Abstract
The cellular envelope of Mycobacterium tuberculosis is highly distinctive and harbors a wealth of unique lipids possessing diverse structural and biological properties. However, the ability to conduct global analyses on the full complement of M. tuberculosis lipids has been missing from the repertoire of tools applied to the study of this important pathogen. We have established methods to detect and identify lipids from all major M. tuberculosis lipid classes through LC/MS lipid profiling. This methodology is based on efficient chromatographic separation and automated ion identification through accurate mass determination and searching of a newly created database (Mtb LipidDB) that contains 2,512 lipid entities. We demonstrate the sensitive detection of molecules representing all known classes of M. tuberculosis lipids from a single crude extract. We also demonstrate the ability of this methodology to identify changes in lipid content in response to cellular growth phases. This work provides a customizable framework and resource to facilitate future studies on mycobacterial lipid biosynthesis and metabolism.
Keywords: lipid profiling, triacylglycerol, phthiocerol dimycocerosate
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis, possesses a cellular envelope rich with a complex milieu of lipids and carbohydrates that have been the subjects of intense research efforts for decades (1–3). Lipids comprise up to 60 percent of the Mtb cellular dry weight, and provide a hydrophobic barrier that enhances cell envelope impermeability. Furthermore, specific lipids of Mtb are shown to play critical roles in virulence and pathogenesis (4–6). The abundance and biological importance of the Mtb lipids has resulted in extensive and elegant studies to elucidate their structures and functions (1–3). In many cases, the lipids of Mtb are unique to this pathogen or shared only with other members of this genus.
Earlier studies demonstrate variability in lipid profiles among different strains of Mtb (7–11) and that minor variations in the structure of individual lipids can occur with changes in the growth environment (12–20). However, targeted and nontargeted assays that monitor changes in Mtb lipid profiles are generally performed by traditional TLC-based methods (21), and global lipidomics analyses in Mtb have been restricted due to limits in the technology to detect and rapidly identify a large number of lipids in a single experiment. Two-dimensional NMR was recently applied to examine global mycobacterial lipid profiles, and this approach allowed for the identification of key lipid differences in 13C-enriched cellular extracts (22). Although this approach easily detects changes in lipid patterns, it is limited by the complexity of the NMR spectra and the overlapping chemical properties of many lipids. Alternatively, MS-based lipidomic strategies allowing simultaneous detection, identification, and quantification of structurally diverse lipid components of Mtb also were evaluated. Leavell and Leary (23) developed an algorithm to analyze high-resolution Fourier transform-ion cyclotron resonance mass data obtained from direct infusions of complex Mtb lipid extracts. The algorithm isolates ions from spectra, and assigns identities from a “user-defined” lipid library based on exact mass. The methodology facilitated rapid comparisons of highly complex spectra and yielded significant findings on the metabolic control of virulence lipids (15). Shui et al. (24) took an alternative strategy and demonstrated the utility of C18 reversed-phase HPLC in combination with ESI-MS to efficiently separate and detect complex lipids of lower abundance. Several anionic Mtb lipid classes were characterized, and lipid profiling was able to identify mycolic acid profile shifts that occurred in response to different physiological growth conditions. However, these previous MS-based analyses were not supported by a complete and portable Mtb lipid database, and did not fully integrate database interrogation with the ability to resolve individual lipids by HPLC and ion data extraction techniques.
Building upon the previous success of MS-based lipidomic approaches for Mtb and to provide tools for comprehensive lipid profiling, we generated a novel database of Mtb lipids, “Mtb LipidDB” that allows for lipid identification using accurate mass measurements obtained in either negative- or positive-ion modes. When coupled to molecular feature (MF) detection from LC/MS spectra, the Mtb LipidDB allows for automated exact mass-based lipid identification. Applying this approach to crude Mtb lipid extracts, we achieved identification of the majority of known extractable lipids from this bacterium. Moreover, we demonstrated the utility of this methodology to rapidly identify quantitative changes that occur in the lipid profile of Mtb in response to growth conditions.
MATERIALS AND METHODS
Bacterial growth and lipid extractions
Mtb strain H37Rv was propagated in glycerol alanine salts medium (25) at 37°C for 14 days, as previously described (26). Cells were harvested by centrifugation (3,000 g for 10 min), washed three times with PBS, pH 7.4, and inactivated by γ irradiation (27). To prepare crude lipid extracts, a modified Bligh and Dyer (28) method was used. Briefly, 200 mg of wet cells were lyophilized in a 13 × 100 mm silanized borosilicate glass tube with a Teflon screw cap. The dried cells were extracted with 6 ml of chloroform-methanol-water (10:10:3; v/v/v) overnight, with constant stirring at room temperature. The sample was centrifuged at 3,000 g for 10 min to remove the delipidated cells. The monophasic lipid extract was transferred to a 13 × 100 mm glass tube, dried under a gentle stream of nitrogen, and stored at −20°C until use. Prior to LC/MS analysis, the dried lipid extract was dissolved in chloroform-methanol-water (10:10:3; v/v/v) to approximate a 4 µg/µl concentration, centrifuged at 3,000 g for 10 min, and transferred to an autosampler vial. To prepare the dilution series, cells were homogenized by bead-beating and serially diluted 2-fold with water in glass tubes. The diluted homogenates were lyophilized, and lipids were extracted as described above. Based on the starting wet weight of the cell pellet, the lipid dry weights were estimated from routinely observed lipid extraction yields.
For Mtb H37Rv growth phase comparisons, three 500 ml Nephelo-sidearm flasks containing 50 mm stirbars and 200 ml Middlebrook 7H9 broth (Difco; Detroit, MI) supplemented with oleic acid-dextrose catalase (Difco) and 0.05% Tween 80 (Sigma-Aldrich; St. Louis, MO) were inoculated with 2 ml of a logarithmic culture to achieve a theoretical optical density at 580 nm (OD580) of 0.004. The flasks were capped tightly and incubated at 37°C under constant stirring at 150 rpm. The OD580 of each culture was measured at regular intervals for 11 days, and aliquots (10 ml) were taken at 73, 108, and 265 h, corresponding to logarithmic, transitionary, and stationary growth phases, respectively. The cell pellets were harvested and washed three times with PBS, pH 7.4. Crude lipid extracts were prepared as described above and randomized for LC/MS analyses.
LC
An Agilent 1200 HPLC (Agilent Technologies; Palo Alto, CA) with a 2.1 inner diameter (ID) × 150 mm, 3.5 µm XBridge C18 column (Waters Corp.; Milford, MA) heated to 45°C was used with a binary solvent system and a flow rate of 320 µl/min. A 2.1 ID × 10 mm, 3.5 µm XBridge C18 guard column (Waters) was placed in series in front of the analytical column. The system was equilibrated with 100% solvent A [5 mM ammonium acetate in methanol-water (99:1; v/v)], and an aliquot of the lipid extract (5 µl = 20 µg dried extract) was applied to the column. Solvent A was maintained at 100% for 2.0 min, followed by a 30.0 min linear gradient to 100% solvent B [5 mM ammonium acetate in n-propanol-hexane-water (79:20:1; v/v/v)], and held at 100% solvent B for 3.0 min. All solvents and chemicals purchased were MS or HPLC grade.
MS
An Agilent 6220 time-of-flight (TOF) mass spectrometer equipped with an Agilent ESI/atmospheric-pressure chemical ionization (APCI) multimode source was used for accurate mass analysis of the LC eluent. Positive- (+) and negative- (−) ion data were generated by operation of the mass spectrometer in a mixed ESI/APCI mode with a capillary voltage of 3000 V, nebulizer of 45 psig, drying gas of 8 l/min, gas temperature of 300°C, vaporizer temperature of 200°C, corona of 2 µA, fragmentor of 120 V, charging voltage of 2000 V, skimmer of 60 V, and octopole radio frequency voltage of 250 V. Mass spectra were acquired in 4 GHz high-resolution mode at a rate of 1.02 spectra/s and 9,700 transients/spectrum, and data were collected as profiled spectra over a mass range of 250 to 3,200 Da. Mass calibration was performed with an Agilent tune mix from 100 to 2,700 Da, and an external reference sprayer introduced mass ions of m/z 922.009798 (+ ion) and m/z 980.016375 (− ion) to enable accurate mass determinations. Data were collected with the Agilent MassHunter WorkStation Data Acquisition software, version B.02.00.
