Abstract
Retinyl esters are the major chemical forms of vitamin A stored in the liver, and can be delivered to peripheral tissues for conversion into biologically active forms. The function and regulation of the hepatic genes that are potentially involved in catalyzing the hydrolysis of retinyl esters remain unclear. Here we show that two lipid hydrolytic genes, pancreatic-related protein 2 (mPlrp2) and procolipase (mClps), expressed specifically in the mouse pancreas, are associated with the ratio of S-adenosylmethionine (AdoMet) to S-adenosylhomocysteine (AdoHcy). Light illumination deficiency or administration of 5′-AMP elevated the ratio of AdoMet to AdoHcy and induced the expression in the liver of mPlrp2 and mClps, which was blocked by all-trans retinoic acid. Mice fed a vitamin A-free diet exhibited increased activation of hepatic mPlrp2 and mClps expression, which was associated with increased methylation of histone H3K4 residues located near the mPlrp2 and mClps promoters. Inhibition of hepatic mPlrp2 and mClps expression by a methylase inhibitor, methylthioadenosine, markedly decreased plasma retinol levels in these mice. The activated hepatic stellate cell (HSC)-T6 cell line specifically expressed mClps and mPlrp2. Inhibition of mClps gene expressions by short hairpin RNA (shRNA) decreased hydrolysis of retinyl esters in the HSC-T6 cell line. These data suggest that the conditional expression of mPlrp2 and mClps is involved in the hydrolysis of retinyl esters in the mouse liver.
Keywords: S-adenosylmethionine, S-adenosylhomocysteine, procolipase, pancreatic-related protein 2, hepatic retinyl ester
Vitamin A and its derivatives (retinoids) play an essential role in differentiation and development by their conversion to active-form retinoic acid, which interacts with the nuclear receptors of the RXR and RAR families, functioning as hormone-activated factors in the regulation of the expression of multiple genes (1, 2). Also, vitamin A mediates vision cycles through its metabolite, 11-cis retinaldehyde, which is the active chromophore in rhodopsin (3, 4). Most of the retinoids in the body are accounted for by the retinyl esters stored in the liver (5, 6). The formation and hydrolysis of retinyl esters are key reactions for maintaining a constant concentration of free retinol in circulation. It has been demonstrated that lecithin:retinol acyltransferase and acylCoA:retinol acyltransferase contribute to the storage of retinoids by the conversion of circulatory free retinol to retinyl esters (7, 8). The enzymes catalyzing the reverse reaction, designated retinyl ester hydrolase (REH), promote the liberation of retinol from stored retinyl esters in the liver into general circulation. Mice fed a vitamin A-deficient diet for 6 weeks decreased the hepatic retinyl esters (9), and in vitamin A-deficient rats, the liver uptake of retinol from blood was obviously impaired (10). However, the identity, function, and regulation of hepatic enzymes potentially involved in catalyzing the hydrolysis of retinyl esters remain unclear (11, 12).
The bile salt-activated carboxylester lipase, an enzyme found in the pancreas and liver cells of several mammalian species, was thought to be the main hepatic REH (13). Mice deficient in carboxylester lipase exhibit normal levels of intestinal and hepatic uptake of retinyl esters and maintain REH activity in the liver and pancreas, suggesting that in the mouse liver and pancreas, there is an REH distinct from carboxylester lipase (14). Pancreatic triglyceride lipase (PTL) and its partner, colipase, have been demonstrated to be major pancreatic REHs in mice. Like classical PTL, the pancreatic lipase-related protein 2 (PLRP2), with strong nucleotide and amino acid sequence homology to PTL, also cleaves triglycerides and retinyl esters, but PLRP2 has broader substrate specificity (15, 16). Although PLRP2 does not absolutely require colipase, colipase may increase PLRP2 protein activity (17). In suckling mice, PLRP2 protein is the major colipase-dependent pancreatic lipase (18).
