Abstract
Peroxisome proliferator-activated receptor-α (PPARα) is a dietary lipid sensor, whose activation results in hypolipidemic effects. In this study, we investigated whether PPARα activation affects energy metabolism in white adipose tissue (WAT). Activation of PPARα by its agonist (bezafibrate) markedly reduced adiposity in KK mice fed a high-fat diet. In 3T3-L1 adipocytes, addition of GW7647, a highly specific PPARα agonist, during adipocyte differentiation enhanced glycerol-3-phosphate dehydrogenase activity, insulin-stimulated glucose uptake, and adipogenic gene expression. However, triglyceride accumulation was not increased by PPARα activation. PPARα activation induced expression of target genes involved in FA oxidation and stimulated FA oxidation. In WAT of KK mice treated with bezafibrate, both adipogenic and FA oxidation-related genes were significantly upregulated. These changes in mRNA expression were not observed in PPARα-deficient mice. Bezafibrate treatment enhanced FA oxidation in isolated adipocytes, suppressing adipocyte hypertrophy. Chromatin immunoprecipitation (ChIP) assay revealed that PPARα was recruited to promoter regions of both adipogenic and FA oxidation-related genes in the presence of GW7647 in 3T3-L1 adipocytes. These findings indicate that the activation of PPARα affects energy metabolism in adipocytes, and PPARα activation in WAT may contribute to the clinical effects of fibrate drugs.
Keywords: PPARα, adipocyte differentiation, insulin resistance, metabolic syndrome
The prevalence of obesity has markedly increased over the last few decades not only in wealthy industrialized countries, but also in poor underdeveloped nations (1). Obesity and overweight have adverse health effects and affect the risk and prognosis of many serious medical conditions, such as type 2 diabetes, coronary heart disease, high blood pressure, and some forms of cancer. Obesity causes excess fat accumulation not only in various tissues, particularly adipose tissues, but also in other insulin-responsive organs such as the skeletal muscle and liver, predisposing one to the development of insulin resistance (2–5). However, the molecular mechanisms underlying insulin resistance and obesity have not been fully clarified, and effective therapeutic approaches are currently of general interest.
Peroxisome proliferator-activated receptors (PPAR) -α, -γ, and -δ are FA-activated nuclear transcription factors that control mRNA expression of numerous genes involved in energy metabolism (6–8). In particular, PPARα is expressed mostly in tissues with high rates of FA oxidation and peroxisomal metabolism (9), such as the liver, brown fat, and heart (10–12). In these tissues, PPARα regulates mRNA expression of genes involved in FA oxidation, and synthetic PPARα agonists, such as fibrates, decrease circulating lipid levels and are commonly used to treat hyperlipidemia and other dyslipidemic states (9, 13). PPARα-deficient (PPARα−/−) mice exhibit higher levels of serum triglyceride (TG) (14). Moreover, these mice show extensive hepatic lipid accumulation and increased gonadal adipose storage and plasma FFA levels (15). In addition, the presence of severe hypoketonemia, hypoglycemia, and hypothermia in PPARα−/− mice clearly supports a function of PPARα in metabolic regulation.
Previous studies showed that adipose tissue dysfunction plays an important role in the development of obesity and obesity-associated diseases (16, 17). Adipose tissue also participates in the regulation of energy homeostasis as an important endocrine organ that secretes many biologically active molecules called adipocytokines (16–19). The secretion pattern of adipocytokines is closely related to adipocyte condition (18). Small adipocytes secrete insulin-sensitizing hormones such as adiponectin, although hypertrophied adipocytes exhibit low expression levels of these factors but increased expression levels of hormones related to insulin resistance, such as such as tumor necrosis factor-α (TNFα), monocyte chemoattractant protein-1 (MCP-1), and interleukin-6 (IL-6), resulting in insulin resistance observed in obesity (19). Indeed, thiazolidinediones (TZD), insulin sensitizers, promote differentiation of preadipocytes by PPARγ activation to increase the number of small adipocytes and to decrease the number of hypertrophied adipocytes by promoting apoptosis (20).
Besides their hypolipidemic effects, PPARα activators attenuate adiposity in animal models of obesity and type-2 diabetes mellitus (21, 22). Recently, several studies have shown that PPARα is expressed in the adipose tissue of humans and rodents, indicating that adipose tissue might also be a target organ of PPARα activators (22–25). Hiuge et al. demonstrated that fibrates directly and transcriptionally increase adiponectin expression level via adipose PPARα (23). Furthermore, it was suggested that PPARα ligands directly stimulate lipolysis in wild-type (WT) adipocytes, but this effect is not observed in PPARα-deficient adipocytes (25). These previous findings indicate that PPARα functions in adipocytes. However, the role of PPARα in adipose tissue is not fully understood.
In this study, we investigated whether PPARα activation affects the energy metabolism in white adipose tissue (WAT). Treatment with bezafibrate, a PPARα agonist, markedly attenuated adiposity in WT mice but not in PPARα−/− mice fed a high-fat diet (HFD). Both genetic and HFD-induced obesity decreased PPARα mRNA expression level, and fasting increased the expression level in WAT. PPARα activation promoted both adipocyte differentiation and FA oxidation in 3T3-L1 adipocytes. These effects were also observed in vivo, which resulted in the enhancement of whole-body oxygen consumption and suppression of adipocyte hypertrophy. These findings indicate that the pharmacological activation of PPARα affects energy metabolism in adipocytes and that the PPARα activation in WAT may contribute to the clinical effects of fibrate drugs.
MATERIALS AND METHODS
Animal experiments
All the mice were maintained in a temperature-controlled (23°C) facility with a constant 12 h light/dark cycle and free access to water. For the analysis of organ PPARα expression levels, 8-week-old male C57BL/6 mice (CLEA Japan, Tokyo, Japan) were fed a standard diet (CRF-1, Charles River Japan, Tokyo, Japan). Non-fasted, fasted (24 h), and fasted (24 h)/refed (24 h) C57BL/6 mice were used before euthanization. After euthanization, their tissues were harvested for RNA isolation. The harvested tissues were immediately frozen in liquid nitrogen and stored at −80°C until use. We used male C57BL/6 mice fed either the standard diet or 60% HFD (D12492 Research Diet, MO) for 12 months for the diet-induced obese model mice. Male ob/ob mice and lean control mice (age 12 weeks from Charles River Japan) received a standard diet for genetically obese mice.
To investigate the effect of the PPARα agonist on adipose tissue metabolism, we used KK mice or WT and PPARα−/− mice with a Sv/129 genetic background. Five-week-old male KK (CLEA Japan) mice were maintained for five to eight weeks either on HFD or on HFD containing 0.2% bezafibrate. The energy intake of all mice was adjusted by pair-feeding. Five to eight weeks after feeding, the mice were subjected to analyses of their oxygen consumption, locomotor activity, oral glucose tolerance test (OGTT), insulin tolerance test (ITT), mRNA expression, FA oxidation in isolated adipocytes, and histological features. Eight-week-old WT and PPARα−/− mice were fed HFD or HFD containing 0.2% bezafibrate for six weeks. The same amount of food was given to all mice. Ten h after fasting, tissues were harvested for RNA analysis. The animal care procedures and methods were approved by the Animal Care Committee of Kyoto University.
To analyze the oxygen consumption of mice, male KK mice fed HFD or HFD containing 0.2% bezafibrate for five weeks were used. The oxygen consumption rate (OCR) of mice under the fed condition was measured using an indirect calorimetric system (Oxymax, Columbus Instruments, OH) every 9 min for 20 h. The measurements started at 9:00 PM and ended at 6:00 PM (both the dark and light phases were 10 h).
Male KK mice were maintained for five weeks either on HFD or HFD containing 0.2% bezafibrate. For OGTT, d-glucose (2 g/kg body weight) was administered through a gastric feeding tube after overnight fasting. For ITT, human insulin (Eli Lilly Japan, Kobe, Japan) was injected intraperitoneally (0.6 units/kg body weight) into nonfasted animals. Blood samples were collected from the tail vein before and 30, 60, 90, and 120 min after injection. Plasma glucose level was determined by the glucose C-test Wako (Wako Pure Chemicals, Osaka, Japan) in accordance with the manufacturer's protocols.
Epididymal WAT was removed from each animal, fixed in 10% formaldehyde/PBS, and maintained at 4°C until use. The fixed samples were embedded in paraffin. They were cut into 12 µm sections using a microtome and mounted on silanized slides. Adipocyte size distribution was determined using National Institutes of Health (NIH) Image J software.
Cell culture and retrovirus-mediated PPARα expression
To prepare stromal-vascular (SV) cells and adipocytes from WAT, epididymal WAT from male mice was removed under sterile conditions and washed in KRH buffer (20 mM HEPES, 140 mM NaCl, 5 mM KCl, 2.5 mM MgSO4, 1 mM CaCl2; pH 7.4). Minced tissue was transferred to a sterile polypropylene tube containing collagenase (1 mg/ml, type II, Sigma) and 1% FA-free BSA (BSA). After incubation at 37°C for 45 min with shaking, the digested tissue was filtered through a sterile 250 μm nylon mesh. The digested tissue was centrifuged at 200 g for 2 min, and mature adipocytes and SV cells were collected. Both cell types were washed twice by centrifugation using KRH buffer containing 1% FA-free BSA. After washing, cells were subjected to each assay.