An Agilent 6520 qTOF was used for MS/MS analyses of the triacylglycerol (TG) lipid-group [M+NH4]+ ions. The instrument setup was the same as described above, except that the OCT RF was set at 750 V. Positive-ion mass spectra were acquired in Auto MS/MS mode, and collision energies with slope of 6.5 V/100 Da and offset 2.0V were used for fragmentation.
Mtb LipidDB development
To enable accurate mass MS-based searching of lipid ions, the novel database “Mtb LipidDB” was created with Microsoft Excel 2007 Pro (Microsoft; Redmond, WA). The database was populated with molecular formulas, structures, and exact masses for the known lipids of Mtb. This information was obtained from a survey of the extensive Mtb biochemical literature describing lipid structures purified from Mtb H37Rv and Mycobacterium bovis Bacillus Calmette-Guérin (BCG) strains (see references in supplementary material). The collective data from these principal publications were used to calculate every theoretically possible fatty acyl combination for each lipid subclass and level 4 class. The reported structural variations were used to generate a representative molecular structure for each Mtb lipid subclass or level 4 class using ChemBioDraw Ultra 11.0 software (Cambridgesoft; Cambridge, MA). Lipids considered to be biosynthetic intermediates, such as phosphatidic acid, hydroxymycolates, and sugar-linked decaprenyl phosphates, were not included in the Mtb LipidDB. Simple free FAs, including tuberculostearic acid, as well as lipid subclasses lacking sufficient detailed structural definition (i.e., definition of the FA types), such as the triacylated trehaloses (29), were also not included in the database. Although absent in the laboratory strain Mtb H37Rv used in this study, the glycosylated phthiodiolone dimycocerosates (DIM Bs) and glycosylated phthiocerol dimycocerosates (DIM As) (phenolic glycolipid) were included in the database because of their biological importance in clinical strains.
In addition to the parent Mtb LipidDB, two searchable database files named “+MH_MtbLipid.csv” and “−MH_MtbLipid.csv” were developed to interface with the Agilent MassHunter software and allow for searching of MS data against the Mtb LipidDB. The +MH_MtbLipid.csv and −MH_MtbLipid.csv files contained entries for each molecular ion that corresponded to a single lipid group in positive- and negative-ion modes, respectively. The MassHunter software provides the option to identify and collapse multiple molecular ions and adducts into a single feature. However, when only a single molecular ion was detected, the software default was to subtract or add the mass of a proton (1.007276 Da) to the experimentally derived molecular ion before interrogation of the +MH_Mtb_Lipid.csv and −MH_MtbLipid.csv database files, respectively. Thus, to allow for the matching of lipid groups that ionized as a single adducted molecular ion other than H+ or H−, (e.g., [M+Na]+ and [M+Ac]− ions), each +MH_Mtb_Lipid.csv and −MH_MtbLipid.csv database file entry contained a mass value equal to the appropriate adduct but with 1.007276 Da subtracted from or added to the calculated molecular ion, respectively. The selection of molecular ions included in each searchable file was an iterative process based on empirically observed ionization properties of each lipid subclass. For instance, in positive-ion mode [M+H]+, [M+NH4]+, [M+Na]+, [2M+Na]+, and [M+2Na-H]+ ions were observed for phosphatidylethanolamines (PEs), whereas TGs were found to primarily generate [M+NH4]+, [M+Na]+, and [2M+Na]+ ions. Thus, with all 2,512 lipid groups represented, the +MH_MtbLipid.csv file contained 9,178 molecular ion entries, and the −MH_MtbLipid.csv file contained 5,311 molecular ion entries. The Mtb LipidDB and the searchable database files are provided as supplementary material, and have been made freely available at http://www.mrl.colostate.edu.
Data processing and analyses
LC/MS data files were processed with the MassHunter Qualitative Analysis Software, version B.02.00 (Agilent Technologies; Santa Clara, CA). MFs were extracted from the raw data using the MF extraction (MFE) algorithm. This algorithm locates related covariant ions (isotopes and charge states) from accurate mass LC/MS data, and combines these ions into a single feature. The MFE parameters used were: extraction algorithm, small molecule; peak filters, ≥500 counts; ion species, +H and −H only; peak spacing tolerance, 0.0025 m/z plus 7.0 ppm; isotope model, common organic molecules; charge state, 1–2; compound filters, none; mass filters, none; and mass defect, none. The resulting MFs were then identified with MassHunter by searching the +MH_MtbLipid.csv (+ ion data) or the −MH_MtbLipid.csv (− ion data) database file with the following search parameters: values to match, mass only; match tolerance, 5 ppm; charge carriers, +H and −H; and charge state range, 1–2. The lists of both identified and unidentified features were exported as an analysis report in Microsoft Excel (Microsoft) format. Further data comparisons were accomplished with Microsoft Excel. For quantitative purposes, lipid groups were compared using the most-dominant molecular ion volumes observed for each lipid subclass as follows: TG, [M+Na]+; DIM A, [M+Na]+; DIM B, [M+Na]+; PE, [M−H]−; phosphatidylinositol (PI), [M−H]−; and cardiolipin (CL), [M−H]−
RESULTS
Development of the novel Mtb LipidDB
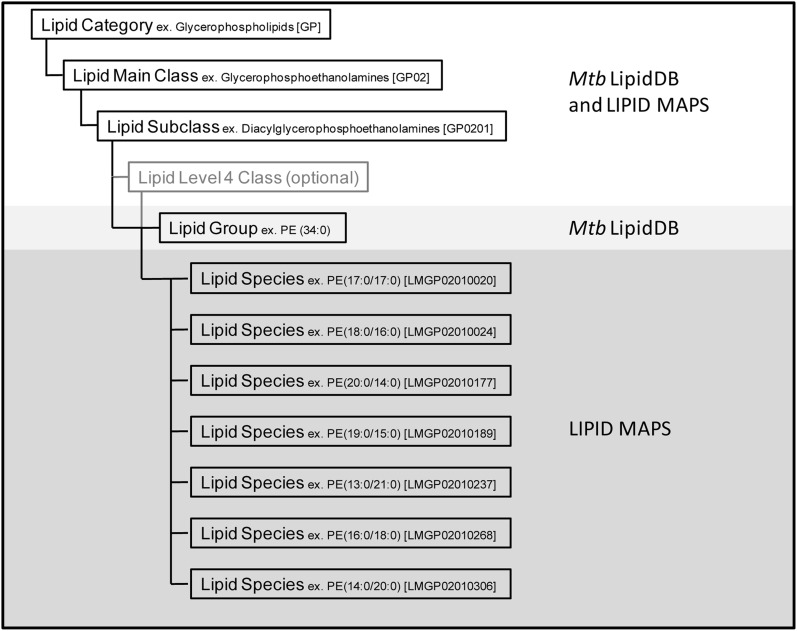
The Mtb LipidDB (supplementary data and summarized in Table 1) was organized in a manner that adhered as closely as possible to the classification hierarchy and structural nomenclature set forth by the LIPID MAPS consortium (LIPID Metabolites and Pathways Strategy; http://www.lipidmaps.org) (30, 31). Six of the eight lipid categories defined by LIPID MAPS are represented in the Mtb LipidDB. The Mtb LipidDB contains 15 lipid main classes, 46 lipid subclasses, and 16 level 4 lipid classes. In instances where Mtb lipids did not fit into the LIPID MAPs classification system, 30 novel lipid subclasses (e.g., diacyltrehaloses) and 16 level 4 classes [e.g., α-mycolic acids (Alpha-MA)] were created. These novel subclasses and level 4 classes were developed for Mtb LipidDB organization purposes only, and have not been accepted by LIPID MAPS as part of their classification hierarchy. A key organizational difference between the Mtb LipidDB and LIPID MAPs occurs at the species level of classification (Fig. 1). In lieu of “lipid species,” the Mtb LipidDB uses “lipid groups,” in which each lipid group possessed a unique chemical formula corresponding to a unique exact mass. Unlike lipid species, an individual lipid group was not distinguished by stereochemistry, unsaturated bond position, or the length and position of individual fatty acyl substituents. However, each lipid group allowed assignment of the head group composition and the sum composition (total fatty acyl carbon number and total degree of fatty acyl unsaturation), and in many cases, represents multiple unique lipid species. The lipid group classification level was designed for the express purpose of simplifying exact mass-based MS identification, in which a successful database query returns a single lipid group that could represent multiple isobaric lipid species. An additional justification for this classification level was the general paucity of sufficient detailed structural information at the lipid species level for many of the mycobacterial lipids reported in the current body of literature. In total, the Mtb LipidDB contains 2,512 lipid groups, and when all potential ion adducts were incorporated, the searchable database files contain 14,489 mass entries.