The murine genes encoding pancreatic-related protein 2 (mPlrp2) and procolipase (mClps) are expressed specifically in the pancreas to hydrolyze fat and retinyl esters in the diet (19). Our recent studies demonstrate that a reduction in light illumination results in upregulated expression of mPlrp2 and mClps in the mouse liver (20), and that the light-modulated all-trans retinoic acid (ATRA) signal plays a key role in the regulation of the expression of these genes (21). In this study, we characterized the molecular mechanism by which mPlrp2 and mClps are expressed specifically in the pancreas by assessing the difference in the ratio of S-adenosylmethionine (AdoMet) to S-adenosylhomocysteine (AdoHcy) between the pancreas and the liver. A temporary, light-modulated change in the ratio of AdoMet to AdoHcy regulates hepatic mPlrp2 and mClps gene expression in mice. Using mice fed a vitamin A-free diet, hepatic mPlrp2 and mClps gene expression was found to be directly related to the level of retinol in the plasma. The results of the studies strongly suggest that hepatic mPlrp2 and mClps gene expression is associated with the hydrolysis of retinyl esters in the mouse liver.
EXPERIMENTAL PROCEDURES
Animals
Female C57BL/6 mice, 6 to 8 weeks old, were used in this work. Mice were maintained in 12 h light/12 h dark (LD) cycles, with lights on at 7:00 AM and off at 7:00 PM, and were allowed free access to food and water. For the vitamin A-free diet experiment, mice were placed on a vitamin A-free diet for 1 week, and the control animals were given normal chow. The vitamin A-free diet (Laboratory, Nanjing, China) for 100 g food by dry weight was composed of vitamin-free casein (18%), sucrose plus maize starch (68.4%), cellulose (2%), hydrogenated coconut oil (4.6%), salt (4.8%), yeast (2%), and a vitamin integration lacking vitamin A (0.2%). A control group of mice was fed using standard laboratory chow. For consistent observations, all samples were collected at 7:00 PM, unless otherwise indicated. All animal care and use procedures were in accordance with the guidelines of the Institutional Animal Care and Use Committee at Nanjing University of Science & Technology.
Light illumination analysis
For light quality experiments, animals were transferred to closed chambers illuminated by light-emitting diode (LED) illuminators (PARA; Nanjing, China). The intensity of colored light was matched to the intensity of the incident white light source by adjusting the distance between the light and the cage covers (about 250 lux). Colored light was generated by the following LEDs (LED-Z, Burbank, CA): blue (470 nm), green (528 nm), yellow (593 nm), and red (625 nm). Mice were maintained in colored L/D (12 h/12 h) or constant darkness at the same schedule as the white cycle for 72 h. For light intensity experiments, animals were transferred from standard light (about 250 lux) to weak light (about 20 lux at the cage cover) from a white light lamp in 12 h/12 h L/D cycles for 72 h. Samples were collected at the time points indicated in the figure legends. Light intensity was measured by Laboratory Digital Light Meter CT-1330B (CETO, Nanjing, China).
RNA isolation and RT-PCR analysis
Total RNA was extracted from fresh liver samples with TriZol Reagent, and the remaining DNA was completely removed by RNase-free DNase treatment. Total RNA (50 ng) was reverse-transcribed with poly(dT)12–18 as a first-strand primer, according to the manufacturer's instructions. RT-PCR was performed and analyzed using an ABI 9700 Detection System. Quantitative real-time RT-PCR was performed, and results were analyzed using an ABI 7300 Detection System utilizing SYBR Green dye. The following primers were designed: Gapdh: 5′-CATCCACTGGTGCTGCCAAGGCTGT-3′ (forward), 5′-ACAACCTGGTC CTCA GTGTAGCCCA-3′ (reverse); mPlrp2: 5′- GGACCAAAGAA GCTGAGT GG-3′(forward), 5′-ATAGTGGG CCGGAA GAGATT-3′(reverse); mClps: 5′-CTTGTGT CCCTCCTTGCAGT-3′ (forward), 5′-GAGCACTCGCTGTTC TCC AT-3′ (reverse). Reaction conditions were as follows: cycle 1 (1×), 50°C for 2 min; cycle 2 (1×), 95°C for 10 min; cycle 3 (35×), 94°C for 15 s, 58°C for 30 s, 72°C for 30 s. All primer sequences were located between more than two exons. Relative gene expression in comparison with Gapdh expression was calculated by the comparative cycle threshold (CT) method.