3T3-L1 murine preadipocytes (from American Type Culture Collection, Manassas, VA) were cultured as previously described (26). Human multipotent adipose tissue-derived stem cells were cultured as previously reported (27). Cells 14 days after the induction of differentiation were used as mature adipocytes.
A PCR fragment coding mouse PPARα obtained by RT-PCR using total liver RNA from C57BL/6 mice was inserted into the site between BamHI and NotI of pMX-puro (a kind gift from Dr. T. Kitamura, University of Tokyo, Tokyo, Japan) (pMX-PPARα). The PCR primers used were as follows: the 5′-primer, TTTGGATCCATGGTGGACACAGAGAGCCCCATC; and the 3′-primer, TTTGCGGCCGCTCAGTACATGTCTCTGTAGATCTCTTGC. Plat-E cells (kindly provided by Dr. Naohito Aoki, Mie University, Tsu, Japan) were cultured in a growth medium at 37°C in 5% CO2. The pMX-PPARα was transfected into Plat-E packaging cells using Lipofectamine2000 (Invitrogen, CA) in accordance with the manufacturer's instructions. Viral supernatants were harvested 48 h after transfection. Viral supernatants were applied to 3T3-L1 cells in the growth medium supplemented with 5 μg/ml polybrene. To eliminate uninfected cells, the medium was replaced with a growth medium containing 2.5 μg/ml puromycin.
Insulin-stimulated 2-deoxy-d-glucose uptake assay
The level of uptake of 2-deoxy-d-[1,2-3H]glucose (2-DG) was measured as previously described (26). Briefly, 10 days after differentiation induction, 3T3-L1 cells were deprived of serum by incubation for 5 h in 12-well plates containing DMEM. The cells were then incubated with 100 nM insulin for 20 min in 450 µl of KRH buffer containing 1% FA-free BSA. Glucose uptake was initiated by adding 50 µl KRH buffer containing 1 mM 2-deoxy-d-[1,2-3H]glucose (1 µCi) (American Radiolabeled Chemicals, MO) to each well, and after 10 min, uptake was terminated by washing the cells three times with ice-cold PBS buffer. The cells were solubilized with 0.1N NaOH, and incorporated radioactivity was measured by liquid scintillation counting. The radioactivity was normalized to protein content determined by the method of Bradford (Bio-Rad Laboratories, CA).
FA oxidation assay
Ten days after differentiation induction, 3T3-L1 cells overexpressing PPARα (L1-PPARα) or control cells (L1-Mock) were incubated in DMEM containing 0.2 mM palmitic acid, 2.5% FA-free BSA, 200 μM l-carnitine, and [14C]palmitic acid (1 μCi/ml) (American Radiolabeled Chemicals) for 16 h. FA oxidation products were assessed as previously described (28) with modification. Briefly, the labeling medium was collected and centrifuged, and the supernatant was transferred to a 50-ml polypropylene tube. An uncapped Eppendorf tube containing a piece of filter paper soaked in benzethonium hydroxide was placed inside a 50 ml tube. After the tube was sealed, 200 μl of 70% perchloric acid was added to the medium sample to release [14C]CO2. The tube was then shaken at 37°C for 1 h. The saturated filter paper containing trapped [14C]CO2 was assessed for radioactivity in a liquid scintillation counter. The acidified medium was centrifuged twice to remove particulate matter, and 200 μl of supernatant was assessed for the amount of [14C]labeled acid-soluble FA metabolites (ASM).
FA oxidation with isolated adipocytes was analyzed as previously described (29). In brief, isolated adipocytes were prepared from epididymal WAT and washed in KRH with 1% BSA. A fresh buffer was added to the cells at a volume twice the packed cell volume. Tubes containing cells and buffer were inverted several times to obtain a homogenous mixture, and equal aliquots of each sample were distributed into 50 ml polystyrene tubes. [14C]palmitic acid was added at a final concentration of 1 μCi/ml. The tubes with a piece of filter paper soaked in benzethonium hydroxide were then sealed and incubated at 37°C for 2 h. The tubes were gently shaken every 30 min during the incubation. After 2 h, 12 M HCl was added to the cells to release [14C]CO2, and each tube was resealed and incubated at 37°C overnight. The amount of [14C]CO2 released was measured by scintillation counting of the filter paper. Radioactivity was normalized to genomic DNA content in each packed cell. Packed cells were incubated overnight with proteinase K (150 μg/ml). Genomic DNA was purified using phenol/chloroform, and its content was quantified by measuring the absorbance at 260 nm.
Measurement of OCR in 3T3-L1 cells
Cellular OCR was measured using a Seahorse Bioscience XF24 analyzer (30) in 24-well plates at 37°C, with correction for positional temperature variations adjusted from 4 empty wells evenly distributed within the plate. L1-Mock or L1-PPARα cells were induced to differentiate and were maintained for 10 days with or without the PPARα agonist GW7647. Immediately before the measurement, cells were washed, and 675 μl of nonbuffered (sodium-carbonate-free) pH 7.4 DMEM medium supplemented with 0.2 mM palmitic acid, 0.2 mM l-carnitine, and 2% FA-free BSA was added to each well. After a 15 min equilibration period, four successive 1.5 min measurements were performed at 7 min intervals with intermeasurement mixing to homogenize the oxygen concentration in the medium. Rotenone (final concentration of 100 nM) was injected to each well using the internal injectors of the cartridge, and four successive 1.5 min measurements were performed at 7 min intervals with intermeasurement mixing.
Measurements of TG content and GPDH activity
To determine intracellular TG amount, 3T3-L1 cells grown in 12-well plates were washed with PBS, and lipids were extracted by hexane-isopropyl alcohol (3:2, v/v) for 1 h at room temperature. Aliquots were transferred and evaporated under a decreased pressure. Samples were resuspended in 10% TritonX-100 in isopropyl alcohol, and TG content was measured enzymatically using triglyceride G test Wako (Wako Pure Chemicals). For Oil Red O staining, 3T3-L1 cells 10 days after the induction of differentiation were fixed with 10% formaldehyde/PBS and stained with Oil Red O solution [0.5% Oil Red O-isopropyl alcohol/H2O (3:2, v/v)].
Cells were washed twice with PBS and collected with a cell scraper into 25 mM Tris-HCl (pH 7.4) containing 1 mM EDTA. The harvested cells were sonicated for 5 s at 40 watts with a microson ULTRASONIC CELL DISRUPTOR (Misonix, Inc., NY). After centrifugation at 12,800 g for 5 min at 4°C, the supernatants were assayed for glycerol-3-phosphate dehydrogenase (GPDH) activity as described elsewhere (31).
RNA analysis
Total RNA was prepared from mouse tissues or cultured 3T3-L1 and human multipotent adipose tissue-derived stem adipocytes using Qiazol lysis reagent (QIAGEN, CA) or Sepasol(R)-RNA I Super (Nacalai Tesuque) in accordance with the manufacturer's protocol. Total RNA was reverse-transcribed using M-MLV reverse transcriptase (Promega, WI) in accordance with the manufacturer's instructions using a thermal cycler (Takara PCR Thermal Cycler SP, Takara, Shiga, Japan). To visualize and compare the expression levels of PPARα and 36B4, semiquantitative PCR was performed. PCR amplification was performed with denaturation at 94°C for 20 s, annealing at 60°C for 40 s, and polymerization at 72°C for 20 s. PCR was performed for 35 cycles. PCR products were electrophoresed on 2% agarose gels and stained with ethidium bromide. To quantify mRNA expression, real-time RT-PCR was performed with a LightCycler System (Roche Diagnostics, Mannheim, Germany) using SYBR Green fluorescence signals as described previously (31–34). The oligonucleotide primers of mouse and human 36B4 and PPARα, mouse adipogenic marker genes, and PPARα target genes were designed using a PCR primer selection program in the website of the Virtual Genomic Center from the GenBank database. All oligonucleotide primer sets used to measure the expression levels of 36B4, adipocyte fatty-acid-binding protein (aP2), PPARγ, and adiponectin were previously described (31–34). The primers used for measurements of the mRNA expression levels of other genes (upstream and downstream, respectively) were TCAGGGTACCACTACGGAGT and CTTGGCATTCTTCCAAAGCG for mouse PPARα mRNA; ACTCCACCTGCAGAGCAACCA and TAGATCTCCTGCAGTAGCGGG for human PPARα mRNA; GCACCATTGCCATTCGATACA and CCACTGCTGTGAGAATAGCCGT for acyl-CoA oxidase (ACO) mRNA; CTGTTAGGCCTCAACACCGAAC and CTGTCATGGCTAGGCTGTACAT for carnitine-palmitoyl transferase-1b (CPT1b) mRNA; and TACCCAACCTTGGCTAGACCTC and GCAACTTCTCCTTGATGATGTCGTA for uncoupling protein 3 (UCP3) mRNA. To compare mRNA expression level among the samples, the copy numbers of all transcripts were divided by that of mouse 36B4 showing a constant expression level in adipocytes. All mRNA expression levels were represented as a ratio relative to that of the control in each experiment.