Fig. 1.
Organizational summary of the Mtb LipidDB and LIPID MAPS classification systems.
TABLE 1.
Summary of the Mtb lipid database
| Fatty acyls (FA) [FA] {745} | |||
| FAs and conjugates [FA01] {641} | |||
| Branched FAs [FA0102] {41} | |||
| Mycolic acids (MAs) [FA0116] {600} | |||
| Alpha mycolic acids (α-MAs) [FA0116z] {21} | |||
| Methoxy mycolic acids (methoxy-MAs) [FA0116z] {21} | |||
| Keto mycolic acids (keto-MAs) [FA0116z] {21} | |||
| Glucose monomycolates (GMMs) [FA0116z] {84} | |||
| Trehalose monomycolates (TMMs) [FA0116z] {84} | |||
| Trehalose dimycolates (TDMs) [FA0116z] {369} | |||
| Fatty esters [FA07] {104} | |||
| Wax diesters [FA0707] {52} | |||
| Phthiocerol dimycocerosates (DIMAs) [FA0707z] {26} | |||
| Phthiodiolone dimycocerosates (DIMBs) [FA0707z] {26} | |||
| Glycosylated wax diesters (GWEs) [FA07y] {52} | |||
| Glycosylated phthiocerol dimycocerosates (PGL-tbs) [FA07yz] {26} | |||
| Glycosylated phthiodiolone dimycocerosates [FA07yz] {26} | |||
| Glycerophospholipids (GPs) [GP] {1295} | |||
| Glycerophosphoethanolamines (PEs) [GP02] {36} | |||
| Diacylglycerolphosphoethanolamines (PEs) [GP0201] {27} | |||
| Monoacylglycerolphosphoethanolamines (lyso-PEs) [GP0205] {9} | |||
| Glycerophosphoglycerols (PGs) [GP04] {50} | |||
| Diacylglycerophosphoglycerols (PGs) [GP0401] {40} | |||
| Monoacylglycerophosphoglycerols (lyso-PGs) [GP0405] {10} | |||
| Glycerophosphoinositols (PIs) [GP06] {34} | |||
| Diacylglycerophosphoinositols (PIs) [GP0601] {26} | |||
| Monoacylglycerophosphoinositols (lyso-PIs) [GP0605] {8} | |||
| Glycerophosphoglycerophosphoglycerols (CLs) [GP12] {138} | |||
| Diacylglycerophosphoglycerophosphodiradylglycerols (CLs) [GP1201] {138} | |||
| Glycerophosphoinositolglycans [GP15s] {1037} | |||
| Monoacylglycerophosphoinositolmonomannosides (lyso-PIM1s) [GP15y] {8} | |||
| Diacylglycerophosphoinositolmonomannosides (PIM1s) [GP15y] {26} | |||
| Monoacylated diacylglycerophosphoinositolmonomannosides (Ac1PIM1s) [GP15y] {63} | |||
| Monoacylglycerophosphoinositoldimannosides (lyso-PIM2s) [GP15y] {8} | |||
| Diacylglycerophosphoinositoldimannosides (PIM2s) [GP15y] {26} | |||
| Monoacylated diacylglycerophosphoinositoldimannosides (Ac1PIM2s) [GP15y] {63} | |||
| Diacylated diacylglycerophosphoinositoldimannosides (Ac2PIM2s) [GP15y] {91} | |||
| Monoacylglycerophosphoinositoltrimannosides (lyso-PIM3s) [GP15y] {8} | |||
| Diacylglycerophosphoinositoltrimannosides (PIM3s) [GP15y] {26} | |||
| Monoacylated diacylglycerophosphoinositoltrimannosides (Ac1PIM3s) [GP15y] {63} | |||
| Diacylated diacylglycerophosphoinositoltrimannosides (Ac2PIM3s) [GP15y] {91} | |||
| Monoacylglycerophosphoinositoltetramannosides (lyso-PIM4s) [GP15y] {8} | |||
| Diacylglycerophosphoinositoltetramannosides (PIM4s) [GP15y] {26} | |||
| Monoacylated diacylglycerophosphoinositoltetramannosides (Ac1PIM4s) [GP15y] {63} | |||
| Diacylated diacylglycerophosphoinositoltetramannosides (Ac2PIM4s) [GP15y] {91} | |||
| Monoacylglycerophosphoinositolpentamannosides (lyso-PIM5s) [GP15y] {8} | |||
| Diacylglycerophosphoinositolpentamannosides (PIM5s) [GP15y] {26} | |||
| Monoacylated diacylglycerophosphoinositolpentamannosides (Ac1PIM5s) [GP15y] {63} | |||
| Diacylated diacylglycerophosphoinositolpentamannosides (Ac2PIM5s) [GP15y] {91} | |||
| Monoacylglycerophosphoinositolhexamannosides (lyso-PIM6s) [GP15y] {8} | |||
| Diacylglycerophosphoinositolhexamannosides (PIM6s) [GP15y] {26} | |||
| Monoacylated diacylglycerophosphoinositolhexamannosides (Ac1PIM6s) [GP15y] {63} | |||
| Diacylated diacylglycerophosphoinositolhexamannosides (Ac2PIM6s) [GP15y] {91} | |||
| Glycerolipids (GLs) [GL] {218} | |||
| Monoradylglycerols [GL01] {18} | |||
| Monoacylglycerols (MGs) [GL0101] {18} | |||
| Diradylglycerols [GL02] {61} | |||
| Diacylglycerols (DGs) [GL0201] {61} | |||
| Triradylglycerols [GL03] {139} | |||
| Triacylglycerols (TGs) [GL0301] {139} | |||
| Prenol lipids (PRs) [PR] {7} | |||
| Quinones and hydroquinones [PR02] {4} | |||
| Ubiquinones [PR0201] {4} | |||
| Polyprenols [PR03] {3} | |||
| Bactoprenols [PR0301] {1} | |||
| Bactoprenol monophosphates [PR0302] {1} | |||
| Bactoprenol diphosphates [PR0303] {1} | |||
| Saccharolipids (SLs) [SL] {226} | |||
| Acyltrehaloses [SL03] {226} | |||
| Diacyltrehaloses (DATs) [SL03y] {13} | |||
| 2,3-di-O-acyltrehaloses (DAT1s) [SL03yz] {6} | |||
| 2,3-di-O-acyltrehaloses (DAT2s) [SL03yz] {7} | |||
| Polyacyltrehaloses (PATs) [SL03y] {50} | |||
| Sulfolipids (SLs) [SL03y] {163} | |||
| Sulfolipid I (SL-Is) [SL03yz] {52} | |||
| Sulfolipid II or II’ (SL-IIs or SL-IIs) [SL03yz] {52} | |||
| Sulfolipid III (SL-IIIs) [SL03yz] {35} | |||
| Diacylated sulfolipid (Ac2SGLs) [SL03yz] {24} | |||
| Polyketides (PKs) [PK] {21} | |||
| Linear polyketides [PK01] {5} | |||
| Mannosyl-b1-phosphomycoketides (MPMs) [PK01y] {5} | |||
| Non-ribosomal peptides/polyketide hybrids [PK14] {16} | |||
| Mycobactins (Mbts) [PK14y] {16} | |||
| LEGEND (indentation represents classification levels as follows) | |||
| Category (abbreviation) [LIPID MAPs identifier] {# of lipid groups in category} | |||
| Main Class (abbreviation) [LIPID MAPs identifier] {# of lipid groups in main class} | |||
| Subclass (abbreviation) [LIPID MAPs identifier, y denotes new proposed subclass] {# of lipid groups in subclass} | |||
| Level 4 class (abbreviation) [LIPID MAPs identifier, z denotes new proposed level 4 class] {# of lipid groups in level 4 class} | |||