Western blotting
The protein samples were separated by 10% homogeneous SDS-PAGE before the proteins were transferred to nitrocellulose membranes. After blocking with milk in PBS-Tween (0.1%) buffer overnight at 4°C, the transferred membranes were incubated with PLRP2 antibody (Santa Cruz Biotechnology) for 2 h at 26°C, followed by HRP-conjugated secondary antibody (Santa Cruz Biotechnology) for 1 h at 26°C. The blotting was developed by using ECL detection reagent (Keygen, China) and visualized by autoradiography. HRP-conjugated monoclonal mouse β-actin was introduced as a reference.
Determination of hepatic levels of AdoMet and AdoHcy
Extracts were separated and quantified by using reverse-phase HPLC (Waters 1525 System; Millipore Corp., Bedford, MA) analysis on a Partisphere bonded phase C18 (reverse phase) cartridge column, according to procedures previously described by She et al. (22). The mobile phase contained 0.1 M sodium acetate, 5 mM heptanesulfonic acid adjusted to pH 4.5 with acetic acid, and 4.2% acetonitrile. The samples were eluted at room temperature with an invariable gradient at a flow rate of 0.8 ml/min. Characteristic peak spectra and retention times compared with those of the standard were used to identify AdoMet and AdoHcy. Quantitation was based on peak areas. AdoMet and AdoHcy standards were purchased from Sigma (St. Louis, MO).
Plasma retinol analysis by HPLC
Analysis of plasma retinol levels was performed by reverse-phase HPLC according to the method described by Teerlink et al. (23). Plasma was collected in EDTA-coated tubes, isolated, and frozen in the dark at −70°C until the determination of retinol concentrations. Retinoid extraction and analytical procedures were carried out in a darkened room under dim yellow light (15 w) to protect retinoids from exposure to light. The extraction procedure involved in single-phase fluid extraction consisted of adding 350 μl of plasma to 50 μl of 1.0 M sodium acetate buffer (pH 4.0). After mixing, 600 μl acetonitrile was added to the mixture, and the samples were immediately vortex-mixed and centrifuged for 5 min at 12,000 g and 4°C, and 450 μl clear supernatant was decanted and diluted with 150 μl water. For HPLC analysis, 400 μl of this solution was injected. The analysis was performed on a Waters HPLC system (Waters 1525) equipped with a vacuum degasser, quaternary pump, autosampler, column heater, and diode array detector. HPLC analysis was conducted on a 5 μm octadecyl silane C18 column (25 cm × 4.6 mm internal diameter; CEL Associates, Pearland, TX). The mobile phase consisted of acetonitrile-methanol-2% ammonium acetate solution-glacial acetic acid (79:2:16:3) with a flow rate of 1.0 ml/min. The temperature of the column was maintained at 30°C. The diode array detector was set at a wavelength of 325 nm to monitor all compounds simultaneously. Characteristic peak spectra and retention times were compared with a retinol standard (Sigma) to identify retinol. Quantitation was based on peak areas.
Treating animals with 5′-AMP, MTA, and ATRA
The indicated dosages of 5′-AMP, methylthioadenosine (MTA), and ATRA (Sigma) were administered by intraperitoneal injections in various groups of mice. After injection, mice were maintained for the desired period of time and then euthanized. Plasma or liver tissue was collected and extracted for analysis of AdoMet and AdoHcy ratios or retinol levels. Total RNA was isolated from liver tissues for RT-PCR analysis.
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation (ChIP) assay was performed as previously described (24, 25), with some modifications for the liver. Briefly, 100 mg of liver tissue was cut into small pieces with a razor blade and cross-linked with 1% formaldehyde for 15 min. Cross-linking was stopped with 0.125 M glycine, and samples were rinsed with 1× PBS containing 1 mM PMSF and disaggregated with a Dounce homogenizer. The nuclei were pelleted and resuspended in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl at pH 8.1, 0.8 μg/ml pepstatin A, 0.6 μg/ml leupeptin, and 1 mM PMSF). The suspension was sonicated using a sonicator (KS-130; Ninbo Kesheng, China) to generate DNA fragments averaging 400–500 bp in length and clarified by centrifugation. For immunoprecipitation, the supernatant was diluted 10-fold with dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, and 20 mM Tris-HCl at pH 8.1) and divided into fractions for control IgG and anti-trimethyl-H3K4 (Millipore). Protein A-Sepharose beads, preblocked with 300 μg/ml of sheared salmon sperm DNA and 0.1% BSA, were used to precipitate antibody-chromatin complexes. Beads were washed sequentially for 5 min each with TSE buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH8.1) with 150 and 500 mM NaCl, buffer III, and TE (10 mM Tris-HCl, 1 mM EDTA PH = 8.0). Immunocomplexes were eluted, the elution was heated to reverse the formaldehyde cross-linking, and DNA was phenol-chloroform extracted. For PCR analysis of the ChIP samples, purified immunoprecipitates were dissolved in 20 μl water.