Immunoblotting
Total cellular proteins were solubilized in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS; pH 7.4) and a protease inhibitor cocktail). The protein concentration of samples was determined using a protein assay kit (Bio-Rad Laboratories). Protein samples (30 μg) were subjected to SDS-PAGE on a 10% gel. Separated proteins were transferred electrophoretically to PVDF membranes (Millipore, MA), which were blocked with 5% nonfat dried milk in phosphate-buffered saline. The membranes were incubated with antibodies to PPARα (Perseus Proteomics, Tokyo, Japan) and β-actin (Cell Signaling Technology, MA), and then with peroxidase-conjugated anti-mouse and anti-rabbit IgG antibodies (Santa Cruz, CA), respectively. Proteins were detected using an ECL Western blotting detection system (GE Healthcare, NJ).
Chromatin immunoprecipitation (ChIP) assay
3T3-L1 cells 14 days after differentiation induction were fixed with 1% formaldehyde in PBS for 10 min. The cells were then rinsed twice with ice-cold PBS containing 2% bovine serum and 0.05% NaN3, centrifuged for 5 min at 3,000 rpm, and resuspended in lysis buffer [1% SDS, 10 mM EDTA, 50 mM Tris-HCl (pH 8.0), and a protease inhibitor cocktail]. Following incubation for 20 min on ice, samples were sonicated on ice. The lysates were centrifuged at 15,000 rpm for 10 min, and the collected supernatant was diluted in buffer [1.1% Triton X-100, 0.11% sodium deoxycholate, 167 mM NaCl, 50 mM Tris-HCl; (pH 8.0) and a protease inhibitor cocktail)]. Samples were precleared with 5 µg of sheared salmon sperm DNA and 50 μl of protein A/G-Sepharose beads (Santa Cruz) for 2 h. Immunoprecipitation with the antibody to PPARα or normal mouse IgG (Santa Cruz) was performed overnight. Samples were then incubated with 20 µl of protein A/G-Sepharose beads for 3 h followed by 10 min sequential washes in RIPA I (0.1% SDS, 1% Triton X-100, 0.1% sodium deoxycholate, 2 mM EDTA, 50 mM Tris-HCl, and 150 mM NaCl; pH 8.0), RIPA II (0.1% SDS, 1% Triton X-100, 0.1% sodium deoxycholate, 2 mM EDTA, 50 mM Tris-HCl, 500 mM NaCl; pH 8.0), buffer III (0.25M LiCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl; pH8.0), and Tris-EDTA buffer. Precipitates were then extracted by incubating with elution buffer (0.5% SDS, 5 mM EDTA, 300 mM NaCl, 10 mM Tris-HCl; pH8.0) at 65°C overnight. After the RNase A and proteinase K treatment, DNA fragments were purified with phenol/chloroform. Purified samples were used as templates for PCR performed for 35 cycles. Oligonucleotide primers composed of the following sequences (upstream and downstream) were used for PCR: aP2, GAATTCCAGCAGGAATCAGG and GCCAAAGAGACAGAGGGCG (300 bp); CPT1b, CCTGTGCTGGTCCCCAACTCACAGC and CTCCTGGTGACCTTTTCCCTACAT T (279 bp).
Statistical analysis
The data were expressed as means ± S.E.M. Statistical significance was evaluated using unpaired Student's t-test for the two groups. The results were considered significant at P < 0.05.
RESULTS
Administration of PPARα agonist attenuated adiposity and improved insulin resistance in mice fed HFD
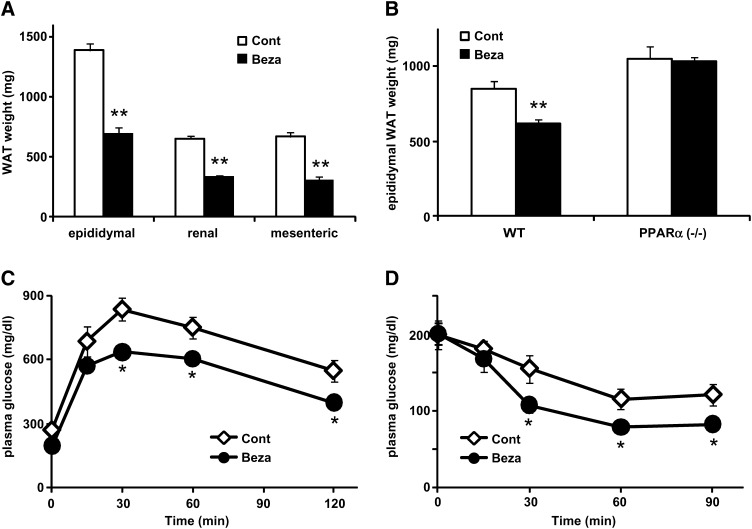
To study the effect of PPARα activation on energy metabolism in WAT, we treated mice fed HFD with bezafibrate, a PPARα agonist. Since PPARα agonists have been shown to reduce food intake in rodents (35), all mice were housed in pair-fed conditions in each experiment. Administration of bezafibrate (0.2%) in mildly obese diabetic KK mice fed HFD for four weeks significantly decreased adiposity (50, 49, and 54% decrease in epididymal, perirenal, and mesenteric WAT, respectively) (Fig. 1A and supplementary Table I). Because bezafibrate has been reported to activate other PPAR subtypes (36), we used PPARα−/− and WT (control) mice, and a similar experiment was performed. For a six-week treatment, bezafibrate significantly reduced adiposity in WT mice similarly to KK mice (Fig. 1B and supplementary Table II). However, this effect was not observed in PPARα−/− mice. Although the WAT weight was slightly higher in PPARα−/− mice than in WT mice, the difference was not statistically significant. These results indicate that pharmacological PPARα activation reduces adiposity independently of its anorexic effect. Next, to examine the effect of a PPARα agonist on insulin sensitivity, we carried out a GTT and ITT. Plasma glucose levels in both tests in the bezafibrate-treated mice were significantly lower than those in the pair-fed control mice (Fig. 1C, D), indicating that a PPARα agonist ameliorates HFD-induced insulin resistance in KK mice.
Fig. 1.
Effects of PPARα activator on adiposity and insulin resistance. A, B: White adipose tissue (WAT) weight from male mildly obese KK mice (A) and wild-type (WT) or PPARα-deficient (PPARα−/−) mice (B) fed high-fat diet control (Cont) or high-fat diet containing 0.2% bezafibrate (Beza) for four and six weeks. In each experiment, the mice were housed under pair-fed conditions. C, D: Plasma glucose levels during oral glucose tolerance test (GTT) (C) and insulin tolerance test (ITT) (D). Both GTT and ITT were performed on KK mice fed high-fat diet (Cont) or high-fat diet containing 0.2% bezafibrate (Beza) for five weeks under pair-fed condition. Plasma glucose level was measured by enzymatic colorimetric assay. Data are means ± SEM of 4-6 animals per group. *P < 0.05, **P < 0.01, compared with control diet group.