LC/MS and automated ion identification allow lipidomic analysis of Mtb
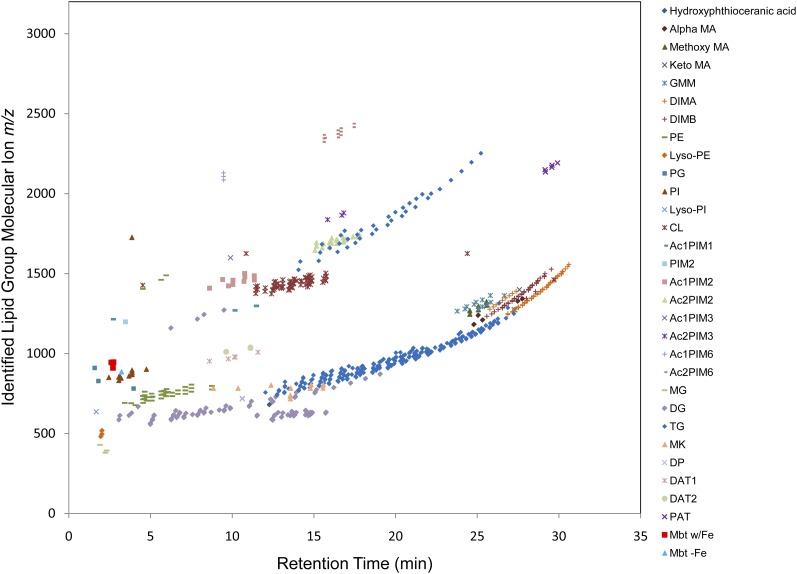
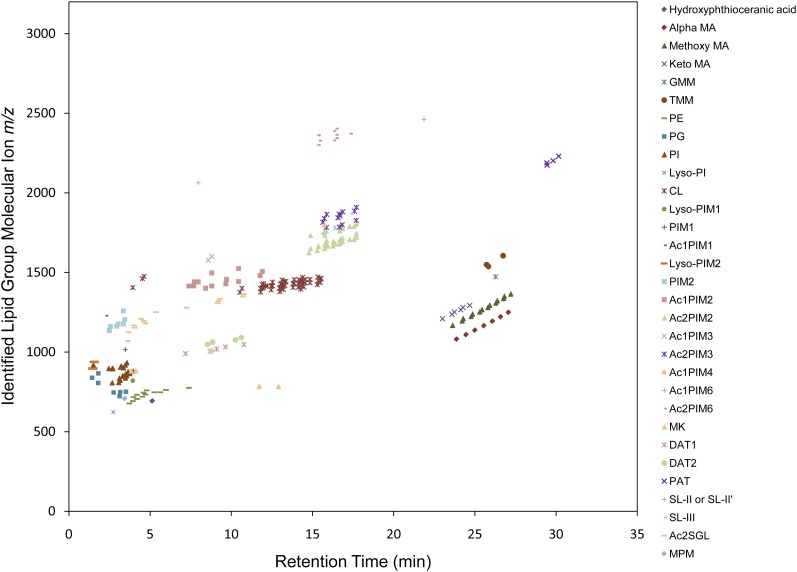
A crude lipid extract from Mtb strain H37Rv grown in standard liquid medium was subjected to C18 reversed-phase chromatography, and the effluent was directly analyzed by accurate-mass (ESI/APCI)-MS. Data collected in the positive- and negative-ion modes were processed using the MFE algorithm of the MassHunter software package, resulting in 1,916 and 744 MFs for the positive- and negative-ion spectra, respectively. The MF lists were searched against the +MH_MtbLipid.csv and −MH_MtbLipid.csv database files with a mass error tolerance of ±5 ppm, yielding database matches for 672 of the 1,916 positive-ion MFs (35.1%) and 248 of the 744 negative-ion MFs (21.8%). Supplementary Table I provides detailed attributes for each identified MF. The identified MFs accounted for 74.8% and 58.4% of the total positive- and negative-ion MF peak abundances (ion volumes), respectively. This indicated that the unassigned MFs were probably low-abundant products representing undefined ion adducts, minor uncharacterized lipid structures, and/or background contaminating molecules. The absolute average mass accuracies were 1.46 (±1.60 SD) ppms for positive-ion MFs and 1.87 (±1.96 SD) ppms for negative-ion MFs. The combined positive- and negative-ion data resulted in the identification of lipid groups from all 6 lipid categories, all 15 lipid main classes, 31 of the 45 lipid subclasses, and 12 of the 14 level 4 classes represented in the Mtb LipidDB. It should be noted that the glycosylated wax ester subclass is not produced by Mtb strain H37Rv and was not included in the number of potential lipid subclasses and level 4 classes. The assigned MFs, grouped by lipid subclass or level 4 class, were plotted by molecular ion m/z and LC retention time (Figs. 2, 3). This demonstrated the clustering of the lipid subclasses or level 4 classes on the basis of retention time and mass. Moreover, the large numbers of plotted MFs revealed the complexity of the ion patterns that must be assigned for each Mtb lipid subclass or level 4 class. To further illustrate the sample complexity and the LC separation, extracted chromatograms for all identified MFs are shown (see supplementary Fig. I).
Fig. 2.
Identified MFs from positive-ion LC/MS analyses of an Mtb total lipid extract. Six hundred seventy-two MF ions were matched to lipid groups representing 25 lipid subclasses from the Mtb LipidDB. See supplementary material for lipid group abbreviations.
Fig. 3.
Identified MFss from negative-ion LC/MS analyses of an Mtb total lipid extract. Two hundred forty-eight MF ions were matched to lipid groups representing 24 lipid subclasses from the Mtb LipidDB. See supplementary material for lipid group abbreviations.
Important observations to consider in MF assignment
As expected, it was possible to observe multiple MFs with nearly identical retention times that matched to different molecular ion entries from the same lipid group, owing to the formation of multiple ion adducts. For instance, there was a preponderance of both [M+Na]+ and [M+NH4]+ ions observed for TGs, whereas there was bias toward both [M+H]+ and [M+Na]+ ions for PEs. However, we also observed instances of multiple MFs with different chromatographic behaviors that matched to the same database entry (i.e., the same molecular ion). Specifically, there were 109 (+ ion mode) and 24 (− ion mode) MFs assigned to the same database entry as that of at least one other MF. This probably resulted from the detection of multiple isobaric lipid species that shared the same sum chemical composition but disparate elution times caused by slightly different fatty acyl repertoires. Thus, subtracting instances in which multiple MFs matched to the same lipid group, a total of 415 unique lipid groups out of the 2,512 present in the database were identified in the Mtb lipid extract. Positive-ion and negative-ion data identified 314 and 193 unique lipid groups, respectively, with an overlap of 92 lipid groups.
The exact mass search window (in this case ±5 ppm) could also impact MF assignment. Specifically, in the Mtb LipidDB, there were 29 instances in which a lipid group mass was within ±5 ppm of another lipid group mass. For example, the mass of the sulfolipid III (SL-III) (C90) lipid group is 0.8 ppm less than the mass of CL (81:1). The issue of mass window overlap was further complicated by the inclusion of multiple molecular ions for each lipid group. For instance, the database mass entry for the diacylated diacylglycerophosphoinositoldimannoside (Ac2PIM2) (70:0) [M+Na−2H]− molecular ion is 1.4 ppm less than the database mass entry for the Ac2PIM2 (72:3) [M−H]− molecular ion. In total, there were 154 out of 9,177 possible instances of mass window overlap in the +MH_MtbLipid.csv file, and 244 out of 5,310 possible instances in the −MH_MtbLipid.csv file. Analyses of the Mtb crude lipid extract resulted in only 29 positive-ion and 28 negative-ion MFs that matched to two lipid entries. However, the MassHunter software provided a scoring algorithm for each database match based on a combination of ppm accuracy and isotopic abundance pattern fit. This scoring mechanism generally appeared to select the correct lipid group, inasmuch as the top candidate displayed a retention time in agreement with the respective parent lipid subclass.
