Abstract
Cystic fibrosis (CF) is associated with abnormal lipid metabolism. We have recently shown variations in plasma levels of several phosphatidylcholine (PC) and lysophopshatidylcholine (LPC) species related to disease severity in CF patients. Here our goal was to search for blood plasma lipid signatures characteristic of CF patients bearing the same mutation (F508del) and different phenotypes, and to study their correlation with forced expiratory volume in 1 s (FEV1) and Pseudomonas aeruginosa chronic infection, evaluated at the time of testing (t = 0) and three years later (t = 3). Samples from 44 F508del homozygotes were subjected to a lipidomic approach based on LC-ESI-MS. Twelve free fatty acids were positively correlated with FEV1 at t = 0 (n = 29). Four of them (C20:3n-9, C20:5n-3, C22:5n-3, and C22:6n-3) were also positively correlated with FEV1 three years later, along with PC(32:2) and PC(36:4) (n = 31). Oleoylethanolamide (OEA) was negatively correlated with FEV1 progression (n = 17). Chronically infected patients at t = 0 showed lower PC(32:2), PC(38:5), and C18:3n-3 and higher cholesterol, cholesterol esters, and triacylglycerols (TAG). Chronically infected patients at t = 3 showed significantly lower levels of LPC(18:0). These results suggest a potential prognostic value for some lipid signatures in, to our knowledge, the first longitudinal study aimed at identifying lipid biomarkers for CF.
Keywords: longitudinal study, multivariate analysis, metabolomics, endocannabinoids, fatty acid amides
Cystic fibrosis (CF) is a genetic disease attributed to mutations in the CFTR gene, coding for a widely expressed chloride channel (1). The disease phenotype is mostly circumscribed to the digestive and respiratory systems. The former mainly consists of intestinal obstruction and meconium ileus, pancreatic malfunction, and intestinal malabsorption. The latter leads to chronic obstructive pulmonary disease with recurrent infections and exacerbations, ultimately resulting in respiratory insufficiency and death (2). In general, there is a weak correlation between genotype and phenotype (3), which strongly suggests either the existence of modifier genes or a strong influence of epigenetic factors, such as environment and methylation/acetylation of DNA and/or proteins. Modifier genes have been the subject of an extensive body of research in the last few years (recently reviewed in Refs. 4 and 5). Exploration of epigenetic factors in CF is in its beginning stage (6). This limited correlation makes it difficult to estimate the prognosis and evolution of the disease, and to optimize treatments and prophylactic actions. The search for predictive or prognostic biomarkers has become capital in the CF field. Biomarkers obtained by rapid, noninvasive methods could be used in the earlier detection of CF exacerbations or as prognostic indicators.
Intense proteomics research to unveil protein markers has resulted in finding associations between myeloperoxidase, interleukin (IL)-8, cleaved α-antitrypsin, and S100A8 (CF antigen) in sputum and the frequency of pulmonary exacerbations (7) or directly in the presence of CFTR mutations (8, 9), in addition to proteomic signatures in serum or corresponding to known inflammation markers (10) or other proteins differentially expressed in target tissues of CF before the appearance of any sign of the disease, like annexin-1 (11), cytosolic phospholipase A2α (11), cytokeratins 8 and 18 (12), and peroxiredoxin 6 (13, 14). More recently, sputum proteomics has revealed that a proline-glycine-proline (PGP) peptide, an extracellular matrix-derived neutrophil attractant, is a marker of inflammatory exacerbation in CF (14). In a recent report, a SELDI-based screening of serum and nasal cells from patients with CF and other respiratory diseases suggested that combinations of peptide ions studied by multivariate analysis have a better predictive value than individual species (15).
In addition to proteins and peptides, smaller molecules like metabolites and particularly lipids may represent an interesting target in the biomarker search. Several pioneering metabolomic profiling studies in CF have been reported very recently. The first revealed the presence of a high content of metabolites, such as amino acids and lactate, as markers of inflammation in broncho-alveolar fluid from CF patients (16). The second unveiled modifications in purinergic signaling, glucose metabolism, and the response to oxidative stress in CF cells (17). The third presented volatile organic compounds as markers of Pseudomonas aeruginosa colonization (18).
With reference to lipids, alterations in the lipid content of cells and fluids of CF patients and models have been reported since the early sixties. These alterations include a decrease in circulating essential fatty acids (19, 20), phospholipids, and lysophospholipids (21); decreased polyunsaturated fatty acids in lung, intestine, and pancreas of mice (22, 23) and nasal cells of patients (24), mostly associated with an imbalanced n-3/n-6 ratio; and decreased linoleic acid (19, 25) and lipoxin A4 in lung exudates from patients (26). These alterations have been attributed largely to malabsorption (19), increased flux through the n-6 biosynthetic pathway (25, 27, 28), and an alteration in the methionine-homocysteine cycle (29, 30).
As in protein biomarker research, we have recently evoked the usefulness of multivariate analysis in lipidomics (31). In a recent paper, we have shown altered phospholipid plasma content associated with both the onset and severity status of the disease (21). In the present work, we have addressed the same question by analyzing the plasma lipid composition of CF patients in a cohort composed exclusively of F508del homozygotes who shared the same treatment modalities and follow-up from one clinical center. On the basis of previous reports, in this study we have covered the main lipid classes by targeting our analysis on phosphatidylcholine (PC) and lysophosphatidylcholine (LPC) species, as well as on free fatty acids, cholesterol, cholesterol esters, triglycerides, and ceramides. We have also addressed for the first time in CF the analysis of fatty acyl amides.
We have compared these lipid signatures at one time point with two clinical parameters. The first parameter was the forced respiratory volume in 1 s (FEV1), considered today the most reliable marker of respiratory function and disease severity in CF. The second parameter was chronic infection by Pseudomonas aeruginosa, the most common bacteria in CF patients, a marker of the inflammation-infection scenario, and one of the main prognostic indicators of worsening in lung disease. Both parameters were measured at two time points, which represent a longitudinal study of disease evolution and a cross-sectional lipid screening. As a result, we show the correlation of the plasma content of a series of lipid species with relevant clinical parameters at both time points. This suggests a potential prognostic value for some of those lipid signatures in relation to the progression of CF disease.
MATERIALS AND METHODS
Ethics statement
All protocols were approved by the Ile-de-France II ethical committee. All patients involved in the study or their parents or legal guardians signed a written consent form.
Patient description and sample collection
Blood plasma samples were collected from CF patients at Hôpital Necker Enfants Malades (Paris, France) using a standard clinical protocol. All patients were at a good nutritional status and out of exacerbation period at the time of collection. All patients were receiving vitamins A, D, E, and K supplements as well as pancreatic enzymes. Sixteen patients were receiving inhaled corticoids. Blood collection was done at fasting. The analysis was performed on samples from 44 patients (description of patients is presented in Table 1). Sample collection was carried out with appropriate ethical committee approval. Samples were collected in VACUETTE® EDTA tubes K3E/EDTA K3 (Greiner Bio-One, Kremsmünster, Austria) and centrifuged at 2800 g for 15 min at 4°C. Plasma was separated and dispensed into 100 µl aliquots so that each aliquot was subjected to a single freezing-thawing cycle. Plasma samples were frozen in liquid nitrogen and stored at −80°C.
TABLE 1.