PPARα expression level increased during adipocyte differentiation in 3T3-L1 cells and changed depending on energy condition in WAT
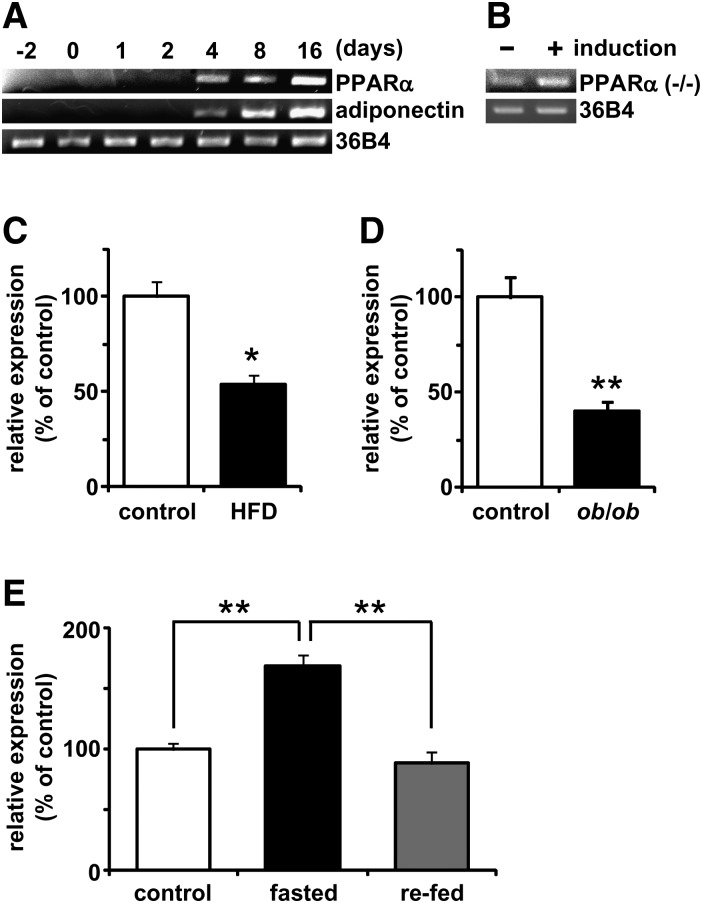
We next examined the expression of PPARα in cultured adipocytes and WAT. As previously reported (22, 23), PPARα mRNA was highly expressed in the liver, brown adipose tissue, and kidney (supplementary Fig. IA, B). The PPARα mRNA expression level in WAT was higher than that in the skeletal muscle, although that in WAT was lower than that in tissues with high mRNA expression level of PPARα (supplementary Fig. IA, B). In WAT, PPARα mRNA was expressed at a higher level in the adipocyte fraction than in the SV cell fraction (supplementary Fig. IC). In cultured 3T3-L1 cells, the mRNA expression level of PPARα increased in parallel with the progression of adipocyte differentiation and in a pattern similar to that of adiponectin mRNA expression (Fig. 2A). In human multipotent adipose tissue-derived stem cells characterized by an extensive proliferative potential and the ability to undergo multilineage differentiation, including adipogenic differentiation (27), adipogenic induction also induced PPARα mRNA expression (Fig. 2B). Because diet-induced obesity and fasting affects the hepatic expression of PPARα (37, 38), we assessed whether PPARα mRNA expression level in WAT changes depending on energy condition. The PPARα mRNA expression in WAT was significantly decreased in both HFD-induced obese mice (12 months) (Fig. 2C) and the genetically obese mouse model (ob/ob) (Fig. 2D) compared with the lean control mice. In the HFD-fed and ob/ob mice, the PPARα mRNA expression level in WAT decreased by 54 and 40%, respectively. Conversely, 24 h fasting increased PPARα mRNA expression level by 1.5-fold (Fig. 2E). The extent of increase was diminished by refeeding after 24 h (Fig. 2E). These findings indicate that PPARα mRNA expression in WAT is changed depending on the energy condition, suggesting that PPARα physiologically and pharmacologically plays an important role in the control of energy balance.
Fig. 2.
mRNA expression levels of PPARα in cultured cells and WAT. A, B: mRNA expression levels of murine PPARα and adiponectin or human PPARα during adipocyte differentiation. 3T3-L1 cells and human multipotent adipose tissue-derived stem cells were harvested at the indicated times after differentiation induction. Semiquantitative PCR conditions are described in “Materials and Methods.” C, D: mRNA expression levels of PPARα in WAT in obese model mice and lean control mice. PPARα mRNA expression levels in C57BL/6 mice fed high-fat diet (HFD) or normal diet (control) for 12 months (C) and in ob/ob mice or lean control mice (D) were quantified by a real-time PCR as described in “Materials and Methods.” E: The mRNA expression levels of PPARα in WAT in C57BL/6 under fed (control), fasted (24 h), and fasted/refed (24 h after refeeding) conditions were quantified by real-time PCR. Data are means ± SEM of 4-6 animals per group. *P < 0.05, **P < 0.01.
PPARα activation entirely correlated with increases in adipogenic gene expression level, GPDH activity, and insulin-stimulated glucose uptake level but not with lipid accumulation level in 3T3-L1 cells
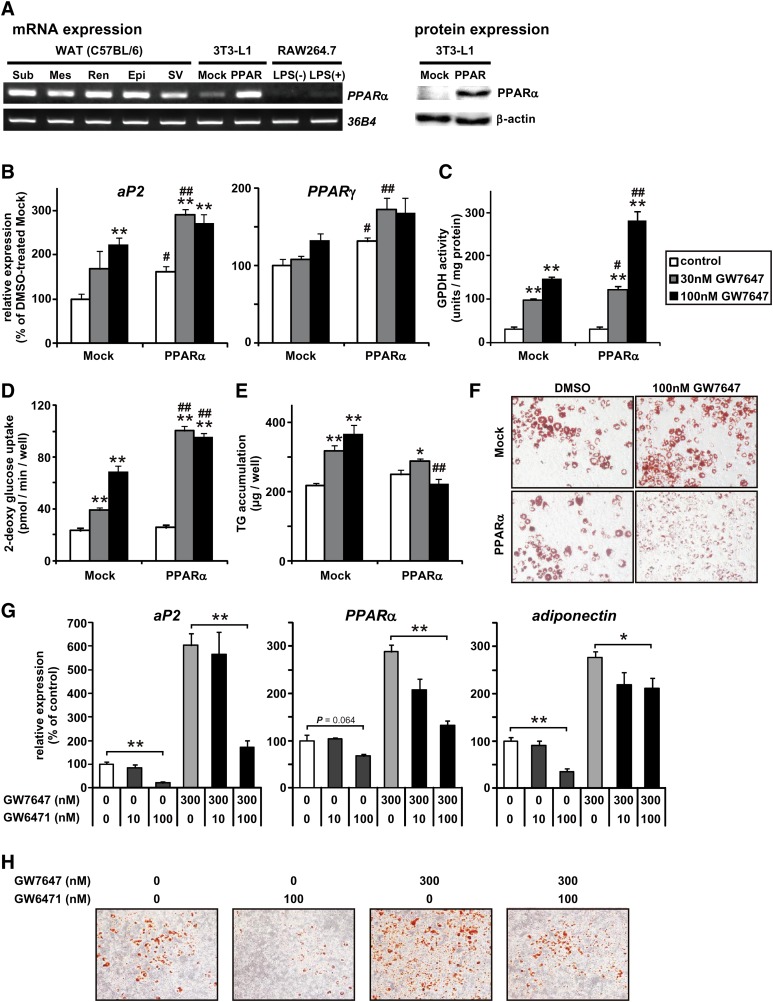
To investigate the direct effect of PPARα activation on adipocyte metabolism, we examined PPARα-agonist-treated 3T3-L1 adipocytes. Because the PPARα mRNA expression level was much lower in 3T3-L1 cells before differentiation induction than in murine SV cells included with preadipocytes (Fig. 3A), we established 3T3-L1 cells overexpressing PPARα mRNA (L1-PPARα) and mock control cells (L1-Mock). The mRNA and protein expression levels of PPARα in L1-PPARα cells were slightly higher than those in SV cells (Fig. 3A). First, we examined the effects of PPARα activation on adipocyte differentiation. GW7647, highly a specific PPARα activator, was added at the time of differentiation induction and kept in the medium throughout the differentiation period. As shown in Fig. 3B, the addition of GW7647 induced the mRNA expression of adipogenic marker genes, such as those encoding aP2 and PPARγ. Furthermore, the GW7647 treatment increased both lipogenic enzyme GPDH activity (on day 6) and insulin-stimulated 2-DG uptake capacity (on day 10) (Fig. 3C, D). The effects of GW7647 depended on GW7647 concentration and exogenous PPARα expression. These results indicate that PPARα activation promotes adipocyte differentiation. However, surprisingly, the TG content was markedly lower in L1-PPARα than in L1-Mock in the presence of 100 nM GW7647 (Fig. 3E, F), although the lipogenic GPDH activity and insulin-stimulated 2-DG uptake capacity were higher in L1-PPARα than in L1-Mock, as shown above. Even in unmanipulated 3T3-L1 cells, GW7647 induced aP2, PPARγ, and adiponectin expression and lipid accumulation during adipocyte differentiation (Fig. 3G, H). In contrast, the addition of the PPARα antagonist GW6471 inhibited them (Fig. 3G, H). Thus, in 3T3-L1 cells, PPARα activation may be associated with adipocyte differentiation physiologically.
Fig. 3.
PPARα activation increased mRNA expression levels of marker factors for adipocyte differentiation but not TG content. A: PPARα expressions in WAT and cultured cells. PPARα mRNA levels in several WATs [subcutaneous (Sub), mesenteric (Mes), perirenal (Rea), and epididymal (Epi) WAT from C57BL/6 mice], stromal-vascular (SV) cells from epididymal WAT in C57BL/6 mice, and cultured cells [3T3-L1 cells overexpressing PPARα (L1-PPARα), mock control (L1-Mock), RAW264.7 macrophages, and LPS-stimulated RAW264.7 macrophages] were visualized by semiquantitative PCR (left). PPARα protein levels in L1-Mock and L1-PPARα were visualized by Western blotting (right). B-F: L1-Mock and L1-PPARα cells were induced to differentiate and cultured with or without GW7647 (30 or 100 nM) for 6-10 days. B-E: The mRNA expression levels of adipogenic marker genes (aP2 and PPARγ) (on day 10) (B), glycerol-3-phosphate dehydrogenase (GPDH) activity (on day 6) (C), the capacity of insulin-stimulated 2-DG transport (on day 10) (D), and intracellular triglyceride (TG) amounts (E) in L1-Mock and L1-PPARα were determined as described in “Materials and Methods.” F: Microscopy views of representative L1-Mock and L1-PPARα cells treated with or without 100 nM GW7647, fixed with formalin, and stained with Oil Red O. The original magnification is 100×. Data are means ± SEM; n = 4-6. *P < 0.05, **P < 0.01 compared with DMSO-treated L1-Mock cells. #P < 0.05, ##P < 0.01 compared with L1-Mock cells treated with the same compounds. G, H: 3T3-L1 cells were induced to differentiate and cultured with or without GW7647 (300 nM) and GW6471 (100 nM) for 10 days. The mRNA expression levels of aP2 and PPARγ (G) and microscopy views of representative cells stained with Oil Red O (H). The original magnification is 100×. Data are means ± SEM; n = 4. *P < 0.05, **P < 0.01.