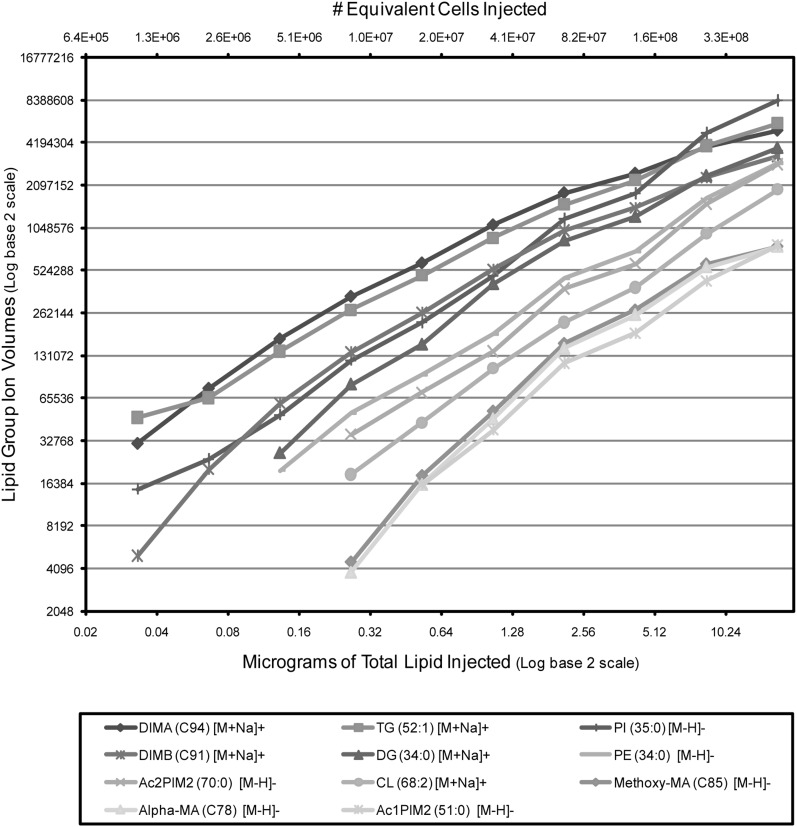
Quantitative and detection limit capabilities
In addition to providing mass and retention time information, the MassHunter MFE algorithm provided a measurement of abundance, termed “volume” for each MF, where volume = [retention time window] × [isotope ion peak heights]. To assess the linearity of this quantitative feature for identified lipid groups, a dilution series of Mtb cells was extracted and the total lipids were analyzed by LC/MS as described above. The MF volumes for representative lipid groups yielded a linear relationship to the quantity of lipid over a dynamic range of approximately three logs. Further, the most abundant lipid subclasses (DIM A, DIM B, PI, TG) could be detected in extracts that were equivalent to <106 Mtb cells (Fig. 4).
Fig. 4.
Ion response scatter plots for an Mtb total lipid extract dilution series. The observed ion volumes were selected from representative lipid group ions detected across a wide dynamic range. Values were plotted against the amount of lipid extract injected (bottom x axis) and the equivalent number of extracted Mtb cells (top x axis).
Assessment of Mtb lipid profiles from different growth phases
Triplicate liquid cultures of Mtb strain H37Rv were sampled at three time points corresponding to logarithmic phase, stationary phase, and an intermediate growth phase between these two termed the “transitionary” phase (see supplementary Fig. II). Crude lipid extracts were prepared from the collected cells and subjected to LC/MS analyses. MF volumes were used to establish relative quantities for each identified lipid group, and comparisons revealed growth phase-dependent differences in lipid class abundance and composition.
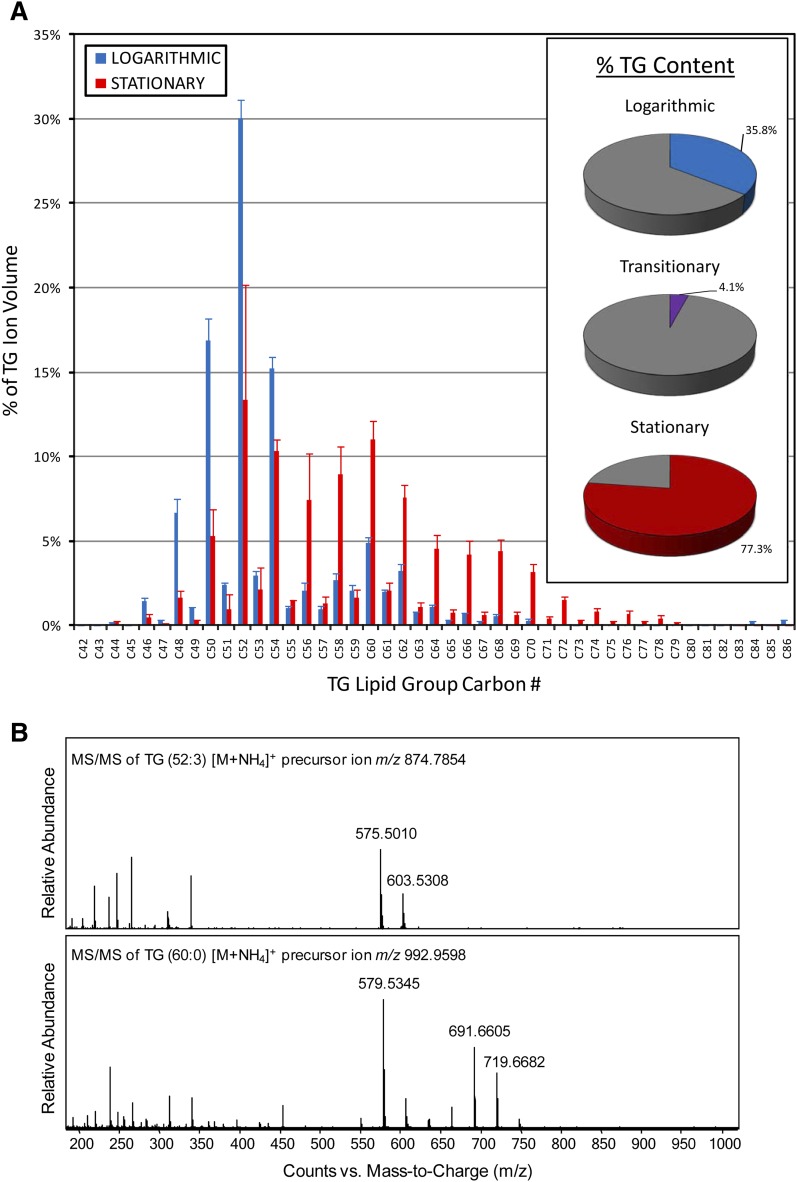
Striking differences in both total TG abundance and TG fatty acyl composition were observed across each of the three growth phases. When expressed as a percentage of all identified lipid groups (+ ion data), TG lipid groups accounted for 35.8%, 4.1%, and 77.3% of the ion volumes in logarithmic, transitionary, and stationary phases, respectively, indicating significant changes in cellular concentrations of TG as the cultures aged (Fig. 5A, inset). Large differences in TG group abundances were also observed from the LC/MS chromatograms (see supplementary Fig. II). The TG FA composition also varied between growth phases, with a shift from predominantly lower mass TG lipid groups in logarithmic phase to higher mass TG groups in stationary phase (Fig. 5A; see supplementary Fig. III). The sum compositions indicated that the smaller TGs were probably esterified with C16, C18, and C19 FAs, whereas longer C22, C24, and C26 acyl functions probably esterified the larger, stationary phase-specific TGs. The latter observation fits well with previous structural reports of mycobacterial TG (13, 32, 33). To further support this structural information, the most-abundant logarithmic and stationary phase TG group [M+NH4]+ ions were targeted for collision-induced dissociation MS/MS (Fig. 5B). Major diacyl product ions were observed that were consistent, with C16:1 and C18:1 being the primary FAs of the major logarithmic TG (52:3) group, and C16:0, C18:0, and C26:0 FAs esterified to the major stationary phase-specific TG (60:0) group.
Fig. 5.
Comparison of TG profiles from Mtb growth phases. A: TG lipid group ion volumes with the same carbon number were combined and normalized to total TG ion volume (for individual TG lipid group profiles, see supplementary Fig. IV). Error bars indicate ±SD (n = 3). The inset shows relative TG abundance as percent total identified lipid group ion volumes. B: MS/MS spectra of the [M+NH4]+ molecular ions for representative TG groups. The major logarithmic-phase TG (52:3) ion produced two major diacyl product ions at m/z 575.5010 and m/z 603.5308 that resulted from neutral losses of C18:1 and C16:1 FAs, respectively. The major stationary phase-specific TG (60:0) ion produced three major diacyl product ions at m/z 579.5345, m/z 691.6605, and m/z 719.6682, which indicated neutral losses of C26:0, C18:0, and C16:0 FAs, respectively.