Preparation of shRNA
The shRNAs targeting rat mClps mRNA were designed and synthesized by GenScript. Sequences used for the shRNAs in the experiments were: shRNA-1, 5′-TGCCAACATGACACCATCCTG-3′; and shRNA-2, 5′-TGCAGTGTAAGAGCAGATGCT-3′. All of these sequences were shown by BLAST search not to share sequence homology with any rat mRNA. Negative control shRNA was used to assess nonspecific gene-silencing effects. The mammalian expression vector pRNA-U6.1 was used to express shRNAs in hepatic stellate cell (HSC)-T6. ShRNAs were introduced into HSC-T6 cells by GenEscort™II, according to the manufacturer's protocol.
Cell culture
HSC-T6 cells were cultured in DMEM (Invitrogen) containing 25 mM glucose, 10% (v/v) FBS, and 100 U/ml penicillin/streptomycin at 37°C/5% CO2. HSC-T6 cells were incubated for 12 h with palmitic acid (100 μM), after which the cells were treated with 5 μM retinol for an additional 12 h, and then shRNAs or the indicated dosages of MTA (4 μM and 12 μM) were introduced into the cells. Retinyl ester concentrations were determined 12 h later.
PLRP2 immunohistochemistry
To detect PLRP2, slides of paraffin-embedded liver sections were deparaffinized and rehydrated. Immunohistochemical staining for PLRP2 with a mouse antibody against PLRP2 (Santa Cruz Biotechnology) was performed according to the manufacturer's protocol.
Statistical analysis
Data are presented as mean ± SEM. Statistical analysis was performed with a Student's t-test. Significance was defined as P < 0.05.
RESULTS
Weak light illumination activated the expression of mPlrp2 and mClps in the liver
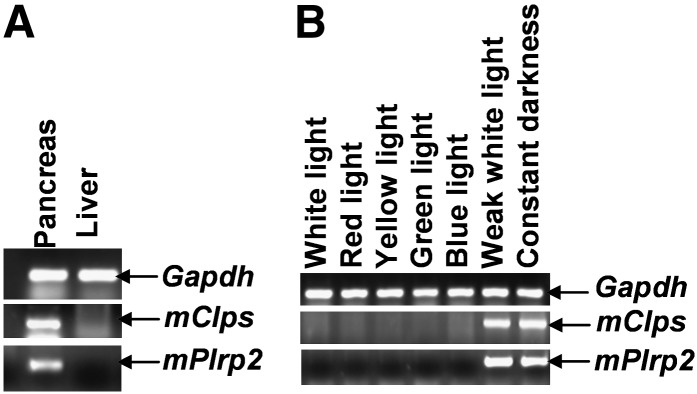
The lipase gene mPlrp2 and its partner, mClps, are expressed specifically in the mouse pancreas under general conditions (26). Our previous work demonstrated that light exposure inhibits both mPlrp2 and mClps gene expression and that darkness induces their expression in the mouse liver (20, 21). We hypothesized that a specific light wavelength or light intensity in illumination was a modulator for mPlrp2 and mClps gene expression. Therefore, we carried out specific wavelength illumination studies to determine whether there was a difference in the responses of these genes to light. Confirming previous observations, no hepatic mPlrp2 and mClps gene expression was observed under LD cycles (Fig. 1A). Mice maintained in similar intensities of various wavelengths, including red light, yellow light, blue light, and green light, failed to exhibit mPlrp2 and mClps gene expression in the liver, indicating that the inhibition of hepatic mPlrp2 and mClps expression by white light was not due to a specific wavelength (Fig. 1B). Next, we investigated whether absolute light deficiency [dark/dark (DD)] was necessary to induce hepatic mPlrp2 and mClps expression. RT-PCR analysis showed that light intensity below 20 lux activated mPlrp2 and mClps gene expression in the liver (Fig. 1B), and no expression of these genes could be determined under high light intensity. These observations suggest that light intensity is a regulatory signal in modulating hepatic mPlrp2 and mClps gene expression. Moreover, Western blotting showed that DD induced PLRP2 protein expression, whereas light exposure for 2 h decreased the protein level, and at 6 h light illumination, PLRP2 almost cannot be detected (see supplementary Fig. I).