PPARα activation enhanced fatty acid oxidation and OCR in 3T3-L1 cells
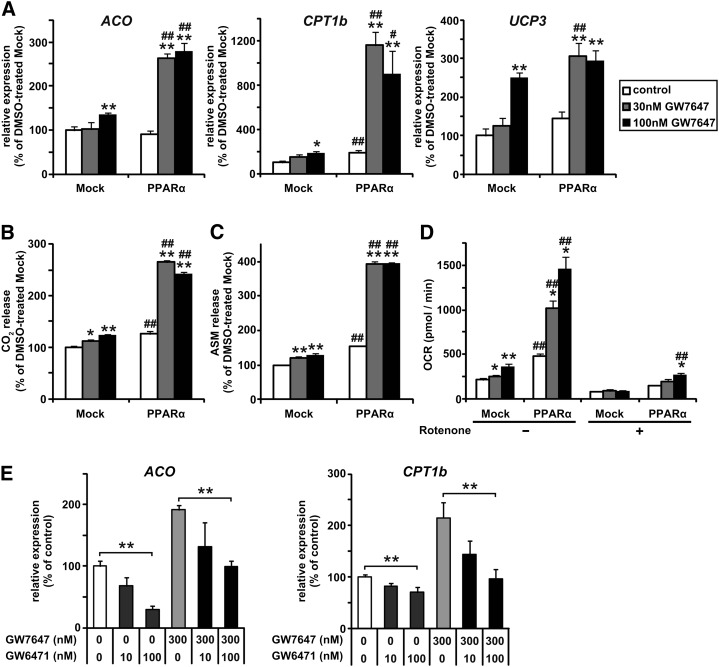
To clarify the inhibitory effect of PPARα activation on lipid accumulation, we examined GW7647-induced FA oxidation in L1-PPARα. Transient GW7647 treatment did not affect the amount of glycerol release or FFA release under our experimental conditions (data not shown). Furthermore, the mRNA expression levels of lipases (hormone-sensitive lipase and adipose triglyceride lipase) related to lipolysis in adipocytes were not changed by the GW7647 treatment (data not shown). On the other hand, chronic GW7647 treatment during adipocyte differentiation induced the mRNA expression of genes involved in FA oxidation, such as those encoding ACO, CPT1b, and UCP3 (Fig. 4A). The induction was observed to be more marked in L1-PPARα than in L1-Mock. When cells were incubated with [14C]palmitic acid for 16 h, the GW7647 treatment enhanced the oxidation of [14C]palmitic acid to CO2 and acid-soluble FA metabolites (Fig. 4B, C). Moreover, OCR determined by extracellular flux analysis (30) was increased by the GW7647 treatment (Fig. 4D). Rotenone, an inhibitor of mitochondrial electron transport, almost completely diminished the increase in OCR. These findings indicate that PPARα activation in adipocytes enhances FA oxidation and increases OCR via the induction of PPAR target genes related to FA oxidation, suggesting that the effects of PPARα activation contribute to the discrepancy between adipogenic activity and lipid accumulation levels induced by PPARα activation. Even in unmanipulated 3T3-L1 cells, GW7647 induced ACO and CPT1b expression (Fig. 4E). In contrast, the addition of a PPARα antagonist GW6471 inhibited it (Fig. 4E). Therefore, PPARα activation might play some physiological roles in fatty acid oxidation in 3T3-L1 cells.
Fig. 4.
Effects of PPARα activation on FA oxidation in cultured adipocytes. A-D: 3T3-L1 cells overexpressing PPARα (L1-PPARα) and mock control (L1-Mock) cells were induced to differentiate and cultured with or without GW7647 (30 or 100 nM) for 10 days. The mRNA expression levels of genes involved in FA oxidation (ACO, CPT1b, and UCP3) (A), oxidation of [14C]palmitic acid to CO2 (B), and acid-soluble FA metabolites (ASM) (C) for 16 h, and oxygen consumption rate (OCR) (D) in L1-Mock and L1-PPARα was determined as described in “Materials and Methods.” Data are means ± SEM; n = 3-6. *P < 0.05, **P < 0.01 compared with DMSO-treated L1-Mock. #P < 0.05, ##P < 0.01 compared with L1-Mock treated with the same compounds. E: 3T3-L1 cells were induced to differentiate and cultured with or without GW7647 (300 nM) and GW6471 (100 nM) for 10 days. The mRNA expression levels of ACO and CPT1b was measured by real-time PCR. Data are means ± SEM; n = 4. **P < 0.01.
PPARα activation induced both adipogenic and #8232FA-oxidation-related genes in WAT and enhanced FA oxidation in isolated adipocytes and the entire body
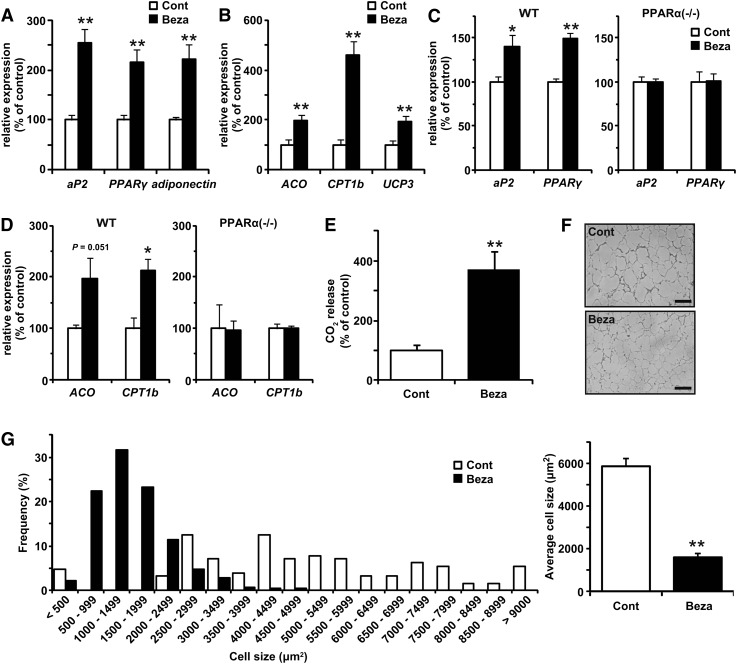
Next, we examined whether the effects of PPARα activation observed in cultured adipocytes were observed in vivo. Treatment of KK mice fed HFD with bezafibrate for four weeks significantly increased the expression levels of both adipogenic marker genes (aP2, PPARγ, and adiponectin) and genes involved in FA oxidation (ACO, CPT1b, and UCP3) (Fig. 5A, B). Similar mRNA inductions by bezafibrate administration for six weeks were observed in WT mice but not in PPARα−/− mice (Fig. 5C, D), suggesting that PPARα plays an essential role in the induction of genes involved in both adipogenesis and FA oxidation in WAT by the bezafibrate treatment. Using an indirect calorimetric system, we found that OCR was higher in the bezafibrate-treated mice than in the control KK mice (supplementary Fig. IIA), despite nearly the same locomotor activity (data not shown). Because these mice were fed HFD, the respiratory exchange ratio (RER) was always low during this measurement; however, the RER of the bezafibrate-treated mice was slightly but significantly lower in the light phase (supplementary Fig. IIB). Moreover, the capacity for the oxidation of [14C]palmitic acid to CO2 was higher in the isolated adipocytes of the bezafibrate-treated mice (Fig. 5E). The size of adipocytes was smaller in epididymal WAT from the bezafibrate-treated mice (Fig. 5F, G). These findings were similar to those in cultured cells and indicate that PPARα activation induced adipogenesis and FA oxidation in WAT, resulting in suppression of adipocyte hypertrophy.
Fig. 5.
Effects of PPARα activator on adipocyte differentiation and FA oxidation in WAT. KK mice (A, B, E-G) and wild-type (WT) or PPARα-deficient (PPARα−/−) mice (C, D) fed HFD (Cont) or HFD containing 0.2% bezafibrate (Beza) were housed under the pair-fed condition for 4-6 weeks. A-D: The mRNA expression levels of adipogenic marker genes (aP2, PPARγ, and adiponectin) (A, C) and genes involved in FA oxidation (ACO, CPT1b, and UCP3) (B, D) in WAT were determined by real-time PCR. E-G: Oxidation of [14C]palmitic acid to CO2 in isolated adipocytes (E), microscopy views of representative histological sections of epididymal WAT (F), and adipocyte size distribution in epididymal WAT (G) from mice fed each experimental diet for 6 weeks were examined. Data are means ± SEM; n = 3-8. *P < 0.05, **P < 0.01 compared with mice fed control diet.