Description of patients
| Patient Code | Age at Inclusion(years) | FEV1%(t = 0) | FEV1%(t = 3) | Chronic Colonization(t = 0) | Chronic Colonization(t = 3) | Infection Evolution Group |
|---|---|---|---|---|---|---|
| 1 | 10 | 99 | 74 | No | No | A |
| 2 | 17 | 97 | 60 | No | ND | |
| 3 | 19 | 67 | 70 | Yes | No | D |
| 4 | 19 | 24 | 27 | No | Yes | B |
| 5 | 3 | ND | ND | No | ND | |
| 6 | 4 | ND | 40 | No | Yes | B |
| 7 | 17 | 79 | 54 | No | No | A |
| 8 | 12 | 85 | 79 | No | Yes | B |
| 9 | 14 | 35 | 30 | Yes | ND | |
| 10 | 17 | 50 | 91 | Yes | Yes | C |
| 11 | 7 | 100 | 88 | Yes | Yes | C |
| 12 | 7 | 98 | 83 | No | No | A |
| 13 | 8 | 58 | 56 | Yes | Yes | C |
| 14 | 1 | ND | ND | Yes | Yes | C |
| 15 | 9 | ND | 39 | No | Yes | B |
| 16 | 18 | 35 | 54 | Yes | Yes | C |
| 17 | 41 | 64 | ND | No | ND | |
| 18 | 13 | 60 | 63 | Yes | ND | |
| 19 | 33 | 20 | ND | Yes | ND | |
| 20 | 14 | 34 | 24 | Yes | Yes | C |
| 21 | 2 | ND | ND | No | Yes | B |
| 22 | 9 | 88 | 72 | No | No | A |
| 23 | 10 | 94 | 95 | No | Yes | B |
| 24 | 17 | 71 | 84 | No | Yes | B |
| 25 | 9 | ND | ND | Yes | ND | |
| 26 | 28 | 31 | ND | Yes | ND | |
| 27 | 2 | ND | ND | No | No | A |
| 28 | 1 | ND | ND | No | Yes | B |
| 29 | 8 | ND | 75 | No | ND | |
| 30 | 15 | 56 | ND | No | No | A |
| 31 | 1 | ND | ND | No | No | A |
| 32 | 13 | 54 | 40 | No | Yes | B |
| 33 | 2 | ND | ND | No | ND | |
| 34 | 5 | ND | ND | No | No | A |
| 35 | 15 | 32 | 22 | No | Yes | B |
| 36 | 7 | 99 | 89 | No | No | A |
| 37 | 8 | 100 | 85 | No | Yes | B |
| 38 | 5 | ND | 53 | No | Yes | B |
| 39 | 9 | 94 | 81 | No | Yes | B |
| 40 | 18 | ND | 49 | Yes | Yes | C |
| 41 | 18 | 54 | 50 | Yes | Yes | C |
| 42 | 13 | 72 | 56 | Yes | Yes | C |
| 43 | 16 | ND | 62 | No | No | A |
| 44 | 8 | 86 | 82 | No | No | A |
ND, not determined.
Genetic and clinical assessment
Patients were chosen according to their CFTR mutation genotype. All patients were homozygous for F508del. Pulmonary function test results (FEV1) were expressed as the percentage of the predicted value (32). FEV1 could only be recorded in 29 patients at the beginning of the study and in 31 patients three years later. In most cases, this was due to the short age of patients. Chronic colonization with Pseudomonas aeruginosa (defined by at least three positive sputum cultures within three months) was recorded for each patient at the beginning of the study and for 34 patients three years later.
Lipid extraction
Aliquots of 100 µl of plasma were subjected to organic extraction by addition of six volumes of chloroform-methanol 2:1 (v/v) containing a mixture of internal standards (all from Avanti Polar Lipids, Alabaster, AL, unless stated otherwise); namely, heptadecanoic acid (Nu-Chek Prep, Elysian, MN), d8-arachidonic acid (Cayman Chemicals, Ann Arbor, MI), heptadecenoylethanolamide (synthesized as previously reported (33)), trinonadecenoin (Nu-Chek Prep), dinonadienoyl-sn-glycerol (Nu-Chek Prep), monoheptadecanoyl-sn-glycerol (Nu-Chek Prep), d8-2-arachidonoyl-sn-glycerol (Cayman Chemicals), 1,2-diheptadecanoyl-sn-glycero-3-phosphoethanolamine, 1,2-diheptadecanoyl-sn-glycero-3-phosphoglycerol, 1,2-diheptadecanoyl-sn-glycero-3-phosphocholine, 1,2-diheptadecanoyl-sn-glycero-3-phosphoserine, 1,2-dipalmitoyl-sn-glycero-3-phosphoinositol, N-lauroyl-ceramide, N-lauroyl-sphingomyelin, and d7-cholesterol.
LC-MS
LC-MS analyses were performed using an Agilent 1200-LC system coupled to a 1946D or Ion-Trap XCT detector interfaced with ESI or APCI (Agilent Technologies, Wilmington, DE). To separate lipids containing one fatty acyl group, a reversed-phase C-18 column packed with conventional porous silica particles of small spherical diameter (Zorbax XDB Eclipse C-18 column from Agilent Technologies, 50 × 4.6 mm id, 1.8 µm particle size, 80 Å pore size) was used. Fatty acyl species were separated by a binary liquid chromatography method. Mobile phase A consisted of methanol containing 0.25% acetic acid and 5 mM ammonium acetate, and mobile phase B corresponded to water containing 0.25% acetic acid and 5 mM ammonium acetate. Separation was performed using a linear gradient from 90% A to 100% B in 2.5 min at a flow rate of 1.5 ml/min with column temperature at 40°C. ESI was set in the negative mode at −4.0 kV capillary voltage and 100V fragmentor voltage. N2 was used as drying gas at a flow rate of 12 l/min with temperature of 350°C and nebulizer pressure of 60 psi. Fatty acids were analyzed monitoring the mass-to-charge ratio (m/z) of the deprotonated molecular ions [M-H]− in selected-ion monitoring mode.
To separate glycerolipids, glycerophospholipids, sphingolipids, and sterol lipids, a reversed-phase Poroshell 300SB C-18 column (2.1 × 75 mm id, coating layer of 0.25 µm on total particle diameter of 5 µm, 300 Å pore size; Agilent Technologies) was used. A linear gradient was applied from 85% A to 100% B in 5 min (75% A to 100% B in 4 min in the case of sterol lipids) at a flow rate of 1.0 ml/min with column temperature set at 50°C. MS detection was performed both in the positive and negative ionization modes. The capillary voltage was set at −4.0 kV, and the skimmer voltage at 40V. N2 was used as drying gas at a flow rate of 10 l/min with temperature of 350°C and nebulizer pressure of 60 psi. Helium was used as collision gas, and fragmentation amplitude was set at 1.2V. For sterol lipids, APCI was set in positive mode, drying gas at 350°C, flow rate of 8 l/min, nebulizer gas pressure at 30 psi, vaporizer temperature at 475°C. Capillary voltage was 300V with the corona current at 5 μA.
Lipids were identified by comparing their LC retention times and MSn fragmentation patterns with those of authentic standards as previously described (33, 34). Because of the coexistence of multiple isobaric and isomeric species, not every individual molecular species of glycerophospholipids could be quantified. For example, the isobars PC(16:1/16:1) and PC(14:0/18:2) were quantified together and expressed as PC(32:2). Detection and analysis were controlled by Agilent/Bruker Daltonics software version 5.2.
Signature comparisons and statistical analyses
Signatures were compared with clinical parameters recorded at the time of sample collection (t = 0) and three years later (t = 3). Patients 2 and 15 showed abnormally high values for most free fatty acids (about 10-fold). Where indicated, these patients were excluded from the analysis. For some patients, the analytical data corresponding to some lipids are missing due to technical problems. Also, some clinical parameters could not be recorded at both time points for some patients. The number of data available is indicated for each test. Data are generally presented as means ± SEM.
Statistical analysis included univariate and multivariate analyses. All variables showed normal distributions, and parametric tests were chosen. Among univariate analyses, Student's t-test was used to compare two groups of patients, ANOVA to compare more than two groups, and Pearson correlation for comparison of numeric variables within the cohort. Multivariate analysis included multiple regression for numeric dependent variables, logistic regression for nominal dependent variables, and principal component analysis (PCA). Analyses were performed with XLStat, PopTools, and GraphPad InStat software.
RESULTS
Free fatty acids and fatty acid ethanolamides are associated with respiratory function
On the basis of our previous results (21, 24) and reports by others (35–37), we first focused our lipidomic screening on 50 lipid molecules or entities. After multiple Pearson correlation analysis of the lipid values at t = 0 with FEV1 measured at sampling time, a series of lipid signatures were found correlated with this respiratory functional parameter (n = 29, p < 0.05). These lipid signatures corresponded to free fatty acids, including saturated, mono-, and polyunsaturated species (Table 2).
TABLE 2.