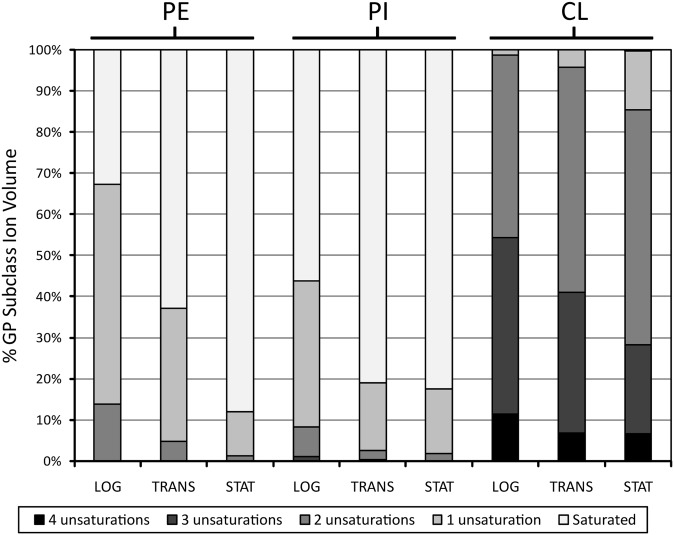
Growth phase-dependent changes in glycerophospholipid (GP) sum compositions were also observed (see supplementary Fig. IVA–C). For instance, PE (34:1) was the most-abundant PE lipid group in the logarithmic phase, whereas the PE (34:0) lipid group dominated in the stationary phase. Interestingly, the sum composition of the PI and PE lipid groups differed significantly from each other, even in the same growth phase, indicating that FA distributions were specific to each GP subclass. Regardless of GP subclass, however, each of the PE, PI, and CL subclasses showed a significant increase in the saturated bond content of their fatty acyl moieties as the cultures aged (Fig. 6).
Fig. 6.
Comparison of GP subclass unsaturated bond content from Mtb growth phases. Percent of each lipid subclass containing lipid groups with fatty acyl sum composition possessing zero to four unsaturated bonds. LOG, logarithmic phase; TRANS, transitionary phase; STAT, stationary phase.
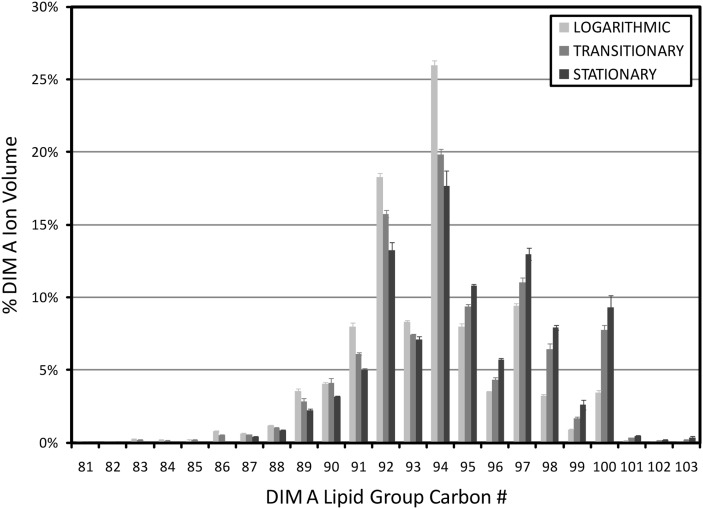
Finally, notable changes were observed within the wax diester subclass. Comparisons of the relative abundances of DIM A lipid groups revealed significant increases in higher-mass DIM A forms as the cultures entered stationary phase (Fig. 7). The average DIM A mass was 1,367.2 (±0.6 SD) atomic mass units (amu) in logarithmic phase, 1,377.4 (±1.5 SD) amu in transitionary phase, and 1,385.9 (±1.8 SD) amu in stationary phase. Comparable levels of mass increases were also observed for DIM B lipid groups (data not shown). In addition to the observed mass increases, the ratio of total DIM A to total DIM B lipid group ion volumes increased as the cultures aged. The total DIM A/B content was 1.31 (±0.08 SD) in logarithmic phase, 1.85 (±0.16 SD) in transitionary phase, and 3.43 (±0.40 SD) in stationary phase.
Fig. 7.
Growth phase-dependent changes in DIM A molecular size. DIM A group ion volumes were normalized to total DIM A ion volumes. Error bars indicate ± SD (n = 3).
DISCUSSION
The global characterization of the Mtb lipid composition presents a significant technical challenge, owing to the structural diversity within this group of macromolecules and the number of complex structures that are unique to Mtb and related species. However, based on the efforts of earlier investigators to fully elucidate the structure of a large majority of these products, it was possible to derive a database of Mtb lipids that possessed 2,512 structures and that could be interrogated with MS data based on the 14,489 mass entries that accounted for multiple ion adducts of both positive- and negative-ion data. The interrogation of this database with MS data of whole-lipid extracts from Mtb allowed for the automated detection and identification of 415 lipid structures encompassing all 15 of the known classes of Mtb lipids. This work represents the largest single survey of Mtb lipid structures from a single study and provides the groundwork for lipidomics studies that are on par with those now being performed with eukaryotic organisms (34).
The Mtb LipidDB was developed to allow lipidomic investigations in an unbiased manner. Thus, lipid group structures and masses were obtained by calculating all potential combinations of the FAs reported in the literature for a specific lipid. This approach resulted in database inclusion of lipid structures containing fatty acyl combinations not previously reported, but that are possible based on the biochemical literature. This expansion of the database was most noted with the mycobacterial lipids commonly referred to as phosphatidylinositolmannosides (PIMs), where 1,037 of the 2,512 lipid structures in the Mtb LipidDB were assigned to a PIM structure. This reflects the variable composition of the PIM structures with one to six mannose residues and one to four acyl moieties (35). Of the 1,037 potential PIM structures included in the Mtb LipidDB, 77 were identified in our analyses. This can be directly compared with other MS-based studies of PIMs isolated from M. bovis BCG (36, 37), where 73 of the possible 1,037 PIM groups included in our database were identified. The inclusion of theoretical lipid structures based on sound biochemical evidence, and more-general descriptions in the literature of acyl composition, are not uncommon. In fact in eukaryotes, it is estimated that there are at least 25,000 TG structures, including those with ether-linked FAs (38).
Beyond the ability to identify individual lipid groups or species, MS offers the potential to perform relative quantitative analyses based on the total ion abundance of individual MFs (39). This approach was used to monitor lipid changes over different phases of Mtb growth in batch culture. TG abundance waned and waxed dramatically as the cultures aged, and TG FA sum compositions also shifted markedly, with the appearance of higher-molecular-weight TG groups in stationary phase. For the purposes of this study, the stationary phase was the point at which the total bacterial growth rate ceased as measured by optical density. Such tightly capped batch cultures grown to high cellular densities have been suggested to be depleted of oxygen (40); thus the observed accumulation of TG in the stationary phase would agree with previous reports demonstrating TG production increases under hypoxic and/or stressful conditions (12–14, 16, 33). Our structural analyses suggested that C26:0 FAs were major components of these TG groups, also in agreement with previous studies on hypoxia-induced TG molecules (13, 14, 33). Alterations in FA composition were not limited to the TGs, inasmuch as the PE, PI, and CL lipid classes also demonstrated similar changes that were class specific. However, common to each class was an observed increase in overall saturation levels as the cultures aged. It should be noted that the presence of polysorbate (Tween 80) in the media may have influenced fatty acyl compositions, considering that this detergent supplies oleic acid esters for growth. The average size of DIM A and DIM B lipids was also shown to increase as the cultures aged. DIM size increases were previously observed in Mtb cells cultivated on odd-chain carbon sources and cholesterol, and also in Mtb cells from infected mice lungs (15, 20). These mass increases were shown to result from addition of methylene units to the esterified mycocerosic acids. These data build on evidence that suggest that Mtb actively modulates its lipid composition in response to the changing microenvironment. Thus, this lipidomic approach provides a working platform for the analyses of more-refined in vitro growth culture models that incorporate defined stress factors (19, 41–48), as well as comparative analyses of lipid profiles from Mtb cells isolated from infected tissues. Such in vivo studies would provide valuable insight into the metabolism and physiology of Mtb. Additionally, combining the two-dimensional 1H-13C heteronuclear single quantum coherence NMR approach applied by Mahrous, Lee, and Lee (22) would potentially provide synergistic methods to produce complementary levels of information. A current limitation to the quantitative lipid profiling is that the true stoichiometric relationships between lipid classes cannot be determined. Even differences between lipid groups belonging to the same lipid subclass could not be used to determine true abundance, considering that acyl chain length and degree of unsaturation are known to influence ionization responses (49). Absolute quantification could be achieved with the incorporation of multiple internal lipid standards to which individual lipid ionization could be normalized. Such approaches are becoming more commonplace for eukaryotic-based lipidomics studies (34). However, appropriate isotopically labeled standards specific for many of the unique Mtb lipids are lacking, and require development to achieve this level of analysis.