PPARα seemed to be recruited to the promoter region of genes involved in both adipogenesis and FA oxidation in 3T3-L1 cells
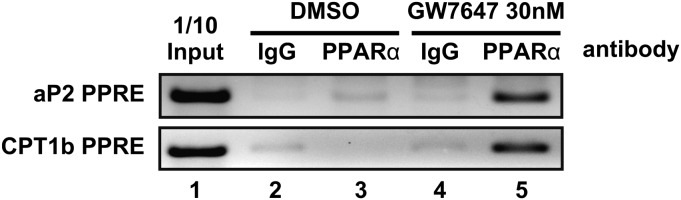
Finally, to confirm whether PPARα was recruited to the promoter region, which contained an identified PPAR-response element (PPRE), of genes related to adipogenesis and FA oxidation in 3T3-L1 cells, we performed ChIP assay. We focused on aP2 as an adipogenic gene and CPT1b as an FA oxidation gene. As shown in Fig. 6, in the presence of GW7647, ChIP assay revealed that PPARα recruitment to the promoter regions of both aP2 and CPT1b was enhanced in 3T3-L1 cells 14 days after differentiation induction. These regions have been shown to contain PPRE (39, 40); it was suggested that PPARα activation by ligand binding enhanced the direct binding of PPARα to PPRE.
Fig. 6.
ChIP assays of aP2 and CPT1b promoter in 3T3-L1 adipocytes treated with or without GW7647. ChIP assays were performed as described in “Materials and Methods.” Soluble chromatin from 3T3-L1 cells, whose differentiation was induced and maintained for 14 days with or without of 30 nM GW7647, was immunoprecipitated with control mouse IgG (lanes 2 and 4) or antibodies against PPARα (lanes 3 and 5). Immunoprecipitates were analyzed by PCR using specific primers for the mouse aP2 or CPT1b promoter region containing potential PPRE. PCR was performed with total chromatin input (lane 1).
DISCUSSION
Several recent studies have demonstrated that PPARα is expressed in WAT of humans and rodents (22–24). WAT plays a key role in regulating whole-body energy metabolism. In this study, we attempted to clarify the functions of PPARα in WAT. We found that the mRNA expression level of PPARα in WAT was decreased by diet-induced and genetic obesity and, conversely, was increased by fasting. These findings suggest that the PPARα activity in WAT plays a physiological role in energy metabolism. PPARα−/− mice exhibited increased adiposity and adipocyte hypertrophy (41). Moreover, these phenotypes seemed to be independent of the hepatic PPARα action (41). The findings in PPARα−/− mice were in contrast to those in 3T3-L1 cells treated with a PPARα agonist in this study.
A number of reports have shown that PPARα is particularly important during fasting (11, 15, 38, 42). Fasted PPARα−/− mice suffer from various metabolic defects, including hypoketonemia, hypothermia, and hypoglycemia. During the fasting period, lipolysis in WAT releases glycerol and FA into the blood. It has been reported that PPARα activators stimulate lipolysis in WAT (25). Therefore, it is suggested that the increase in PPARα mRNA expression level in WAT during fasting is important for adaptation to fasting.
Moreover, several studies have shown that the number of mitochondrial DNA copies and the expression levels of mitochondrial genes are decreased in WAT of obese animal models and humans (29, 43, 44). In the diabetic state, the mitochondrial β-oxidation of FA is attenuated. In addition, mitochondria show abnormal morphologies (44). In this study, the increase in OCR (Fig. 4D) was almost diminished in the presence of rotenone, an inhibitor of mitochondrial electron transport, in the PPARα-transfected 3T3-L1 adipocytes, suggesting that PPARα activity is important for mitochondrial FA oxidation in adipocytes. Thus, it is suggested that the downregulation of PPARα in obese WAT is involved in obesity-induced mitochondrial dysfunction and metabolic disorders.
WAT has not been considered as a main target tissue of PPARα agonists thus far. However, we detected PPARα mRNA expression at higher levels in WAT than in the skeletal muscle, where PPARα agonists stimulate FA oxidation (45), although the PPARα mRNA expression level in adipocytes was much lower than those in the liver and brown adipose tissue (BAT). In addition, several reports have recently indicated the effects of PPARα agonists on adipocytes. For example, PPARα agonists transcriptionally induce the mRNA expression of adiponectin and its receptors via PPARα activation both in vivo and in vitro (23, 22). It has also been reported that Wy-14643 directly enhances lipolysis in isolated adipocytes (25). In this study, we showed that PPARα agonists promoted FA oxidation and adipocyte differentiation in cultured adipocytes and WAT, both of which are closely related to the pathogenesis of metabolic disorders. Therefore, the data from our and other groups suggest that the PPARα activation in adipocytes is pharmacologically significant for the regulation of lipid metabolism to maintain whole-body energy balance, although to clarify details of the significance of PPARα activation in adipocytes, further investigation is necessary, such as experiments using PPARα WAT-specific knockout mice.
Several studies have demonstrated that adipocytes contain large amounts of mitochondria in their tiny cytosolic compartment (46, 47). Several genetic and pharmacological studies indicate that an increase in FA oxidation capacity in adipocytes leads to the attenuation of adiposity and improvement of obesity-induced metabolic disorders (48–53). Moreover, intense hyperleptinemia induced by means of adenovirus-mediated transfer of the leptin gene causes a rapid disappearance of WAT, at least in part through mechanisms other than decreased food intake (54). Leptin induces the expression of genes involved in FA oxidation partly through a process dependent on PPARα activation (55). The hyperleptinemia-induced WAT loss was not observed in PPARα−/− mice (56). Because this WAT loss is not accompanied by any increase in plasma free FA and ketone body levels or ketonuria (57), it was suggested that FA derived from adipocyte TG is oxidized inside adipocytes via PPARα activation. Indeed, in this study, the treatment with bezafibrate significantly attenuated adiposity and enhanced FA oxidation in adipocytes, accompanied by the upregulation of genes related to FA oxidation in WAT. These findings indicate that bezafibrate acts on PPARα in WAT directly, suggesting that WAT is a target of pharmacological treatments with PPARα agonists.
Interestingly, PPARα activation promoted not only FA oxidation but also adipocyte differentiation. Adipocyte differentiation is closely related to obesity and obesity-induced metabolic disorders, such as insulin resistance (18–20). PPARγ plays a central role in the regulation of adipocyte differentiation, and its synthesized agonists, thiazolidinediones, potently facilitate adipocyte differentiation to improve insulin resistance (18–20). In this study, the treatment with a PPARα agonist induced the mRNA expression of adipogenic genes, such as aP2 and PPARγ, at higher levels in PPARα-expressing 3T3-L1 cells (L1-PPARα) than in control 3T3-L1 cells (L1-Mock). In addition, the induction of these genes was observed only in WT mice but not in PPARα−/− mice. These observations suggest that PPARα activation can promote adipocyte differentiation. The effect of PPARα on adipocyte differentiation was confirmed by ChIP assay, showing that PPARα was recruited to PPRE in each promoter region of adipogenic genes directly regulated by PPARγ in WAT. A previous study has also shown that PPARα activation induces the mRNA expression of glycerol kinase (58), which catalyzes the direct conversion of glycerol to glycerol-3-phosphate in human adipocytes. Glycerol-3-phosphate is an important material for FA esterification to form TG in adipocytes. Thus, it is suggested that a PPARα-specific adipogenic pathway exists, although the effect of PPARα seems to be partially shared with that of PPARγ.
We provided experimental evidence that PPARα activation in adipocytes leads to the promotion of both adipocyte differentiation and the FA oxidation pathway. These effects of PPARα activation are caused by the direct binding of PPARα to PPRE located in the promoter regions of genes involved in adipocyte differentiation and FA oxidation. We also demonstrated that PPARα expression level in WAT is changed by several nutrient conditions, such as obesity and fasting. Because adipocytes are closely related to whole-body carbohydrate and lipid metabolism, PPARα activity in WAT may play an important role in regulating whole-body energy metabolism. Furthermore, PPARα in WAT may contribute to the improvement of obesity-induced metabolic disorders, such as insulin resistance, in combination with fibrate drugs.
The authors thank S. Sasaka (Primetech Corp., Tokyo, Japan) for technical support in experiments using XF24 Extracellular Flux Analyzer; and S. Shinoto and Y. Tada for technical assistance.