Lipid species correlated significantly with FEV1 values obtained at t = 0
| Lipid Species | r | p |
|---|---|---|
| C16:1n-7 | 0.3919 | 0.0432 |
| C16:0 | 0.4702 | 0.0133 |
| C18:1n-9 | 0.3873 | 0.0459 |
| C18:0 | 0.4857 | 0.0102 |
| C18:3n-3 | 0.3817 | 0.0494 |
| C18:2n-6 | 0.3732 | 0.0461 |
| C20:3n-6 | 0.5208 | 0.0059 |
| C20:3n-9 | 0.5735 | 0.0018 |
| C22:5n-6 | 0.4148 | 0.0314 |
| C22:5n-3 | 0.5633 | 0.0022 |
| C20:5n-3 | 0.4035 | 0.0369 |
| C22:6n-3a | 0.3744 | 0.0497 |
The analysis was performed by linear regression (p < 0.05).
Patient 2 was excluded due to abnormally high values.
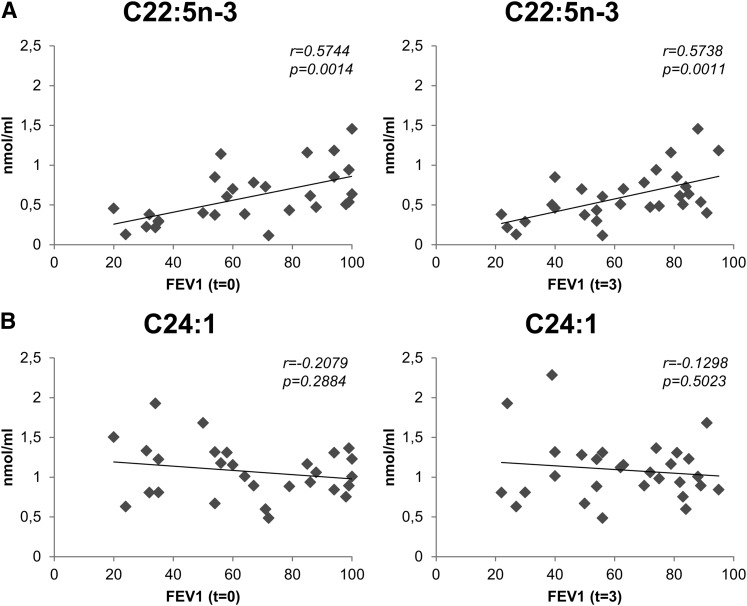
Interestingly, selected PUFA were significantly correlated with FEV1 at t = 3 (n = 31), which confers them a predictive potential on this parameter. This included C20:5n-3, C22:5n-3, and C22:6n-3, known as anti-inflammatory molecules, and C20:3n-9, the endpoint of the n-9 series. Also, two PC species [PC(36:4) and PC(32:2)] were also positively correlated with FEV1 at t = 3 (n = 31). These lipids and their respective correlation values are presented in Table 3. In addition, the desaturation index, defined by the C16:1/C16:0 ratio, was also positively correlated with FEV1 at t = 3 (n = 31, r = 0.4540, p = 0.0117). Fig. 1 shows the scatter plots corresponding to the plasma concentration of two fatty acids, one PUFA (C22:5n-3; Fig. 1A) that is normally esterified in glycerophospholipids and one monounsaturated (C24:1; Fig. 1B) that is usually found in sphingolipids, against FEV1 data at both t = 0 and t = 3. C22:5n-3 is shown as an example of those free fatty acids (mostly PUFA) that present a significant correlation with FEV1 (Fig. 1A) in contrast to C24:1, which shows no correlation (Fig. 1B).
TABLE 3.
Lipid species and ratios significantly correlated with FEV1 values measured at t = 3 (p < 0.05)
| Lipid Species | r | p |
|---|---|---|
| PC(32:2) | 0.3373 | 0.0269 |
| PC(36:4) | 0.3668 | 0.0424 |
| C20:3n-9a | 0.5342 | 0.0028 |
| C20:5n-3a | 0.3979 | 0.0325 |
| C22:5n-3a | 0.5738 | 0.0011 |
| C22:6n-3b | 0.5156 | 0.0050 |
| C16:1/C16:0 (unsaturation index) | 0.4540 | 0.0117 |
Patient 2 was excluded.
Patients 2 and 15 were excluded due to abnormally high values.
Fig. 1.
Plasma free fatty acids are correlated with FEV1. Scatter plots corresponding to the comparison between plasma levels of two free fatty acids (C22:5n-3 in panel A and C24.1 in panel B), and FEV1 values at t = 0 (left panels) and t = 3 (right panels). C22:5n-3 is positively correlated with FEV1 at both time points, while C24:1 presents no significant correlation (n = 28 for t = 0; n = 30 for t = 3; data corresponding to patient 2 were removed in both cases).
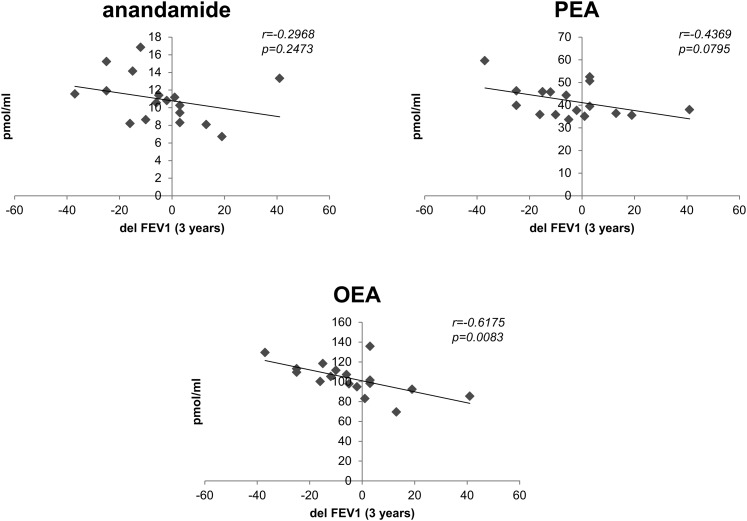
The potential association of lipid signatures with FEV1 variation between t = 0 and t = 3 (delFEV1) was also calculated. In this case, the plasma levels of fatty acid ethanolamides, including anandamide, palmitoylethanolamide (PEA) and oleoylethanolamide (OEA) were negatively correlated with delFEV1 (Fig. 2). Only the correlation of OEA and delFEV1 could be considered significant (n = 17, r = 0.618, p = 0.008).
Fig. 2.
Plasma endocannabinoids are correlated with delFEV1. Scatter plots corresponding to the comparison between plasma levels of three endocannabinoids (anandamide, PEA, and OEA) and the variation in FEV1 values between t = 0 and t = 3 (delFEV1). All three endocannabinoids are negatively correlated with delFEV1. This correlation is statistically significant (p < 0.05) for OEA (n = 17).