A central element to our MS-based lipidomics strategy was automated product identification based on accurate mass matching. This approach has become more commonplace for metabolomics-based experiments that utilize high-resolution mass spectrometers (50, 51). For global lipid characterization, however, this methodology can be complicated by differential acylation of the same lipid, resulting in isobaric lipid species. Thus, the Mtb LipidDB was designed such that the output returned only FA sum composition, thus limiting potential mass overlap between database entries. In this study, we demonstrated a relatively small number of instances in which mass overlap (±5 ppm) occurs between lipid groups in the Mtb LipidDB. Indeed, it may be advantageous that Mtb does not produce appreciable levels of phosphatidic acid, phosphatidylserine, or phosphatidylcholine, because these would overlap in mass with other Mtb phospholipids, as observed in eukaryotes (52). Furthermore, we note that the unique large, apolar molecular characteristics of many Mtb lipids provide distinguishing mass features for these molecules in a eukaryotic lipid background (unpublished observations). We also view the automated identification process described in this study as a screening method, and believe that database identifications based on accurate mass alone should be regarded as tentative. Lipid identifications of biological interest should be subjected to MS/MS characterization and structural confirmation, as was done with select TG groups in this study. Interpretation of the resulting data can be aided by well-documented Mtb lipid collision-induced dissociation fragmentation patterns. Indeed we have obtained confirmatory MS/MS product ion spectra for representatives of many of the lipid subclasses identified in this study (data not shown). Identification confidence has also been shown to be enhanced with the incorporation of retention time criteria into the database searching process in the presence of internal standards that allow for normalization of retention times (53–56). Thus, performing MS/MS analyses in a spot-check fashion would allow for further retention time curation of the searchable database files, and this could easily be developed through an iterative process within a laboratory. Finally, the accuracy of the database identifications will also improve with the increasing resolution of newly developed mass spectrometers.
The limitations of the current Mtb LipidDB also need to be recognized. First, the database is constrained by the reported lists of FAs considered for the various lipid group repertoires. For example, only the FAs observed by Walker, Barakat, and Hung (32) were used to calculate GP lipid group masses, yet other atypical FAs may be esterified to mycobacterial GPs and would thus escape identification from the database search. The Mtb LipidDB does not take into consideration mass shifts due to oxidation or degradation of lipids, which may be keys to a complete understanding of Mtb lipid metabolism. Only well-defined lipids were included in the database; a novel and recently described wax ester lipid identified from Mtb cultivated under iron-limiting conditions was not included because of its partial structural elucidation (48). Biosynthetic lipid intermediates, such as phosphatidic acid, acyl-CoA, and sugar-linked decaprenol phosphates, were also not included in the database. Finally, the database in general is restricted to the lipid composition of the Mtb H37Rv strain used in this study, and does not contain lipids found in other mycobacterial species. Keeping these limitations in mind, the flexible data format of the database should allow users to easily amend and add lipid entries according to their specific research interests.
Supplementary Material
Footnotes
Abbreviations:
- Ac2PIM2
- diacylated diacylglycerophosphoinositoldimannoside
- Alpha-MA
- α mycolic acid
- amu
- atomic mass units
- APCI
- atmospheric pressure chemical ionization
- BCG
- Bacillus Calmette-Guérin
- CL
- cardiolipin
- DAT
- diacyltrehalose
- DIM A
- phthiocerol dimycocerosate
- DIM B
- phthiodiolone dimycocerosate
- GP
- glycerophospholipid
- MF
- molecular feature
- MFE
- molecular feature extraction
- Mtb
- Mycobacterium tuberculosis
- OD580
- optical density at 580 nm
- PE
- phosphatidylethanolamine
- PI
- phosphatidylinositol
- PIM
- phosphatidylinositolmannoside
- TG
- triacylglycerol
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one table and four figures.
REFERENCES
- 1.Brennan P. J., Nikaido H. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64: 29–63. [DOI] [PubMed] [Google Scholar]
- 2.Daffe M., Draper P. 1998. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microb. Physiol. 39: 131–203. [DOI] [PubMed] [Google Scholar]
- 3.Crick D. C., Brennan P. J., McNeil M. R. 2004. The cell wall of Mycobacterium Tuberculosis. Tuberculosis. 2nd edition Rom W. N., Garay S. M., Lippincott Williams & Wilkins, Philadelphia: 115–134. [Google Scholar]
- 4.Cox J. S., Chen B., McNeil M., Jacobs W. R., Jr 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 402: 79–83. [DOI] [PubMed] [Google Scholar]
- 5.Reed M. B., Domenech P., Manca C., Su H., Barczak A. K., Kreiswirth B. N., Kaplan G., Barry C. E., III 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 431: 84–87. [DOI] [PubMed] [Google Scholar]
- 6.Rao V., Fujiwara N., Porcelli S. A., Glickman M. S. 2005. Mycobacterium tuberculosis controls host innate immune activation through cyclopropane modification of a glycolipid effector molecule. J. Exp. Med. 201: 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goren M. B., Brokl O., Schaefer W. B. 1974. Lipids of putative relevance to virulence in Mycobacterium tuberculosis: correlation of virulence with elaboration of sulfatides and strongly acidic lipids. Infect. Immun. 9: 142–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daffe M., Laneelle M. A. 1988. Distribution of phthiocerol diester, phenolic mycosides and related compounds in mycobacteria. J. Gen. Microbiol. 134: 2049–2055. [DOI] [PubMed] [Google Scholar]
- 9.Daffe M., McNeil M., Brennan P. J. 1991. Novel type-specific lipooligosaccharides from Mycobacterium tuberculosis. Biochemistry. 30: 378–388. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe M., Aoyagi Y., Ridell M., Minnikin D. E. 2001. Separation and characterization of individual mycolic acids in representative mycobacteria. Microbiology. 147: 1825–1837. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe M., Aoyagi Y., Mitome H., Fujita T., Naoki H., Ridell M., Minnikin D. E. 2002. Location of functional groups in mycobacterial meromycolate chains; the recognition of new structural principles in mycolic acids. Microbiology. 148: 1881–1902. [DOI] [PubMed] [Google Scholar]
- 12.Garton N. J., Christensen H., Minnikin D. E., Adegbola R. A., Barer M. R. 2002. Intracellular lipophilic inclusions of mycobacteria in vitro and in sputum. Microbiology. 148: 2951–2958. [DOI] [PubMed] [Google Scholar]
- 13.Daniel J., Deb C., Dubey V. S., Sirakova T. D., Abomoelak B., Morbidoni H. R., Kolattukudy P. E. 2004. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J. Bacteriol. 186: 5017–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sirakova T. D., Dubey V. S., Deb C., Daniel J., Korotkova T. A., Abomoelak B., Kolattukudy P. E. 2006. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology. 152: 2717–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain M., Petzold C. J., Schelle M. W., Leavell M. D., Mougous J. D., Bertozzi C. R., Leary J. A., Cox J. S. 2007. Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc. Natl. Acad. Sci. USA. 104: 5133–5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garton N. J., Waddell S. J., Sherratt A. L., Lee S. M., Smith R. J., Senner C., Hinds J., Rajakumar K., Adegbola R. A., Besra G. S., et al. 2008. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 5: e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsunaga I., Naka T., Talekar R. S., McConnell M. J., Katoh K., Nakao H., Otsuka A., Behar S. M., Yano I., Moody D. B., et al. 2008. Mycolyltransferase-mediated glycolipid exchange in Mycobacteria. J. Biol. Chem. 283: 28835–28841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ojha A. K., Baughn A. D., Sambandan D., Hsu T., Trivelli X., Guerardel Y., Alahari A., Kremer L., Jacobs W. R., Jr., Hatfull G. F. 2008. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol. Microbiol. 69: 164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deb C., Lee C. M., Dubey V. S., Daniel J., Abomoelak B., Sirakova T. D., Pawar S., Rogers L., Kolattukudy P. E. 2009. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS ONE. 4: e6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X., Nesbitt N. M., Dubnau E., Smith I., Sampson N. S. 2009. Cholesterol metabolism increases the metabolic pool of propionate in Mycobacterium tuberculosis. Biochemistry. 48: 3819–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobson G., Minnikin D. E., Minnikin S. M., Parlett M., Goodfellow M., Ridell M., Magnusson M. 1985. Systematic analysis of complex mycobacterial lipids. Chemical Methods in Bacterial Systematics. Goodfellow M., Minnikin D. E., Academic Press, London: 237–265. [Google Scholar]
