties of mouse lipases. To help reconcile data in mice with human physiology, we expressed recombinant mPTL and mPLRP2 in Pichia pastoris GS115 and characterized their biochemical properties in vitro.
Several conclusions about mPTL and mPLRP2 pertinent to their role in dietary fat digestion can be made from the reported investigations. First, mPLRP2 has significant activity against TAGs, comparable to that of mPTL. Second, unlike mPTL, which is inhibited by bile salt micelles, mPLRP2 has activity in the presence of bile salt micelles even when colipase is absent. Third, colipase stimulates the activity of mPLRP2 at all concentrations of bile salts. In fact, the apparent affinity of mPLRP2 for colipase is greater than that of mPTL. Differences in interfacial absorption and in activity in the presence of BSA suggest that the role in the molecular mechanism of lipolysis for colipase may differ between mPLRP2 and mPTL. Lastly, the physiological concentration of mPLRP2 is sufficient to compensate for the loss of mPTL activity.
Both mPTL and mPLRP2 exhibit robust lipase activity against various TAGs in the presence of colipase, and their lipase activities are comparable. The activity of mPTL against substrates with varying acyl chain length ranged from 2- to 4-fold higher than the activity of mPLRP2 at all bile salt concentrations. Still, the activity of mPLRP2 is much higher than CEL activity, suggesting that mPLRP2 can contribute to TAG digestion in the mouse (23). We provided additional evidence for a physiological role of mPLRP2 in vivo by performing assays with physiological ratios of mPTL, mPLRP2, and colipase. Under these conditions, TAG hydrolysis by either mPTL or mPLRP2 alone was nearly as efficient as when both lipases were included in the assay. These results provide an explanation for the absence of significant effects on dietary fat digestion in adult mice deficient in either mPLRP2 or mPTL (6, 30). Under physiological conditions, mPTL and mPLRP2 compete for the limited colipase to boost their lipase activity. The deficiency of mPTL or mPLRP2 increased the molar ratio of available colipase for the remaining lipase, a situation that would increase the activity of the remaining lipase. In each case, the activity of the remaining lipase should be sufficient to prevent steatorrhea, even without a compensatory increase in mRNA (6). The somewhat lower activity of mPLRP2 would also explain the observation that fat digestion was delayed in mPTL-deficient mice (6).
Mouse PTL and PLRP2 had distinctive responses to bile salts, protein, and colipase. Bile salt micelles completely inhibited mPTL, and colipase was required for activity. Although colipase increased mPLRP2 activity, it was not required; mPLRP2 had significant activity in the absence of colipase. Mouse PLRP2 efficiently absorbed onto lipid and bile salt emulsions without colipase, whereas mPTL lacked the ability to absorb onto the emulsion without colipase. Furthermore, in the presence of colipase, BSA substantively inhibited mPTL lipase activity, but it had no apparent impact on mPLRP2 lipase activity. Collectively, these results suggest that the role of colipase in the ternary complex with the lipid-water interface and mPLRP2 is distinct from that of mPTL. With mPTL, colipase has a primary role in anchoring mPTL at the interface. Colipase may also stabilize the lid domain of mPTL in an active conformation (31). Through these two functions, colipase facilitates the activity of PTL and is, in fact, required for PTL activity in vivo, as evidenced by studies with colipase-deficient mice (2). In contrast, colipase is not required for mPLRP2 absorption to the interface. Even so, colipase stimulates the activity of mPLRP2. It may do this by stabilizing the lid domain in an active conformation or by orienting mPLRP2 on the interface in a more productive complex. Unexpectedly, the interaction between mPLRP2 and colipase appears stronger than the interaction between hPTL and colipase, and mPTL appears to have weaker affinity for colipase than does either hPTL or mPLRP2.
The structural basis for the differences in behavior among mPTL, hPTL, and mPLRP2 is not evident from examining the structures. Comparison of the residues implicated in colipase binding by the crystal structure of the PTL-colipase complex shows that they are well conserved. The only exception is the substitution of the tyrosine at position 406 in mPTL and hPTL with an asparagine in mPLRP2. The same substitution is present in hPLRP2, which is not activated by colipase, making it unlikely this change contributes significantly to the differences between the lipases. Even though the presumed colipase interacting residues in the lid domain of the three lipases are conserved, there are important structural differences in the lid domain that increase the mobility of the human and rat PLRP2 lid domains compared with the domain in PTL (11, 32). Residues in the lid of hPTL form polar interactions with residues in the core of the protein. These residues are conserved between hPTL and mPTL. The residues in the lid are not conserved in mPLRP2. Any alteration in the orientation of the lid domain could influence the interaction among the lipase, colipase, and the interface as the structure of the lid is known to mediate interfacial and colipase binding of PTL and PLRP2 (22). Data with hPTL demonstrated that the residues in the β5′ loop contribute to the interaction of lipase and colipase in the presence of bile salt micelles (33). There are several differences in the sequence of these 10 residues between mouse and human PTL and mPLRP2, notably leucine for proline at position 411, and arginine for a leucine at position 413. Overall, the hydrophilic to lipophilic balance changes from 0.8 in mPTL and hPTL to 1.0 in mPLRP2. The significance of the difference is unclear. It remains likely that other regions of the proteins contribute to the interaction as well.