Some lipid signatures are associated with Pseudomonas aeruginosa colonization
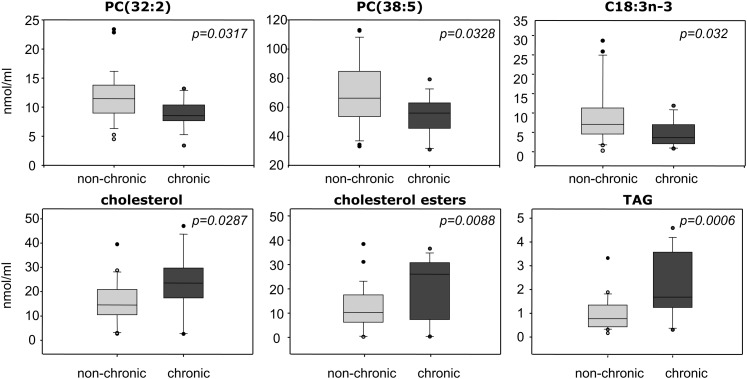
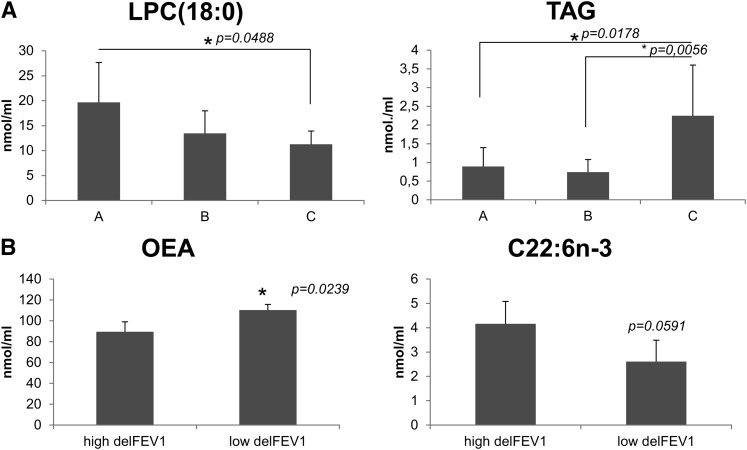
At t = 0, those patients colonized by Pseudomonas aeruginosa showed significantly lower levels of PC(38:5), PC(32:2) and C18:3n-3, and higher levels of cholesterol, cholesterol esters, and total triacylglycerols (TAG) than noncolonized patients (n = 44, Fig. 3). Differences between colonized and noncolonized patients at t = 3 (n = 34) were limited to LPC(18:0), which was significantly lower in patients that were colonized at t = 3 (12.6 versus 19.6 nmol/ml, p = 0.013). Interestingly, this lipid was also differentially displayed depending on the evolution of infection. The latter was studied by grouping patients according to their infection status at both t = 0 and t = 3 (A: noncolonized, n = 11; B: evolved from noncolonized at t = 0 to colonized at t = 3, n = 13; C: colonized at both time points, n = 9). As shown in Fig. 4A (left panel), LPC(18:0) was significantly lower in patient group C than in noncolonized patient group A. Accordingly, patients noncolonized at t = 0 but colonized at t = 3 (group B) presented intermediate values, although not statistically different from groups A and C. This result suggested for LPC(18:0) a predictive value concerning chronic infection. Another differentially displayed lipid signature was TAG, which was significantly increased in group C compared with groups A and B (Fig. 4A, right panel), strongly suggesting an association of this lipid class with chronic infection.
Fig. 3.
Plasma lipids associated with chronic Pseudomonas aeruginosa colonization. Box plots corresponding to those lipid species differentially displayed in patients presenting with chronic versus nonchronic Pseudomonas aeruginosa infection (p < 0.05; n = 44).
Fig. 4.
A: Plasma lipids associated with the evolution of Pseudomonas aeruginosa chronic infection. Patients were grouped according to their colonization by Pseudomonas aeruginosa at t = 0 and t = 3. A: noncolonized (n = 11). B: noncolonized at t = 0 but colonized at t = 3 (n = 13). C: colonized at both time points (n = 9). ANOVA analysis showed that the three groups were significantly different in terms of LPC(18:0) and TAG content (p = 0.0492 and p = 0.0026, respectively). Post-test showed significant differences between A and C, and between B and C for TAG. No post-test differences were found for LPC(18:0) (*p < 0.05). B: Differences in DHA and OEA between extremes of delFEV1. The five patients showing the highest and lowest values for delFEV1 were chosen for analysis (n = 5; p = 0.0591 for DHA; p = 0.0239 for OEA).
Extremes of phenotype confirm differences in free fatty acids and OEA
An additional study was performed by selecting extremes of phenotype based on FEV1 values. For this analysis the five patients who showed the highest and the five patients who showed the lowest FEV1 at each time point were chosen.
At t = 0, significant differences were found in several free fatty acids, including saturated (C16:0 and C18:0), middle-chain monounsaturated (C16:1n-7, C18:1n-9), n-6 polyunsaturated (C20:3n-6, C20:4n-6, C22:5n-6), n-3 polyunsaturated (C20:5n-3, C22:5n-3, C22:6n-3), and C20:3n-9. In all cases, the best phenotypes presented a 2-fold increased content with respect to the worst phenotypes. No changes were observed in saturated and monounsaturated long-chain fatty acids. At t = 3, the same free fatty acids were significantly different in the same sense and amplitude (2-fold), except for three fatty acids (C18:0, C18:1n-9, C20:3n-6). When delFEV1 between t = 3 and t = 0 was considered, only OEA was significantly different (Fig. 4B, left panel), showing higher values in the worst phenotype group, which is in agreement with the negative correlation found with the same parameter. Concerning the rest of molecules, free C22:6n-3 was higher in the best phenotype group and close to significance (p = 0.05; Fig. 4B, right panel).
Phenotype prediction by multivariate analysis
To establish predictive models for FEV1 and bacterial colonization, multiple regression and logistic regression approaches were used, respectively. The number of independent variables was restricted to four, given the limited number of individuals and missing data for some of the lipids. Only lipids showing the most significant associations with any of the clinical parameters by univariate analysis were chosen for multiple regression, namely, LPC(18:0), C22:5n-3, total TAG, and OEA (designed as B, C, D, and E, respectively). Alternatively, a three-variable system was considered by excluding OEA, due to the smaller number of observations for this lipid (n = 28).
Multiple regression models were used to evaluate potential prediction of FEV1 at t = 0 and t = 3, as well as delFEV1. None of the three models was significantly predictive (p = 0.0809, p = 0.0881, p = 0.1608, respectively). Nevertheless, C22:5n-3 contributed significantly to the prediction of FEV1 at both t = 0 (p = 0.0434) and t = 3 (p = 0.0193), and OEA contributed significantly to the prediction of delFEV1 (p = 0.0379), in accordance with the results of univariate analysis. FEV1 at t = 0 and t = 3 were predicted significantly by the following two-variable models composed of C22:5n-3 and TAG (p = 0.0370 and p = 0.0323, respectively), with a significant contribution of C22:5n-3 in both cases (p = 0.0418 and p = 0.0106, respectively): A[FEV1 (t = 3)] = 32.877 + (45.564 × C) + (1.908 × D) and A[FEV1 (t = 0)] = 45.187 + (40.201 × C) − (2.543 × D). The rest of parameters are presented in Table 4.
TABLE 4.
Predictive multiple regression models
| Predicted Variable | p | R (%) | n | Independent Variable | Coefficient | SE | 95% CI | t Ratio | Individual p |
|---|---|---|---|---|---|---|---|---|---|
| FEV1 (t = 0) | 0.0370 | 25.90 | 25a | A | 45.187 | 15.765 | 12.490 to 77.883 | 2.866 | 0.0090 |
| C | 40.201 | 18.602 | 1.621 to 78.782 | 2.161 | 0.0418 | ||||
| D | −2.543 | 4.430 | −11.731 to 6.646 | 0.5740 | 0.5718 | ||||
| FEV1 (t = 3) | 0.0323 | 25.88 | 27a | A | 32.877 | 12.380 | 7.325 to 58.429 | 2.656 | 0.0138 |
| C | 45.564 | 16.427 | 11.659 to 79.469 | 2.774 | 0.0106 | ||||
| D | 1.908 | 3.515 | −5.348 to 9.164 | 0.5427 | 0.5923 | ||||
| Infection (t = 0) | 0.0120 | 49.31 | 23a | A | 0.3889 | 0.2835 | −0.2066 to 0.9844 | 1.372 | 0.1869 |
| B | −0.0078 | 0.0116 | −0.0322 to 0.0167 | 0.6679 | 0.5126 | ||||
| C | −0.2071 | 0.2172 | −0.6635 to 0.2493 | 0.9532 | 0.3531 | ||||
| D | 0.2546 | 0.0847 | 0.07647 to 0.4327 | 3.003 | 0.0076 | ||||
| E | −0.0004 | 0.0004 | −0.0013 to 0.0004 | 1.031 | 0.3163 | ||||
| Infection evolution | 0.0106 | 34.52 | 30 | A | 1.269 | 0.4110 | 0.4235 to 2.114 | 3.086 | 0.0048 |
| B | −0.0289 | 0.0152 | −0.0601 to 0.0023 | 1.902 | 0.0683 | ||||
| C | −0.3470 | 0.3629 | −1.093 to 0.3990 | 0.9564 | 0.3477 | ||||
| D | 0.2483 | 0.1300 | −0.0189 to 0.5156 | 1.911 | 0.0671 |
A: Predicted variable; B: LPC(18:0); C: C22:5n-3; D: Total TAG; E: OEA.
Patient 2 excluded due to abnormally high values.