Fig. 1.
Light intensity, but not specific wavelength, influenced expression of mPlrp2 and mClps in the liver. A: Specific expression of mPlrp2 and mClps was detected in the pancreas in LD mice by RT-PCR, whereas no expression of both genes was found in the liver in LD mice. B: Weak white light (<20 lux) induced mPlrp2 and mClps gene expression in the liver in LD mice. The similar high intensity (about 250 lux) of white, red, yellow, green, and blue light had the same inhibitory function on mPlrp2 and mClps expression. DD mice were examined as a positive control. Results are representative of three independent experiments. Gapdh mRNA was monitored as an internal control.
Specific expression of mPlrp2 and mClps was associated with the ratio of AdoMet-AdoHcy in the pancreas
Although light intensity plays a dominant role in inducing mPlrp2 and mClps gene expression in the liver, light exposure or darkness failed to induce a significant difference in the expression pattern of either gene in the pancreas. Under normal conditions for maintaining animals, the expression of the mPlrp2 and mClps genes was tissue specific and restricted to the pancreas; both gene products functioned in hydrolyzing lipid from diet fat and retinyl esters (19). The conditional expression of mPlrp2 and mClps in the liver by weak light illumination or darkness was reversible. To identify whether various states of energy metabolism in tissues could result in the specific expression of mPlrp2 and mClps, we examined the difference between the pancreas and liver tissues. First, nucleotides were analyzed from extracts of both tissues. There was no significant difference in the levels of ATP, ADP, and AMP between the pancreas and the liver, excluding the possibility that nucleotide metabolism results in the specific expression of mPlrp2 and mClps. Then, we compared the ratio of AdoMet to AdoHcy, which represents the potential of methylation in tissue (27). HPLC profiles showed that the peak of AdoMet was much higher than that of AdoHcy in the pancreas (Fig. 2A), whereas liver tissues had no obvious differences in the peaks of AdoMet and AdoHcy (Fig. 2B). Next, we quantified the levels of the peaks obtained from the liver and pancreas (Fig. 2C, n = 6). This analysis revealed that the ratio of AdoMet to AdoHcy was markedly higher in the pancreas, compared with that found in the liver.
Fig. 2.
Increased ratios of AdoMet and AdoHcy present in the pancreas compared with the liver under LD conditions. A: Representative HPLC profiles of AdoMet and AdoHcy extracted from the pancreas. B: HPLC profiles from the liver. Peak 1 is AdoHcy and peak 2 is AdoMet. The detection wavelength was 260 nm. C: Quantification of AdoMet and AdoHcy in the pancreas and liver with respect to their internal protein concentration. Data are means ± SEM; *P < 0.05; n = 6.
Darkness or weak light illumination elevated the ratio of AdoMet to AdoHcy in livers
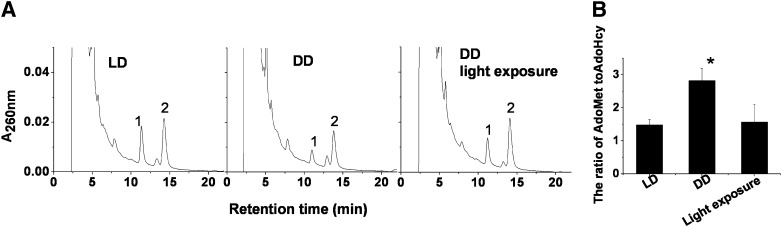
To demonstrate that the ratio of Adomet to AdoHcy was associated with hepatic mPlrp2 and mClps expression, we measured AdoMet and AdoHcy in the mouse livers under darkness and weak light illumination. HPLC profiles showed that peak AdoHcy declined and peak AdoMet rose in mouse livers in the DD cycle compared with the normal LD cycle (Fig. 3A). A similar change was observed in mice maintained under weak light illumination. Moreover, in DD animals, light exposure elevated the AdoHcy peak to a level comparable to that of LD animals (Fig. 3A). Quantitative analysis of peak levels revealed that the ratio of AdoMet to AdoHcy was markedly elevated in mouse livers under darkness compared with light illumination; the elevated level of the ratio in the liver was close to that found in the pancreas, and light exposure in DD mice for 4 h reversibly returned this ratio to the normal levels found in LD animals (Fig. 3B). Previous observations demonstrated that light exposure in DD mice inhibited mPlrp2 and mClps expression in the liver (20, 21).