The kinetic behavior of mPTL and mPLRP2 aids in the interpretation of published data in colipase-deficient and PLRP2-deficient mice. In adult mice, colipase deficiency would be expected to eliminate the contribution of PTL activity completely and also substantively reduce PLRP2 activity. Consequently, the intestinal TAG lipase activity derived from pancreas is likely attributable to CEL and PLRP2. Thus, colipase-deficient adult mice exhibit normal TAG digestion when fed a low-fat diet and severe fat maldigestion when fed a high-fat diet. Likely, the combination of gastric lipase, CEL, and PLRP2, even with diminished activity, is sufficient when dietary intake of fat is low but not when intake is increased. In newborns, our earlier report showed that colipase is required for fat digestion through an unclear PTL-independent mechanism, as PTL is not expressed until near weaning, although indirect data suggested that PLRP2 accounted for the colipase-stimulated TAG lipase activity in the newborn (2, 5, 9, 34–37). Our in vitro characterization supports that conclusion. In this scenario, the diminished activity of mPLRP2 in the absence of colipase was not sufficient to digest the high concentrations of dietary fat in mouse breast milk, resulting in steatorrhea.
Importantly, our findings have important implications in extrapolating results in mice to humans. The biochemical properties of mouse and human PTL are similar, but there are important differences in the properties of mouse and human PLRP2. Human PLRP2 is completely inhibited by bile salt micelles when trioctanoin or triolein is the substrate. Colipase restores activity to human PLRP2 in 4 mM NaTDC, although the activity is significantly lower than the activity of mPLRP2 (10). Based on in vitro data, it has been suggested that hPLRP2 is predominantly a galactolipase and, perhaps, a monoacylglycerol lipase. The situation differs from the mouse, where available data suggest mPLRP2 can make a significant contribution to TAG digestion. These major differences between mouse and human PLPR2 question the reliability of extrapolating results in mice to humans, unless the behavior of hPLRP2 is completely different in vivo than in vitro. This caveat should be kept in mind because the conditions of the assay may have a large effect on activity. For instance, the inclusion of gum arabic in the assay significantly increased the activity of hPLRP2 against triolein, compared with emulsification with bile salts (10). The activity of hPLRP2 in the duodenal lumen with a test meal is unknown at present. Interestingly, hPLRP2 had no activity against bovine milk, a finding that has implications for the role of hPLRP2 in human newborns (10). Even though humans have delayed expression of PTL similar to the pattern described in mice, further studies in human newborns are required before it is concluded that PLRP2 is essential for efficient fat digestion in human newborns, as is the case in mice (37).
In summary, we have expressed and characterized recombinant mPTL and mPLRP2 in vitro. Unlike hPLRP2, mPLRP2 possesses high lipase activity against various TAG substrates with specific activities comparable to those of mPTL. Mouse PLRP2 also exhibits excellent abilities to interact with colipase and to absorb onto the lipid-interface, and to resist inhibition by proteins. The unique biochemical characteristics of mPLRP2 indicate that PLRP2 may contribute significantly to TAG digestion in mice and may be able to compensate when PTL is absent or inhibited. Because the properties of mPLRP2 differ from those of hPLRP2, results regarding dietary fat digestion in mice may not be applicable to humans. Consideration of these differences should be kept in mind when interpreting data obtained in mouse models.