As shown in Table 4, Pseudomonas colonization at t = 0 was significantly predicted by the model A = 0.3889 − (0.00778 × B) − (0.2071 × C) + (0.2546 × D) − (0.0004225 × E) (R2 = 49.31%, p = 0.0120), with the significant contribution of TAG (p = 0.0076). Prediction of bacterial colonization at t = 3 was not significant (p = 0.1699). These results were confirmed by logistic regression (p < 0.0001; Table 5). Prediction of bacterial colonization evolution by grouping patients in groups A, B, and C as shown above was not significant in a four-variable model (p = 0.1112), but it was significant in a three-variable model (p = 0.0106; Table 4) as follows: A[infection evolution] = 1.269 − (0.02890 × B) − (0.3470 × C) + (0.2483 × D). No colinearity was observed among the four variables in any of the models.
TABLE 5.
Predictive logistic regression model for Pseudomonas aeruginosa colonization at t = 0
| Variable | Coefficient | SE | P | Odds Ratio |
|---|---|---|---|---|
| B | −3.4947 | 3185.8691 | 0.9991 | 0.0304 |
| C | −46.1798 | 20288. 3208 | 0.9982 | 0.0000 |
| D | 305.5965 | 25552.1129 | 0.9981 | 5.2344 |
| E | −10.2913 | 4265.7739 | 0.9981 | 0.0000 |
A: Predicted variable; B: LPC(18:0); C: C22:5n-3; D: Total TAG; E: OEA (p < 0.0001, n = 24).
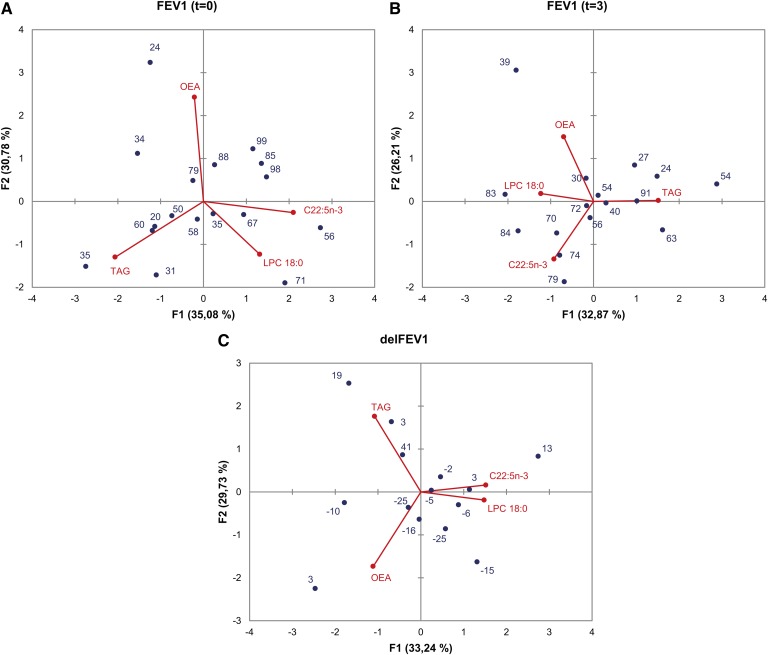
A PCA analysis was performed on the same four independent variables that were used in multiple regression analyses [LPC(18:0), C22:5n-3, TAG, and OEA]. The results of PCA are visualized in Figs. 5 and 6, which illustrate the ensemble of results obtained in other tests and presented above. In Fig. 5, the dependent variables are FEV1 at t = 0 (Fig. 5A), FEV1 at t = 3 (Fig. 5B), and delFEV1 (Fig. 5C). The highest FEV1 values at t = 0 (Fig. 5A) correspond to positive F1, with a major contribution of C22:5n-3 and, to a lesser extent, LPC(18:0). These two variables are also associated with higher FEV1 at t = 3, as they contribute negatively to the eigenvectors represented by F1 and F2 in Fig. 5B. The axis determined by F2 (Fig. 5C) makes a fair separation of positive and negative delFEV1. The major contribution corresponds to TAG and C22:5n-3 for positive F2, and OEA for negative F2.
Fig. 5.
Principal component analysis of FEV1 values and plasma lipids. PCA biplots correspond to FEV1 at t = 0 (A), FEV1 at t = 3 (B), and delFEV1 (C) as dependent variables. Blue spots and numbers represent FEV1 values. Red spots and lines represent the four independent variables selected, namely LPC(18:0), C22:5n-3, TAG, and OEA. In each case, variables are represented by the first and second principal components (F1 and F2). Percent contribution of each principal component to total variance is indicated in the respective axis title.
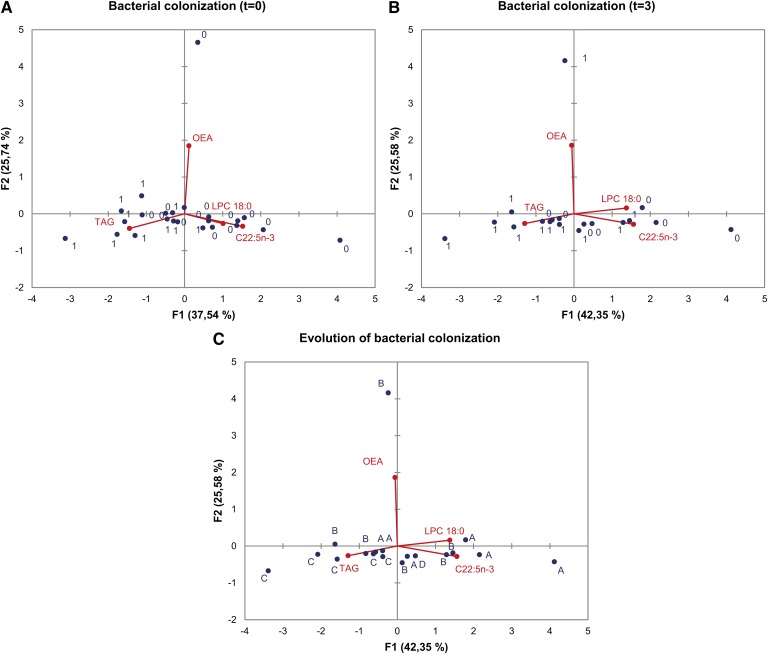
Fig. 6.
Principal component analysis of Pseudomonas aeruginosa colonization and plasma lipids. PCA biplots correspond to bacterial colonization at t = 0 (A), bacterial colonization at t = 3 (B), and the evolution of bacterial colonization (C) as dependent variables. Spots labeled “0” denote noncolonized individuals; spots labeled “1” denote colonized individuals; spots labeled “A” denote noncolonized individuals at either t = 0 or t = 3; spots labeled “B” denote noncolonized individuals at t = 0 that are colonized at t = 3; spots labeled “C” denote colonized individuals at both t = 0 and t = 3; the spot labeled “D” represents one individual colonized at t = 0 and noncolonized at t = 3. Red spots and lines represent the four independent variables selected, namely LPC(18:0), C22:5n-3, TAG, and OEA. In each case, variables are represented by the first and second principal components (F1 and F2). Percent contribution of each principal component to total variance is indicated in the respective axis title.
Concerning bacterial infection (Fig. 6), most of the colonized individuals at t = 0 and t = 3 are represented by negative values of F1 (Fig. 6A, B, respectively), with major participation of TAG. Fig. 6C shows a PCA in which the dependent variable is the evolution of bacterial colonization represented by three groups (A, B, and C) as described for Fig. 4A. One individual, represented by D, evolved from a colonized to a noncolonized status. F1 separates A and C individuals fairly well, while B individuals are randomly distributed with respect to principal components F1 and F2.
DISCUSSION
In the present work, we have explored the potential of lipid signatures as biomarkers for CF prognosis by comparing the blood plasma lipidome of F508del homozygotes obtained at one single time point, with a follow-up of their longitudinal clinical phenotype after three years. Our approach, applied for the first time in CF, included two novel aspects with respect to previous plasma lipid studies. First, it has been performed on a group of individuals presenting the same disease genotype. This approach addresses the capital issue of the lack of correlation between genotype and phenotype characteristic of CF. Second, the additional time point for clinical outcome parameters, established three years after blood sampling, constitutes a first attempt to evaluate the predictive potential of plasma lipids toward CF disease progression.