Supplementary Material
Footnotes
Abbreviations:
- 2-DG
- 2-deoxy-d-glucose
- ACO
- acyl-CoA oxidase
- aP2
- adipocyte fatty-acid-binding protein
- BAT
- brown adipose tissue
- ChIP
- chromatin immunoprecipitation
- CPT1b
- carnitine-palmitoyl transferase-1b
- GPDH
- glycerol-3-phosphate dehydrogenase
- HFD
- high-fat diet
- IL-6
- interleukin-6
- ITT
- insulin tolerance test
- MCP-1
- monocyte chemoattractant protein-1
- OCR
- oxygen consumption rate
- OGTT
- oral glucose tolerance test
- PPAR
- peroxisome proliferator-activated receptor
- PPRE
- PPAR-response element
- RER
- respiratory exchange ratio
- SV
- stromal vascular
- TG
- triglyceride
- TNFα
- tumor necrosis factor-α
- TZD
- thiazolidinedione
- UCP3
- uncoupling protein 3
- WAT
- white adipose tissue
- WT
- wild-type
This work was supported by Grants-in-Aid for Scientific Research 19780096, 19.4826, 22228001, and 22780116 from the Ministry of Education, Culture, Sport, Science and Technology of Japan and by the Uehara Memorial Foundation.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures and two tables.
REFERENCES
- 1.Ogden C. L., Yanovski S. Z., Carroll M. D., Flegal K. M. 2007. The epidemiology of obesity. Gastroenterology. 132: 2087–2102. [DOI] [PubMed] [Google Scholar]
- 2.Collins S. 2005. Overview of clinical perspectives and mechanisms of obesity. Birth Defects Res. A Clin. Mol. Teratol. 73: 470–471. [DOI] [PubMed] [Google Scholar]
- 3.Mokdad A. H., Ford E. S., Bowman B. A., Dietz W. H., Vinicor F., Bales V. S., Marks J. S. 2003. Prevalence of obesity, diabetes, and obesity-related health risk factors, 2001. JAMA. 289: 76–79. [DOI] [PubMed] [Google Scholar]
- 4.Unger R. H., Orci L. 2001. Diseases of liporegulation: new perspective on obesity and related disorders. FASEB J. 15: 312–321. [DOI] [PubMed] [Google Scholar]
- 5.van Herpen N. A., Schrauwen-Hinderling V. B. 2008. Lipid accumulation in non-adipose tissue and lipotoxicity. Physiol. Behav. 94: 231–241. [DOI] [PubMed] [Google Scholar]
- 6.Issemann I., Green S. 1990. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 347: 645–650. [DOI] [PubMed] [Google Scholar]
- 7.Evans R. M., Barish G. D., Wang Y. X. 2004. PPARs and the complex journey to obesity. Nat. Med. 10: 355–361. [DOI] [PubMed] [Google Scholar]
- 8.Cho M. C., Lee K., Paik S. G., Yoon D. Y. 2008. Peroxisome proliferators-activated receptor (PPAR) modulators and metabolic disorders. PPAR Res. Vol. 2008, 14 pages; doi:10.1155/2008/679137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lefebvre P., Chinetti G., Fruchart J. C., Staels B. 2006. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J. Clin. Invest. 116: 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kersten S., Desvergne B., Wahli W. 2000. Roles of PPARs in health and disease. Nature. 405: 421–424. [DOI] [PubMed] [Google Scholar]
- 11.Hashimoto T., Cook W., Qi C., Yeldandi A., Reddy J., Rao M. 2000. Defect in peroxisome proliferator-activated receptor alpha-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J. Biol. Chem. 275: 28918–28928. [DOI] [PubMed] [Google Scholar]
- 12.Minnich A., Tian N., Byan L., Bilder G. 2001. A potent PPAR agonist stimulates mitochondrial fatty acid-oxidation in liver and skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 280: E270–E279. [DOI] [PubMed] [Google Scholar]
- 13.Schoonjans K., Staels B., Auwerx J. 1996. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J. Lipid Res. 37: 907–925. [PubMed] [Google Scholar]
- 14.Akiyama T. E., Nicol C. J., Fievet C., Staels B., Ward J. M., Auwerx J., Lee S. S., Gonzalez F. J., Peters J. M. 2001. Peroxisome proliferator-activated receptor-alpha regulates lipid homeostasis, but is not associated with obesity: studies with congenic mouse lines. J. Biol. Chem. 276: 39088–39093. [DOI] [PubMed] [Google Scholar]
- 15.Kersten S., Seydoux J., Peters J. M., Gonzalez F. J., Desvergne B., Wahli W. 1999. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J. Clin. Invest. 103: 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cawthorn W. P., Sethi J. K. 2008. TNF-alpha and adipocyte biology. FEBS Lett. 582: 117–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stern N., Osher E., Greenman Y. 2007. Hypoadiponectinemia as a marker of adipocyte dysfunction–part II: the functional significance of low adiponectin secretion. J. Cardiometab. Syndr. 2: 288–294. [DOI] [PubMed] [Google Scholar]
- 18.Matsuzawa Y. 2006. The metabolic syndrome and adipocytokines. FEBS Lett. 580: 2917–2921. [DOI] [PubMed] [Google Scholar]
- 19.Kadowaki T., Hara K., Yamauchi T., Terauchi Y., Tobe K., Nagai R. 2003. Molecular mechanism of insulin resistance and obesity. Exp. Biol. Med. (Maywood). 228: 1111–1117. [DOI] [PubMed] [Google Scholar]
- 20.Yamauchi T., Kamon J., Waki H., Murakami K., Motojima K., Komeda K., Ide T., Kubota N., Terauchi Y., Tobe K., et al. 2001. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor gamma (PPARgamma) deficiency and PPARgamma agonist improve insulin resistance. J. Biol. Chem. 276: 41245–41254. [DOI] [PubMed] [Google Scholar]
- 21.Guerre-Millo M., Gervois P., Raspé E., Madsen L., Poulain P., Derudas B., Herbert J. M., Winegar D. A., Willson T. M., Fruchart J. C., et al. 2000. Peroxisome proliferator-activated receptor alpha activators improve insulin sensitivity and reduce adiposity. J. Biol. Chem. 275: 16638–16642. [DOI] [PubMed] [Google Scholar]
- 22.Tsuchida A., Yamauchi T., Takekawa S., Hada Y., Ito Y., Maki T., Kadowaki T. 2005. Peroxisome proliferator-activated receptor (PPAR)alpha activation increases adiponectin receptors and reduces obesity-related inflammation in adipose tissue: comparison of activation of PPARalpha, PPARgamma, and their combination. Diabetes. 54: 3358–3370. [DOI] [PubMed] [Google Scholar]
- 23.Hiuge A., Tenenbaum A., Maeda N., Benderly M., Kumada M., Fisman E. Z., Tanne D., Matas Z., Hibuse T., Fujita K., et al. 2007. Effects of peroxisome proliferator-activated receptor ligands, bezafibrate and fenofibrate, on adiponectin level. Arterioscler. Thromb. Vasc. Biol. 27: 635–641. [DOI] [PubMed] [Google Scholar]
- 24.Loviscach M., Rehman N., Carter L., Mudaliar S., Mohadeen P., Ciaraldi T. P., Veerkamp J. H., Henry R. R. 2000. Distribution of peroxisome proliferator-activated receptors (PPARs) in human skeletal muscle and adipose tissue: relation to insulin action. Diabetologia. 43: 304–311. [DOI] [PubMed] [Google Scholar]
- 25.Guzman M., Lo Verme J., Fu J., Oveisi F., Blazquez C., Piomelli D. 2004. Oleoylethanolamide stimulates lipolysis by activating the nuclear receptor peroxisome proliferator-activated receptor alpha (PPAR-alpha). J. Biol. Chem. 279: 27849–27854. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi N., Goto T., Taimatsu A., Egawa K., Kato S., Kusudo T., Sakamoto T., Ohyane C., Lee J. Y., Kim Y. I., et al. 2009. Bixin regulates mRNA expression involved in adipogenesis and enhances insulin sensitivity in 3T3–L1 adipocytes through PPARgamma activation. Biochem. Biophys. Res. Commun. 390: 1372–1376. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez A. M., Elabd C., Delteil F., Astier J., Vernochet C., Saint-Marc P., Guesnet J., Guezennec A., Amri E. Z., Dani C., et al. 2004. Adipocyte differentiation of multipotent cells established from human adipose tissue. Biochem. Biophys. Res. Commun. 315: 255–263. [DOI] [PubMed] [Google Scholar]
- 28.Lewin T. M., Wang S., Nagle C. A., Van Horn C. G., Coleman R. A. 2005. Mitochondrial glycerol-3-phosphate acyltransferase-1 directs the metabolic fate of exogenous fatty acids in hepatocytes. Am. J. Physiol. Endocrinol. Metab. 288: E835–E844. [DOI] [PubMed] [Google Scholar]