Footnotes
Abbreviations:
- CEL
- carboxyl ester lipase
- NaTC
- sodium taurocholate
- NaTDC
- sodium taurodeoxycholate
- PLRP2
- pancreatic lipase-related protein-2
- hPLRP2
- human PLRP2
- mPLRP2
- mouse PLRP2
- rPLRP2
- rat PLRP2
- PTL
- pancreatic triglyceride lipase
- hPTL
- human PTL
- mPTL
- mouse PTL
- TAG
- triacylglycerol
This work was supported by National Institutes of Health Grant DK-080820. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Carriere F., Barrowman J. A., Verger R., Laugier R. 1993. Secretion and contribution to lipolysis of gastric and pancreatic lipases during a test meal in humans. Gastroenterology. 105: 876–888. [DOI] [PubMed] [Google Scholar]
- 2.D'Agostino D., Cordle R. A., Kullman J., Erlanson-Albertsson C., Muglia L. J., Lowe M. E. 2002. Decreased postnatal survival and altered body weight regulation in procolipase deficient mice. J. Biol. Chem. 277: 7170–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghishan F. K., Moran J. R., Durie P. R., Greene H. L. 1984. Isolated congenital lipase-colipase deficiency. Gastroenterology. 86: 1580–1582. [PubMed] [Google Scholar]
- 4.Howles P. N., Carter C. P., Hui D. Y. 1996. Dietary free and esterified cholesterol absorption in cholesterol esterase (bile salt-stimulated lipase) gene-targeted mice. J. Biol. Chem. 271: 7196–7202. [DOI] [PubMed] [Google Scholar]
- 5.Lowe M. E., Kaplan M. H., Jackson-Grusby L., D'Agostino D., Grusby M. J. 1998. Decreased neonatal dietary fat absorption and T cell cytotoxicity in pancreatic lipase-related protein 2-deficient mice. J. Biol. Chem. 273: 31215–31221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huggins K. W., Camarota L. M., Howles P. N., Hui D. Y. 2003. Pancreatic triglyceride lipase deficiency minimally affects dietary fat absorption but dramatically decreases dietary cholesterol absorption in mice. J. Biol. Chem. 278: 42899–42905. [DOI] [PubMed] [Google Scholar]
- 7.Lowe M. E. 1994. Pancreatic triglyceride lipase and colipase: Insights into dietary fat digestion. Gastroenterology. 107: 1524–1536. [DOI] [PubMed] [Google Scholar]
- 8.Gilham D., Labonte E. D., Rojas J. C., Jandacek R. J., Howles P. N., Hui D. Y. 2007. Carboxyl ester lipase deficiency exacerbates dietary lipid absorption abnormalities and resistance to diet-induced obesity in pancreatic triglyceride lipase knockout mice. J. Biol. Chem. 282: 24642–24649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Agostino D., Lowe M. E. 2004. Pancreatic lipase-related protein 2 is the major colipase-dependent pancreatic lipase in suckling mice. J. Nutr. 134: 132–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eydoux C., De Caro J., Ferrato F., Boullanger P., Lafont D., Laugier R., Carriere F., De Caro A. 2007. Further biochemical characterization of human pancreatic lipase-related protein 2 expressed in yeast cells. J. Lipid Res. 48: 1539–1549. [DOI] [PubMed] [Google Scholar]
- 11.Eydoux C., Spinelli S., Davis T. L., Walker J. R., Seitova A., Dhe-Paganon S., De Caro A., Cambillau C., Carriere F. 2008. Structure of human pancreatic lipase-related protein 2 with the lid in an open conformation. Biochemistry. 47: 9553–9564. [DOI] [PubMed] [Google Scholar]
- 12.Sias B., Ferrato F., Grandval P., Lafont D., Boullanger P., De Caro A., Leboeuf B., Verger R., Carriere F. 2004. Human pancreatic lipase-related protein 2 is a galactolipase. Biochemistry. 43: 10138–10148. [DOI] [PubMed] [Google Scholar]
- 13.Jayne S., Kerfelec B., Foglizzo E., Granon S., Hermoso J., Chapus C., Crenon I. 2002. Activation of horse PLRP2 by bile salts does not require colipase. Biochemistry. 41: 8422–8428. [DOI] [PubMed] [Google Scholar]
- 14.Amara S., Lafont D., Fiorentino B., Boullanger P., Carriere F., De Caro A. 2009. Continuous measurement of galactolipid hydrolysis by pancreatic lipolytic enzymes using the pH-stat technique and a medium chain monogalactosyl diglyceride as substrate. Biochim Biophys Acta. 1791: 983–990. [DOI] [PubMed] [Google Scholar]
- 15.Andersson L., Carriere F., Lowe M. E., Nilsson A., Verger R. 1996. Pancreatic lipase-related protein 2 but not classical pancreatic lipase hydrolyzes galactolipids. Biochim. Biophys. Acta. 1302: 236–240. [DOI] [PubMed] [Google Scholar]
- 16.Jennens M. L., Lowe M. E. 1995. Rat GP-3 is a pancreatic lipase with kinetic properties that differ from colipase-dependent pancreatic lipase. J. Lipid Res. 36: 2374–2381. [PubMed] [Google Scholar]
- 17.Cordle R. A., Lowe M. E. 1998. Purification and characterization of human procolipase expressed in yeast cells. Protein Expr. Purif. 13: 30–35. [DOI] [PubMed] [Google Scholar]