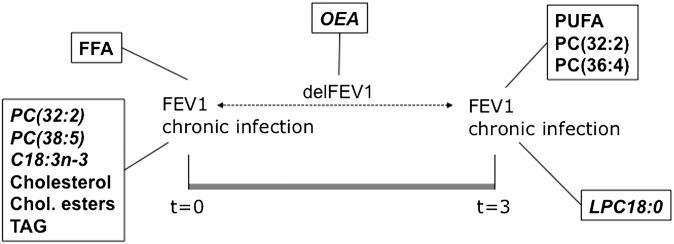
Concerning the lipid species covered by our study, a targeted analysis on 50 chemical entities has been performed. We have addressed PC and LPC species, because they were found differentially displayed in the plasma from patients and associated with severity status in our previous study (21). We also included free fatty acids, as they represent in plasma an important component of what has been traditionally analyzed in fatty acid profiling. Two ceramide species particularly abundant in plasma were analyzed, as well as neutral lipid groups. We also addressed for the first time the analysis of fatty acid ethanolamides, including the endocannabinoid anandamide, in CF. The results obtained are summarized in Fig. 7. Concerning the potential impact of treatments on the plasma content in these lipids, it must be pointed out that none of the patients recruited were receiving systemic anti-inflammatory therapy. All of them were receiving the same nutritional supplements and pancreatic enzyme replacement. The only potential source of variability was the treatment by inhaled corticoids of 16 of them. However, no statistically significant impact of this medication was found on any of the lipid species analyzed.
Fig. 7.
Summary of results. Lipids showing significant correlations with any clinical parameters are shown in bold. Plain text denotes positive correlations, and italic text denotes negative correlations.
Our findings concerning free fatty acids can be described at several levels. First, a positive correlation was found between most of the free fatty acids analyzed and the severity status in CF and the evolution of the disease. Second, the highest and most significant correlations corresponded to n-3 fatty acids. Saturated, monounsaturated, and n-6 PUFA presented similar, but not significant, trends when compared with the FEV1 outcome three years after blood testing. Third, the content in Mead acid (C20:3n-9), an endpoint of biosynthesis through the n-9 pathway characteristic of essential fatty acid deficiency, was also correlated with FEV1 at both time points. This finding is intriguing, as Mead acid and the 16:1/16:0 ratio are usually increased when PUFA are decreased, unlike in this case. Fourth, there is a pool of fatty acids that does not present any significant variations associated with the phenotype of patients. The latter includes saturated and monounsaturated 20- to 26-carbon fatty acids, which are usually esterified in sphingolipid molecules. It must be pointed out that most of these correlations were obtained after excluding patients 2 and 15, who presented remarkably high levels of most free fatty acids (about 10-fold). These two patients were characterized by a mild phenotype; no other biochemical or physiological markers were abnormal in them. The ensemble of observed alterations point to a particular contribution of free fatty acids originating from glycerophospholipids, not from sphingolipids. This would suggest a differential phospholipase activity of the type A1 and/or A2, as this activity would cleave selectively glycerolipid-bound fatty acids if a concomitant variation in lysophospholipids was found, which is not the case. Phospholipase activity and expression in CF tissues has been addressed but not comprehensively (11, 38–40). In general, CF is associated with higher phospholipase A2 activity and expression, but this has been only examined in cell lines (39) and mouse models (11, 38–40). When considering plasma levels of free fatty acids, lipoprotein lipase (LPL) cannot be excluded from the main picture. In fact, LPL is known to present phospholipase A1 activity, although its main substrate is the TAG molecule.
In our study, TAG and other neutral lipids, such as cholesterol and cholesterol esters, are significantly increased in patients presenting with chronic colonization by Pseudomonas aeruginosa. In a recent report, patients of acute infection showed lower levels of total cholesterol and HDL-cholesterol, as well as higher, though not significant, levels of TAG compared with noninfected patients with diverse pathologies (41). In a chronic infection scenario like CF, higher TAG and lower cholesterol levels have been found in patients compared with the general population (42). In the same study, a high prevalence of hypertriglyceridemia in CF patients was shown, which has been attributed to the chronic infection status. Nevertheless, in our study, TAG correlated significantly with age at inclusion (r = 0.4492, p = 0.0032). In fact, TAG was the only lipid species in our study showing an association with age. This somehow challenges the association of TAG with clinical status, but it discards the potential bias of age when addressing lipid analysis. Nevertheless, in the evolution of bacterial colonization (groups A, B, and C), the other parameter significantly associated with TAG was not significantly associated with age (p = 0.2954). The fact that all neutral lipids were increased suggests a potential role of lipid mobilization from adipose tissue. A high and significant correlation was found between total cholesterol and TAG levels (r = 0.7336, p < 0.0001), in agreement with the data shown by Figueroa et al. (42). High levels of neutral lipids in plasma could be due to greater synthesis rate in the liver, to decreased clearance, or both. Clearance of TAG from circulation is mainly due to LPL activity. As evoked by several authors (42, 43), one of the factors that can lead to increased TAG and cholesterol synthesis is higher production of pro-inflammatory cytokines, such as TNF-α that is associated with chronic inflammation and severe stress, which inhibit LPL activity and potentiate hepatic lipogenesis (44). In our study, which nonetheless represents a different scenario, LPL inhibition by TNF-α could account, at least in part, for the higher TAG and the lower FFA levels in the most severe patients.
Another potential factor, although independent of infection, that could contribute to the suggested increase in lipogenesis in CF arises from a recent report (45), where CFTR silencing in the intestinal cell model CaCo2/15 led to enhanced lipogenesis and delta-7 desaturase activity through an increase in the expression of SREBP-1c. Those data directly challenged the contribution of CFTR dysfunction to intestinal malabsorption.
One of the goals of the present work was to evaluate the prognostic potential of plasma lipid signatures by assessing their association with the clinical outcome three years after lipid analysis. As commented above, n-3 PUFA and Mead acid were positively correlated with FEV1 values at t = 3, in addition to two unsaturated chain-containing phospholipids, PC(32:2) and PC(36:4). At t = 3, neutral lipids (cholesterol, cholesterol esters, and TAG) were no longer associated with chronic infection, unlike LPC(18:0), which was significantly increased in individuals noncolonized by Pseudomonas aeruginosa. In our previous study of plasma from CF patients and healthy siblings, PC and LPC species, including LPC(18:0) and PC(36:4), were suggested to be associated with the severity status of patients (21). Our findings in the current work are somehow in accordance with those previous data and represent a new dimension in that prognostic value is suggested.
Time-dependent variation in FEV1 values (delFEV1) can be arguably considered the best determinant of disease progression in CF. Negative values correspond to a degradation of respiratory function, while positive values indicate the opposite. In our targeted study, after multiple correlation analysis, OEA presented a significant negative correlation with delFEV1, while PEA and the endocannabinoid anandamide showed a similar trend although not statistically significant. To our knowledge, this is the first report that addresses the plasma content in endocannabinoids and fatty acid ethanolamides in CF. Fatty acid ethanolamides are bioactive lipids that activate G-protein-coupled receptors to modulate multiple processes in living organisms, including inflammation (34). OEA derives from the metabolism of oleic acid, which, through phospholipid remodeling, is transformed into the OEA-precursor N-oleoyl phosphatidylethanolamine (NAPE) and subsequently cleaved by a NAPE-phospholipase D (NAPE-PLD) into OEA (33). An increase in OEA could be due to either increased NAPE-PLD or to decreased fatty-acid amide hydrolase (FAAH), the catabolizing enzyme. Synthesis takes place mainly in the liver and adipose tissue and is increased in starvation and cold exposure (46), while nothing is known about its potential synthesis or metabolism in the lung. Therefore, it can be argued that in severe CF a harsh stress-like scenario may lead to an increase of OEA in plasma. OEA binds and activates PPAR-α, TRPV1, and GPR119, leading to lipolysis stimulation and ceramidase inhibition (47). Interestingly, cannabinoid derivatives have been suggested as potentially therapeutic in CF (48), but they have never been tested in this context. While OEA has been used as an anti-inflammatory molecule (49), according to our present data, it can be speculated that the presence of higher levels in blood might be taken as a marker of an inflammatory process.