- 29.Wilson-Fritch L., Nicoloro S., Chouinard M., Lazar M. A., Chui P. C., Leszyk J., Straubhaar J., Czech M. P., Corvera S. 2004. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J. Clin. Invest. 114: 1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu M., Neilson A., Swift A. L., Moran R., Tamagnine J., Parslow D., Armistead S., Lemire K., Orrell J., Teich J., et al. 2007. Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am. J. Physiol. Cell Physiol. 292: C125–C136. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi N., Kawada T., Yamamoto T., Goto T., Taimatsu A., Aoki N., Kawasaki H., Taira K., Yokoyama K. K., Kamei Y., et al. 2002. Overexpression and ribozyme-mediated targeting of transcriptional coactivators CREB-binding protein and p300 revealed their indispensable roles in adipocyte differentiation through the regulation of peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 277: 16906–16912. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi N., Kawada T., Goto T., Yamamoto T., Taimatsu A., Matsui N., Kimura K., Saito M., Hosokawa M., Miyashita K., et al. 2002. Dual action of isoprenols from herbal medicines on both PPARgamma and PPARalpha in 3T3–L1 adipocytes and HepG2 hepatocytes. FEBS Lett. 514: 315–322. [DOI] [PubMed] [Google Scholar]
- 33.Goto T., Takahashi N., Kato S., Egawa K., Ebisu S., Moriyama T., Fushiki T., Kawada T. 2005. Double dioxygenation by mouse 8S-lipoxygenase: specific formation of a potent peroxisome proliferator-activated receptor alpha agonist. Biochem. Biophys. Res. Commun. 338: 136–143. [DOI] [PubMed] [Google Scholar]
- 34.Kuroyanagi K., Kang M. S., Goto T., Hirai S., Ohyama K., Kusudo T., Yu R., Yano M., Sasaki T., Takahashi N., et al. 2008. Citrus auraptene acts as an agonist for PPARs and enhances adiponectin production and MCP-1 reduction in 3T3–L1 adipocytes. Biochem. Biophys. Res. Commun. 366: 219–225. [DOI] [PubMed] [Google Scholar]
- 35.Fu J., Gaetani S., Oveisi F., Lo Verme J., Serrano A., Rodríguez De Fonseca F., Rosengarth A., Luecke H., Di Giacomo B., Tarzia G., et al. 2003. Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature. 425: 90–93. [DOI] [PubMed] [Google Scholar]
- 36.Krey G., Braissant O., L'Horset F., Kalkhoven E., Perroud M., Parker M. G., Wahli W. 1997. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol. Endocrinol. 11: 779–791. [DOI] [PubMed] [Google Scholar]
- 37.Svegliati-Baroni G., Candelaresi C., Saccomanno S., Ferretti G., Bachetti T., Marzioni M., De Minicis S., Nobili L., Salzano R., Omenetti A., et al. 2006. A model of insulin resistance and nonalcoholic steatohepatitis in rats: role of peroxisome proliferator-activated receptor-alpha and n-3 polyunsaturated fatty acid treatment on liver injury. Am. J. Pathol. 169: 846–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patsouris D., Mandard S., Voshol P. J., Escher P., Tan N. S., Havekes L. M., Koenig W., März W., Tafuri S., Wahli W., et al. 2004. PPARalpha governs glycerol metabolism. J. Clin. Invest. 114: 94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graves R. A., Tontonoz P., Ross S. R., Spiegelman B. M. 1991. Identification of a potent adipocyte-specific enhancer: involvement of an NF-1-like factor. Genes Dev. 5: 428–437. [DOI] [PubMed] [Google Scholar]
- 40.Brandt J. M., Djouadi F., Kelly D. P. 1998. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 273: 23786–23792. [DOI] [PubMed] [Google Scholar]
- 41.Knauf C., Rieusset J., Foretz M., Cani P. D., Uldry M., Hosokawa M., Martinez E., Bringart M., Waget A., Kersten S., et al. 2006. Peroxisome proliferator-activated receptor-alpha-null mice have increased white adipose tissue glucose utilization, GLUT4, and fat mass: role in liver and brain. Endocrinology. 147: 4067–4078. [DOI] [PubMed] [Google Scholar]
- 42.Leone T. C., Weinheimer C. J., Kelly D. P. 1999. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc. Natl. Acad. Sci. USA. 96: 7473–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bogacka I., Xie H., Bray G. A., Smith S. R. 2005. Pioglitazone induces mitochondrial biogenesis in human subcutaneous adipose tissue in vivo. Diabetes. 54: 1392–1399. [DOI] [PubMed] [Google Scholar]
- 44.Choo H. J., Kim J. H., Kwon O. B., Lee C. S., Mun J. Y., Han S. S., Yoon Y. S., Yoon G., Choi K. M., Ko Y. G. 2006. Mitochondria are impaired in the adipocytes of type 2 diabetic mice. Diabetologia. 49: 784–791. [DOI] [PubMed] [Google Scholar]
- 45.Muoio D. M., Way J. M., Tanner C. J., Winegar D. A., Kliewer S. A., Houmard J. A., Kraus W. E., Dohm G. L. 2002. Peroxisome proliferator-activated receptor-alpha regulates fatty acid utilization in primary human skeletal muscle cells. Diabetes. 51: 901–909. [DOI] [PubMed] [Google Scholar]
- 46.Loncar D., Afzelius B. A., Cannon B. 1988. Epididymal white adipose tissue after cold stress in rats. I. Nonmitochondrial changes. J. Ultrastruct. Mol. Struct. Res. 101: 109–122. [DOI] [PubMed] [Google Scholar]
- 47.Chen C. H., Lin E. C., Cheng W. T., Sun H. S., Mersmann H. J., Ding S. T. 2006. Abundantly expressed genes in pig adipose tissue: an expressed sequence tag approach. J. Anim. Sci. 84: 2673–2683. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y. X., Lee C. H., Tiep S., Yu R. T., Ham J., Kang H., Evans R. M. 2003. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell. 113: 159–170. [DOI] [PubMed] [Google Scholar]
- 49.Saha P. K., Kojima H., Martinez-Botas J., Sunehag A. L., Chan L. 2004. Metabolic adaptations in the absence of perilipin: increased beta-oxidation and decreased hepatic glucose production associated with peripheral insulin resistance but normal glucose tolerance in perilipin-null mice. J. Biol. Chem. 279: 35150–35158. [DOI] [PubMed] [Google Scholar]
- 50.Wong K. E., Szeto F. L., Zhang W., Ye H., Kong J., Zhang Z., Sun X. J., Li Y. C. 2009. Involvement of the vitamin D receptor in energy metabolism: regulation of uncoupling proteins. Am. J. Physiol. Endocrinol. Metab. 296: E820–E828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagase I., Yoshida T., Kumamoto K., Umekawa T., Sakane N., Nikami H., Kawada T., Saito M. 1996. Expression of uncoupling protein in skeletal muscle and white fat of obese mice treated with thermogenic beta 3-adrenergic agonist. J. Clin. Invest. 97: 2898–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mercader J., Ribot J., Murano I., Felipe F., Cinti S., Bonet M. L., Palou A. 2006. Remodeling of white adipose tissue after retinoic acid administration in mice. Endocrinology. 147: 5325–5332. [DOI] [PubMed] [Google Scholar]
- 53.Crowe S., Turpin S. M., Ke F., Kemp B. E., Watt M. J. 2008. Metabolic remodeling in adipocytes promotes ciliary neurotrophic factor-mediated fat loss in obesity. Endocrinology. 149: 2546–2556. [DOI] [PubMed] [Google Scholar]
- 54.Chen G., Koyama K., Yuan X., Lee Y., Zhou Y. T., O'Doherty R., Newgard C. B., Unger R. H. 1996. Disappearance of body fat in normal rats induced by adenovirus-mediated leptin gene therapy. Proc. Natl. Acad. Sci. USA. 93: 14795–14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki A., Okamoto S., Lee S., Saito K., Shiuchi T., Minokoshi Y. 2007. Leptin stimulates fatty acid oxidation and peroxisome proliferator-activated receptor alpha gene expression in mouse C2C12 myoblasts by changing the subcellular localization of the alpha2 form of AMP-activated protein kinase. Mol. Cell. Biol. 27: 4317–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee Y., Yu X., Gonzales F., Mangelsdorf D. J., Wang M. Y., Richardson C., Witters L. A., Unger R. H. 2002. PPAR alpha is necessary for the lipopenic action of hyperleptinemia on white adipose and liver tissue. Proc. Natl. Acad. Sci. USA. 99: 11848–11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimabukuro M., Koyama K., Chen G., Wang M. Y., Trieu F., Lee Y., Newgard C. B., Unger R. H. 1997. Direct antidiabetic effect of leptin through triglyceride depletion of tissues. Proc. Natl. Acad. Sci. USA. 94: 4637–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mazzucotelli A., Viguerie N., Tiraby C., Annicotte J. S., Mairal A., Klimcakova E., Lepin E., Delmar P., Dejean S., Tavernier G., et al. 2007. The transcriptional coactivator peroxisome proliferator activated receptor (PPAR)gamma coactivator-1 alpha and the nuclear receptor PPAR alpha control the expression of glycerol kinase and metabolism genes independently of PPAR gamma activation in human white adipocytes. Diabetes. 56: 2467–2475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.