- 18.Lowe M. E. 1997. New pancreatic lipases: gene expression, protein secretion, and the newborn. Methods Enzymol. 284: 285–297. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y., Lowe M. E. 1998. Human pancreatic triglyceride lipase expressed in yeast cells: purification and characterization. Protein Expr. Purif. 13: 36–40. [DOI] [PubMed] [Google Scholar]
- 20.Cordle R. A., Lowe M. E. 1998. The hydrophobic surface of colipase influences lipase activity at an oil-water interface. J. Lipid Res. 39: 1759–1767. [PubMed] [Google Scholar]
- 21.Crenon I., Jayne S., Kerfelec B., Hermoso J., Pignol D., Chapus C. 1998. Pancreatic lipase-related protein type 1: a double mutation restores a significant lipase activity. Biochem. Biophys. Res. Commun. 246: 513–517. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y., Lowe M. E. 2000. The open lid mediates pancreatic lipase function. J. Lipid Res. 41: 48–57. [PubMed] [Google Scholar]
- 23.Hansson L., Blackberg L., Edlund M., Lundberg L., Stromqvist M., Hernell O. 1993. Recombinant human milk bile salt-stimulated lipase. Catalytic activity is retained in the absence of glycosylation and the unique proline-rich repeats. J. Biol. Chem. 268: 26692–26698. [PubMed] [Google Scholar]
- 24.Crandall W. V., Lowe M. E. 2001. Colipase residues Glu64 and Arg65 are essential for normal lipase-mediated fat digestion in the presence of bile salt micelles. J. Biol. Chem. 276: 12505–12512. [DOI] [PubMed] [Google Scholar]
- 25.Chahinian H., Bezzine S., Ferrato F., Ivanova M. G., Perez B., Lowe M. E., Carriere F. 2002. The beta 5′ loop of the pancreatic lipase C2-like domain plays a critical role in the lipase-lipid interactions. Biochemistry. 41: 13725–13735. [DOI] [PubMed] [Google Scholar]
- 26.Bezzine S., Ferrato F., Ivanova M. G., Lopez V., Verger R., Carriere F. 1999. Human pancreatic lipase: colipase dependence and interfacial binding of lid domain mutants. Biochemistry. 38: 5499–5510. [DOI] [PubMed] [Google Scholar]
- 27.Eydoux C., Aloulou A., De Caro J., Grandval P., Laugier R., Carriere F., De Caro A. 2006. Human pancreatic lipase-related protein 2: tissue localization along the digestive tract and quantification in pancreatic juice using a specific ELISA. Biochim. Biophys. Acta. 1760: 1497–1504. [DOI] [PubMed] [Google Scholar]
- 28.Gaskin K. J., Durie P. R., Lee L., Hill R., Forstner G. G. 1984. Colipase and lipase secretion in childhood-onset pancreatic insufficiency. Delineation of patients with steatorrhea secondary to relative colipase deficiency. Gastroenterology. 86: 1–7. [PubMed] [Google Scholar]
- 29.Lowe M. E. 2002. The triglyceride lipases of the pancreas. J. Lipid Res. 43: 2007–2016. [DOI] [PubMed] [Google Scholar]
- 30.Miller R., D'Agostino D., Erlanson-Albertsson C., Lowe M. E. 2009. Enterostatin deficiency increases serum cholesterol but does not influence energy homeostasis in mice. Am. J. Physiol. Endocrinol. Metab. 297: E856–E865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lowe M. E. 1997. Colipase stabilizes the lid domain of pancreatic triglyceride lipase. J. Biol. Chem. 272: 9–12. [DOI] [PubMed] [Google Scholar]
- 32.Roussel A., Yang Y., Ferrato F., Verger R., Cambillau C., Lowe M. 1998. Structure and activity of rat pancreatic lipase-related protein 2. J. Biol. Chem. 273: 32121–32128. [DOI] [PubMed] [Google Scholar]
- 33.Freie A. B., Ferrato F., Carriere F., Lowe M. E. 2006. Val-407 and Ile-408 in the beta5′-loop of pancreatic lipase mediate lipase-colipase interactions in the presence of bile salt micelles. J. Biol. Chem. 281: 7793–7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X., Lindquist S., Lowe M., Noppa L., Hernell O. 2007. Bile salt-stimulated lipase and pancreatic lipase-related protein 2 are the dominating lipases in neonatal fat digestion in mice and rats. Pediatr. Res. 62: 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller R., Lowe M. E. 2008. Carboxyl ester lipase from either mother's milk or the pancreas is required for efficient dietary triglyceride digestion in suckling mice. J. Nutr. 138: 927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Payne R. M., Sims H. F., Jennens M. L., Lowe M. E. 1994. Rat pancreatic lipase and two related proteins: enzymatic properties and mRNA expression during development. Am. J. Physiol. 266: G914–G921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y., Sanchez D., Figarella C., Lowe M. E. 2000. Discoordinate expression of pancreatic lipase and two related proteins in the human fetal pancreas. Pediatr. Res. 47: 184–188. [DOI] [PubMed] [Google Scholar]