An ultimate objective of our study was to provide a lipid signature or a set of lipid signatures that may be used as a predictive model for CF progression. This is a complex endeavor that should be achieved ideally by the integration of multidisciplinary datasets arising from proteomics, transcriptomics, and metabolomics studies. Nevertheless, we made an attempt assuming the limited number of individuals enrolled in the study. We first selected the four variables showing the greatest association with clinical parameters in univariate analyses, namely, LPC(18:0), C22:5n-3, TAG and OEA, and we subsequently constructed potentially predictive models by multiple regression and logistic regression. We have shown some models that are able to predict with statistical significance the outcome of FEV1 at both time points, as well as Pseudomonas colonization at t = 0, and the evolution of the latter parameter along the three-year period. In all cases, the contribution of each variable to a respective clinical parameter found in univariate analysis was confirmed. Interestingly, the addition of age as an independent variable did not modify individual contribution (not shown), which confirms that our results are not biased by this parameter. PCA analyses illustrate the findings, confirm the trends, and provide a visual perspective of the contributions of different lipid variables.
In summary, even though our results must be taken cautiously due to the limited number of individuals enrolled, they represent a proof of concept for larger multicentric studies that address lipid alterations in CF patients from a novel perspective. Our study also opens new clues for further research in CF physiopathology, where the role of fatty acid ethanolamides may constitute an exciting new field.
Acknowledgments
The authors thank Diane-Lore Vieu and Delphine Roussel for their technical support.
Footnotes
Abbreviations:
- CF
- cystic fibrosis
- FEV1
- forced expiratory volume in 1 second
- IL
- interleukin
- LPC
- lysophosphatidylcholine
- OEA
- oleoylethanolamide
- PC
- phosphatidylcholine
- PCA
- principal component analysis
- PEA
- palmitoylethanolamide
- TAG
- triacylglycerol
This work was supported by European Commission Grant NEUPROCF (FP6), by the associations Vaincre la Mucoviscidose and ABCF2 Protéines, and by Legs Poix-University of Paris 5.
REFERENCES
- 1.Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L., et al. 1989. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 245: 1066–1073. [DOI] [PubMed] [Google Scholar]
- 2.Ollero M., Brouillard F., Edelman A. 2006. Cystic fibrosis enters the proteomics scene: new answers to old questions. Proteomics. 6: 4084–4099. [DOI] [PubMed] [Google Scholar]
- 3.Sermet-Gaudelus I., Dechaux M., Vallee B., Fajac A., Girodon E., Nguyen-Khoa T., Marianovski R., Hurbain I., Bresson J. L., Lenoir G., et al. 2005. Chloride transport in nasal ciliated cells of cystic fibrosis heterozygotes. Am. J. Respir. Crit. Care Med. 171: 1026–1031. [DOI] [PubMed] [Google Scholar]
- 4.Accurso F. J., Sontag M. K. 2008. Gene modifiers in cystic fibrosis. J. Clin. Invest. 118: 839–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collaco J. M., Cutting G. R. 2008. Update on gene modifiers in cystic fibrosis. Curr. Opin. Pulm. Med. 14: 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shuto T., Furuta T., Oba M., Xu H., Li J. D., Cheung J., Gruenert D. C., Uehara A., Suico M. A., Okiyoneda T., et al. 2006. Promoter hypomethylation of Toll-like receptor-2 gene is associated with increased proinflammatory response toward bacterial peptidoglycan in cystic fibrosis bronchial epithelial cells. FASEB J. 20: 782–784. [DOI] [PubMed] [Google Scholar]
- 7.Sloane A. J., Lindner R. A., Prasad S. S., Sebastian L. T., Pedersen S. K., Robinson M., Bye P. T., Nielson D. W., Harry J. L. 2005. Proteomic analysis of sputum from adults and children with cystic fibrosis and from control subjects. Am. J. Respir. Crit. Care Med. 172: 1416–1426. [DOI] [PubMed] [Google Scholar]
- 8.Gray R. D., MacGregor G., Noble D., Imrie M., Dewar M., Boyd A. C., Innes J. A., Porteous D. J., Greening A. P. 2008. Sputum proteomics in inflammatory and suppurative respiratory diseases. Am. J. Respir. Crit. Care Med. 178: 444–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkinson M. M., Busuttil A., Hayward C., Brock D. J., Dorin J. R., Van Heyningen V. 1988. Expression pattern of two related cystic fibrosis-associated calcium-binding proteins in normal and abnormal tissues. J. Cell Sci. 91: 221–230. [DOI] [PubMed] [Google Scholar]
- 10.Srivastava M., Eidelman O., Jozwik C., Paweletz C., Huang W., Zeitlin P. L., Pollard H. B. 2006. Serum proteomic signature for cystic fibrosis using an antibody microarray platform. Mol. Genet. Metab. 87: 303–310. [DOI] [PubMed] [Google Scholar]
- 11.Bensalem N., Ventura A. P., Vallee B., Lipecka J., Tondelier D., Davezac N., Dos Santos A., Perretti M., Fajac A., Sermet-Gaudelus I., et al. 2005. Down-regulation of the anti-inflammatory protein annexin A1 in cystic fibrosis knock-out mice and patients. Mol. Cell. Proteomics. 4: 1591–1601. [DOI] [PubMed] [Google Scholar]
- 12.Davezac N., Tondelier D., Lipecka J., Fanen P., Demaugre F., Debski J., Dadlez M., Schrattenholz A., Cahill M. A., Edelman A. 2004. Global proteomic approach unmasks involvement of keratins 8 and 18 in the delivery of cystic fibrosis transmembrane conductance regulator (CFTR)/deltaF508-CFTR to the plasma membrane. Proteomics. 4: 3833–3844. [DOI] [PubMed] [Google Scholar]
- 13.Trudel S., Kelly M., Fritsch J., Nguyen-Khoa T., Therond P., Couturier M., Dadlez M., Debski J., Touqui L., Vallee B., et al. 2009. Peroxiredoxin 6 fails to limit phospholipid peroxidation in lung from cftr-knockout mice subjected to oxidative challenge. PLoS ONE. 4: e6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaggar A., Jackson P. L., Noerager B. D., O'Reilly P. J., McQuaid D. B., Rowe S. M., Clancy J. P., Blalock J. E. 2008. A novel proteolytic cascade generates an extracellular matrix-derived chemoattractant in chronic neutrophilic inflammation. J. Immunol. 180: 5662–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomes-Alves P., Imrie M., Gray R. D., Nogueira P., Ciordia S., Pacheco P., Azevedo P., Lopes C., de Almeida A. B., Guardiano M., et al. 2010. SELDI-TOF biomarker signatures for cystic fibrosis, asthma and chronic obstructive pulmonary disease. Clin. Biochem. 43: 168–177. [DOI] [PubMed] [Google Scholar]
- 16.Wolak J. E., Esther C. R., Jr, O'Connell T. M. 2009. Metabolomic analysis of bronchoalveolar lavage fluid from cystic fibrosis patients. Biomarkers. 14: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wetmore D. R., Joseloff E., Pilewski J., Lee D. P., Lawton K. A., Mitchell M. W., Milburn M. V., Ryals J. A., Guo L. 2010. Metabolomic profiling reveals biochemical pathways and biomarkers associated with pathogenesis in cystic fibrosis cells. J. Biol. Chem. 285: 30516–30522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robroeks C. M., van Berkel J. J., Dallinga J. W., Jobsis Q., Zimmermann L. J., Hendriks H. J., Wouters M. F., van der Grinten C. P., van de Kant K. D., van Schooten F. J., et al. 2010. Metabolomics of volatile organic compounds in cystic fibrosis patients and controls. Pediatr. Res. 68: 75–80. [DOI] [PubMed] [Google Scholar]
- 19.Farrell P. M., Mischler E. H., Engle M. J., Brown D. J., Lau S. M. 1985. Fatty acid abnormalities in cystic fibrosis. Pediatr. Res. 19: 104–109. [DOI] [PubMed] [Google Scholar]