- 22.Mahrous E. A., Lee R. B., Lee R. E. 2008. A rapid approach to lipid profiling of mycobacteria using 2D HSQC NMR maps. J. Lipid Res. 49: 455–463. [DOI] [PubMed] [Google Scholar]
- 23.Leavell M. D., Leary J. A. 2006. Fatty acid analysis tool (FAAT): an FT-ICR MS lipid analysis algorithm. Anal. Chem. 78: 5497–5503. [DOI] [PubMed] [Google Scholar]
- 24.Shui G., Bendt A. K., Pethe K., Dick T., Wenk M. R. 2007. Sensitive profiling of chemically diverse bioactive lipids. J. Lipid Res. 48: 1976–1984. [DOI] [PubMed] [Google Scholar]
- 25.Takayama K., Schnoes H. K., Armstrong E. L., Boyle R. W. 1975. Site of inhibitory action of isoniazid in the synthesis of mycolic acids in Mycobacterium tuberculosis. J. Lipid Res. 16: 308–317. [PubMed] [Google Scholar]
- 26.Sonnenberg M. G., Belisle J. T. 1997. Definition of Mycobacterium tuberculosis culture filtrate proteins by two-dimensional polyacrylamide gel electrophoresis, N-terminal amino acid sequencing, and electrospray mass spectrometry. Infect. Immun. 65: 4515–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Covert B. A., Spencer J. S., Orme I. M., Belisle J. T. 2001. The application of proteomics in defining the T cell antigens of Mycobacterium tuberculosis. Proteomics. 1: 574–586. [DOI] [PubMed] [Google Scholar]
- 28.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 29.Munoz M., Laneelle M. A., Luquin M., Torrelles J., Julian E., Ausina V., Daffe M. 1997. Occurrence of an antigenic triacyl trehalose in clinical isolates and reference strains of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 157: 251–259. [DOI] [PubMed] [Google Scholar]
- 30.Fahy E., Subramaniam S., Brown H. A., Glass C. K., Merrill A. H., Jr, Murphy R. C., Raetz C. R., Russell D. W., Seyama Y., Shaw W., et al. 2005. A comprehensive classification system for lipids. J. Lipid Res. 46: 839–861. [DOI] [PubMed] [Google Scholar]
- 31.Fahy E., Subramaniam S., Murphy R. C., Nishijima M., Raetz C. R., Shimizu T., Spener F., van Meer G., Wakelam M. J., Dennis E. A. 2009. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 50 (Suppl.): 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker R. W., Barakat H., Hung J. G. 1970. The positional distribution of fatty acids in the phospholipids and triglycerides of Mycobacterium smegmatis and M. bovis BCG. Lipids. 5: 684–691. [DOI] [PubMed] [Google Scholar]
- 33.Low K. L., Rao P. S., Shui G., Bendt A. K., Pethe K., Dick T., Wenk M. R. 2009. Triacylglycerol utilization is required for regrowth of in vitro hypoxic nonreplicating Mycobacterium bovis bacillus Calmette-Guerin. J. Bacteriol. 191: 5037–5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ejsing C. S., Sampaio J. L., Surendranath V., Duchoslav E., Ekroos K., Klemm R. W., Simons K., Shevchenko A. 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA. 106: 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nigou J., Gilleron M., Brando T., Puzo G. 2004. Structural analysis of mycobacterial lipoglycans. Appl. Biochem. Biotechnol. 118: 253–267. [DOI] [PubMed] [Google Scholar]
- 36.Hsu F. F., Turk J., Owens R. M., Rhoades E. R., Russell D. G. 2007. Structural characterization of phosphatidyl-myo-inositol mannosides from Mycobacterium bovis Bacillus Calmette Guerin by multiple-stage quadrupole ion-trap mass spectrometry with electrospray ionization. I. PIMs and lyso-PIMs. J. Am. Soc. Mass Spectrom. 18: 466–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu F. F., Turk J., Owens R. M., Rhoades E. R., Russell D. G. 2007. Structural characterization of phosphatidyl-myo-inositol mannosides from Mycobacterium bovis Bacillus Calmette Guerin by multiple-stage quadrupole ion-trap mass spectrometry with electrospray ionization. II. Monoacyl- and diacyl-PIMs. J. Am. Soc. Mass Spectrom. 18: 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McAnoy A. M., Wu C. C., Murphy R. C. 2005. Direct qualitative analysis of triacylglycerols by electrospray mass spectrometry using a linear ion trap. J. Am. Soc. Mass Spectrom. 16: 1498–1509. [DOI] [PubMed] [Google Scholar]
- 39.Sandra K., Pereira Ados S., Vanhoenacker G., David F., Sandra P. 2010. Comprehensive blood plasma lipidomics by liquid chromatography/quadrupole time-of-flight mass spectrometry. J. Chromatogr. A. 1217: 4087–4099. [DOI] [PubMed] [Google Scholar]
- 40.Wayne L. G. 2001. In vitro model of hypoxically induced nonreplicating persistence of Mycobacterium tuberculosis. Mycobacterium tuberculosis Protocols. Parish T., Stoker N. G., Humana Press, Totowa, NJ: 247–268. [DOI] [PubMed] [Google Scholar]
- 41.Wayne L. G., Hayes L. G. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64: 2062–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Betts J. C., Lukey P. T., Robb L. C., McAdam R. A., Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43: 717–731. [DOI] [PubMed] [Google Scholar]
- 43.Fisher M. A., Plikaytis B. B., Shinnick T. M. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184: 4025–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hampshire T., Soneji S., Bacon J., James B. W., Hinds J., Laing K., Stabler R. A., Marsh P. D., Butcher P. D. 2004. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis (Edinb.). 84: 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muttucumaru D. G., Roberts G., Hinds J., Stabler R. A., Parish T. 2004. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis (Edinb.). 84: 239–246. [DOI] [PubMed] [Google Scholar]
- 46.Voskuil M. I. 2004. Mycobacterium tuberculosis gene expression during environmental conditions associated with latency. Tuberculosis (Edinb.). 84: 138–143. [DOI] [PubMed] [Google Scholar]
- 47.Voskuil M. I., Visconti K. C., Schoolnik G. K. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb.). 84: 218–227. [DOI] [PubMed] [Google Scholar]
- 48.Bacon J., Dover L. G., Hatch K. A., Zhang Y., Gomes J. M., Kendall S., Wernisch L., Stoker N. G., Butcher P. D., Besra G. S., et al. 2007. Lipid composition and transcriptional response of Mycobacterium tuberculosis grown under iron-limitation in continuous culture: identification of a novel wax ester. Microbiology. 153: 1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koivusalo M., Haimi P., Heikinheimo L., Kostiainen R., Somerharju P. 2001. Quantitative determination of phospholipid compositions by ESI-MS: effects of acyl chain length, unsaturation, and lipid concentration on instrument response. J. Lipid Res. 42: 663–672. [PubMed] [Google Scholar]
- 50.Ding J., Sorensen C. M., Zhang Q., Jiang H., Jaitly N., Livesay E. A., Shen Y., Smith R. D., Metz T. O. 2007. Capillary LC coupled with high-mass measurement accuracy mass spectrometry for metabolic profiling. Anal. Chem. 79: 6081–6093. [DOI] [PubMed] [Google Scholar]
- 51.Sana T. R., Roark J. C., Li X., Waddell K., Fischer S. M. 2008. Molecular formula and METLIN Personal Metabolite Database matching applied to the identification of compounds generated by LC/TOF-MS. J. Biomol. Tech. 19: 258–266. [PMC free article] [PubMed] [Google Scholar]
- 52.Schwudke D., Hannich J. T., Surendranath V., Grimard V., Moehring T., Burton L., Kurzchalia T., Shevchenko A. 2007. Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra. Anal. Chem. 79: 4083–4093. [DOI] [PubMed] [Google Scholar]
- 53.Kurvinen J. P., Aaltonen J., Kuksis A., Kallio H. 2002. Software algorithm for automatic interpretation of mass spectra of glycerolipids. Rapid Commun. Mass Spectrom. 16: 1812–1820. [DOI] [PubMed] [Google Scholar]
- 54.Haimi P., Uphoff A., Hermansson M., Somerharju P. 2006. Software tools for analysis of mass spectrometric lipidome data. Anal. Chem. 78: 8324–8331. [DOI] [PubMed] [Google Scholar]
- 55.Yetukuri L., Katajamaa M., Medina-Gomez G., Seppanen-Laakso T., Vidal-Puig A., Oresic M. 2007. Bioinformatics strategies for lipidomics analysis: characterization of obesity related hepatic steatosis. BMC Syst. Biol. 1: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ding J., Sorensen C. M., Jaitly N., Jiang H., Orton D. J., Monroe M. E., Moore R. J., Smith R. D., Metz T. O. 2008. Application of the accurate mass and time tag approach in studies of the human blood lipidome. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 871: 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.