- 20.Roulet M., Frascarolo P., Rappaz I., Pilet M. 1997. Essential fatty acid deficiency in well nourished young cystic fibrosis patients. Eur. J. Pediatr. 156: 952–956. [DOI] [PubMed] [Google Scholar]
- 21.Guerrera I. C., Astarita G., Jais J. P., Sands D., Nowakowska A., Colas J., Sermet-Gaudelus I., Schuerenberg M., Piomelli D., Edelman A., et al. 2009. A novel lipidomic strategy reveals plasma phospholipid signatures associated with respiratory disease severity in cystic fibrosis patients. PLoS ONE. 4: e7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freedman S. D., Blanco P. G., Shea J. C., Alvarez J. G. 2002. Analysis of lipid abnormalities in CF mice. Methods Mol. Med. 70: 517–524. [DOI] [PubMed] [Google Scholar]
- 23.Freedman S. D., Katz M. H., Parker E. M., Laposata M., Urman M. Y., Alvarez J. G. 1999. A membrane lipid imbalance plays a role in the phenotypic expression of cystic fibrosis in cftr(−/−) mice. Proc. Natl. Acad. Sci. USA. 96: 13995–14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freedman S. D., Blanco P. G., Zaman M. M., Shea J. C., Ollero M., Hopper I. K., Weed D. A., Gelrud A., Regan M. M., Laposata M., et al. 2004. Association of cystic fibrosis with abnormalities in fatty acid metabolism. N. Engl. J. Med. 350: 560–569. [DOI] [PubMed] [Google Scholar]
- 25.Ollero M., Laposata M., Zaman M. M., Blanco P. G., Andersson C., Zeind J., Urman Y., Kent G., Alvarez J. G., Freedman S. D. 2006. Evidence of increased flux to n-6 docosapentaenoic acid in phospholipids of pancreas from cftr−/− knockout mice. Metabolism. 55: 1192–1200. [DOI] [PubMed] [Google Scholar]
- 26.Karp C. L., Flick L. M., Park K. W., Softic S., Greer T. M., Keledjian R., Yang R., Uddin J., Guggino W. B., Atabani S. F., et al. 2004. Defective lipoxin-mediated anti-inflammatory activity in the cystic fibrosis airway. Nat. Immunol. 5: 388–392. [DOI] [PubMed] [Google Scholar]
- 27.Andersson C., Al-Turkmani M. R., Savaille J. E., Alturkmani R., Katrangi W., Cluette-Brown J. E., Zaman M. M., Laposata M., Freedman S. D. 2008. Cell culture models demonstrate that CFTR dysfunction leads to defective fatty acid composition and metabolism. J. Lipid Res. 49: 1692–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strandvik B., Gronowitz E., Enlund F., Martinsson T., Wahlstrom J. 2001. Essential fatty acid deficiency in relation to genotype in patients with cystic fibrosis. J. Pediatr. 139: 650–655. [DOI] [PubMed] [Google Scholar]
- 29.Innis S. M., Davidson A. G. 2008. Cystic fibrosis and nutrition: linking phospholipids and essential fatty acids with thiol metabolism. Annu. Rev. Nutr. 28: 55–72. [DOI] [PubMed] [Google Scholar]
- 30.Innis S. M., Hasman D. 2006. Evidence of choline depletion and reduced betaine and dimethylglycine with increased homocysteine in plasma of children with cystic fibrosis. J. Nutr. 136: 2226–2231. [DOI] [PubMed] [Google Scholar]
- 31.Brulet M., Seyer A., Edelman A., Brunelle A., Fritsch J., Ollero M., Laprevote O. 2010. Lipid mapping of colonic mucosa by cluster TOF-SIMS imaging and multivariate analysis in cftr knockout mice. J. Lipid Res. 51: 3034–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knudson R. J., Lebowitz M. D., Holberg C. J., Burrows B. 1983. Changes in the normal maximal expiratory flow-volume curve with growth and aging. Am. Rev. Respir. Dis. 127: 725–734. [DOI] [PubMed] [Google Scholar]
- 33.Astarita G., Ahmed F., Piomelli D. 2008. Identification of biosynthetic precursors for the endocannabinoid anandamide in the rat brain. J. Lipid Res. 49: 48–57. [DOI] [PubMed] [Google Scholar]
- 34.Astarita G., Piomelli D. 2009. Lipidomic analysis of endocannabinoid metabolism in biological samples. J Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2755–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guilbault C., De Sanctis J. B., Wojewodka G., Saeed Z., Lachance C., Skinner T. A., Vilela R. M., Kubow S., Lands L. C., Hajduch M., et al. 2008. Fenretinide corrects newly found ceramide deficiency in cystic fibrosis. Am. J. Respir. Cell Mol. Biol. 38: 47–56. [DOI] [PubMed] [Google Scholar]
- 36.Teichgraber V., Ulrich M., Endlich N., Riethmuller J., Wilker B., De Oliveira-Munding C. C., van Heeckeren A. M., Barr M. L., von Kurthy G., Schmid K. W., et al. 2008. Ceramide accumulation mediates inflammation, cell death and infection susceptibility in cystic fibrosis. Nat. Med. 14: 382–391. [DOI] [PubMed] [Google Scholar]
- 37.White N. M., Jiang D., Burgess J. D., Bederman I. R., Previs S. F., Kelley T. J. 2007. Altered cholesterol homeostasis in cultured and in vivo models of cystic fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 292: L476–L486. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y. Z., Abolhassani M., Ollero M., Dif F., Uozumi N., Lagranderie M., Shimizu T., Chignard M., Touqui L. 2010. Cytosolic phospholipase A2alpha mediates Pseudomonas aeruginosa LPS-induced airway constriction of CFTR-/- mice. Respir. Res. 11: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medjane S., Raymond B., Wu Y., Touqui L. 2005. Impact of CFTR DeltaF508 mutation on prostaglandin E2 production and type IIA phospholipase A2 expression by pulmonary epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 289: L816–L824. [DOI] [PubMed] [Google Scholar]
- 40.Dif F., Wu Y. Z., Burgel P. R., Ollero M., Leduc D., Aarbiou J., Borot F., Garcia-Verdugo I., Martin C., Chignard M., et al. 2010. Critical role of cytosolic phospholipase A2{alpha} in bronchial mucus hypersecretion in CFTR-deficient mice. Eur. Respir. J. 36: 1120–1130. [DOI] [PubMed] [Google Scholar]
- 41.Luthold S., Berneis K., Bady P., Muller B. 2007. Effects of infectious disease on plasma lipids and their diagnostic significance in critical illness. Eur. J. Clin. Invest. 37: 573–579. [DOI] [PubMed] [Google Scholar]
- 42.Figueroa V., Milla C., Parks E. J., Schwarzenberg S. J., Moran A. 2002. Abnormal lipid concentrations in cystic fibrosis. Am. J. Clin. Nutr. 75: 1005–1011. [DOI] [PubMed] [Google Scholar]
- 43.Fon Tacer K., Pompon D., Rozman D. 2010. Adaptation of cholesterol synthesis to fasting and TNF-alpha: profiling cholesterol intermediates in the liver, brain, and testis. J. Steroid Biochem. Mol. Biol. 121: 619–625. [DOI] [PubMed] [Google Scholar]
- 44.Feingold K. R., Grunfeld C. 1987. Tumor necrosis factor-alpha stimulates hepatic lipogenesis in the rat in vivo. J. Clin. Invest. 80: 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mailhot G., Rabasa-Lhoret R., Moreau A., Berthiaume Y., Levy E. 2010. CFTR depletion results in changes in fatty acid composition and promotes lipogenesis in intestinal Caco 2/15 cells. PLoS ONE. 5: e10446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LoVerme J., Guzman M., Gaetani S., Piomelli D. 2006. Cold exposure stimulates synthesis of the bioactive lipid oleoylethanolamide in rat adipose tissue. J. Biol. Chem. 281: 22815–22818. [DOI] [PubMed] [Google Scholar]
- 47.Hansen H. S. 2010. Palmitoylethanolamide and other anandamide congeners. Proposed role in the diseased brain. Exp. Neurol. 224: 48–55. [DOI] [PubMed] [Google Scholar]
- 48.Fride E., Ponde D., Breuer A., Hanus L. 2005. Peripheral, but not central effects of cannabidiol derivatives: mediation by CB(1) and unidentified receptors. Neuropharmacology. 48: 1117–1129. [DOI] [PubMed] [Google Scholar]
- 49.Lo Verme J., Fu J., Astarita G., La Rana G., Russo R., Calignano A., Piomelli D. 2005. The nuclear receptor peroxisome proliferator-activated receptor-alpha mediates the anti-inflammatory actions of palmitoylethanolamide. Mol. Pharmacol. 67: 15–19. [DOI] [PubMed] [Google Scholar]