Fig. 3.
Darkness and weak light illumination elevated the ratio of AdoMet to AdoHcy in livers of mice. A: Representative HPLC profiles of AdoMet and AdoHcy extracted from mouse livers under LD, DD, and light exposure in DD for 4 h. Peak 1, AdoHcy; peak 2, AdoMet. B: Quantification of AdoMet and AdoHcy in mouse livers under various light illuminations. Data are means ± SEM; n = 5; *P < 0.05, compared with LD.
Administration of ATRA lowered the ratio of AdoMet to AdoHcy in DD livers, and administration of 5′-AMP increased the ratio of AdoMet to AdoHcy in LD livers
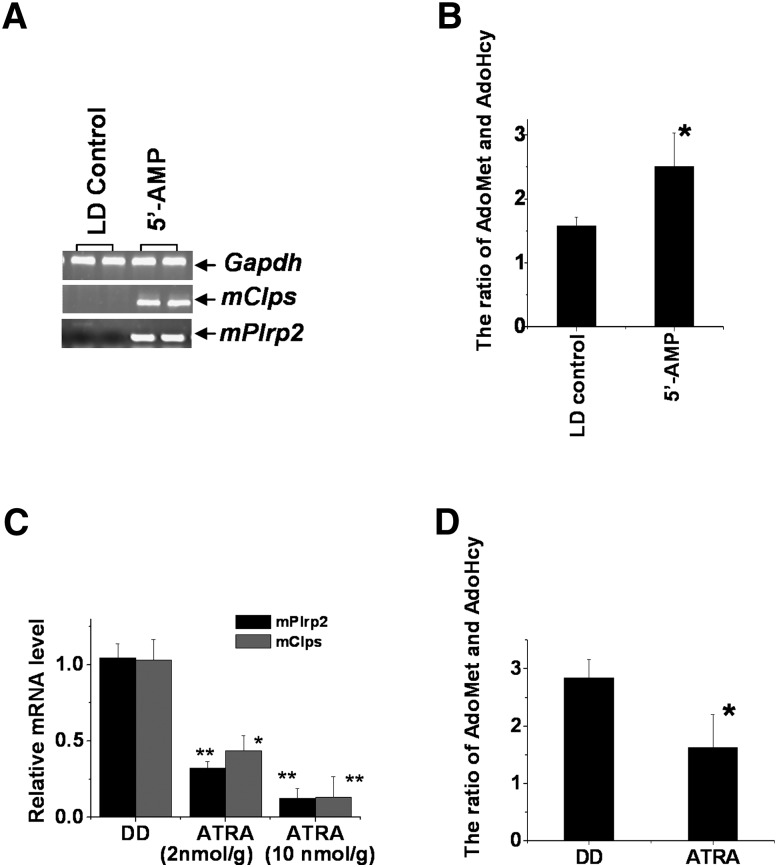
Our recent studies demonstrate that ATRA has a feedback inhibition to hepatic mPlrp2 and mClps. The light-modulated ATRA signal regulated ecto-5′-nucleotidase (CD73) gene expression by the RARα receptor, affecting the circulatory levels of 5′-AMP, resulting in mPlrp2 and mClps expression (21). Therefore, we carried out experiments to analyze the alteration of hepatic AdoMet and AdoHcy in ATRA- and 5′-AMP-treated animals. RT-PCR analysis showed that administration of 5′-AMP in LD animals activated mPlrp2 and mClps gene expression in the liver (Fig. 4A), confirming a previous Northern analysis; expectedly, LD livers indicated that the ratio of AdoMet to AdoHcy increased notably after 5′-AMP injection (Fig. 4B). Next, ATRA (2 nmol/g and 10 nmol/g)-treated mice exhibited reduced mPlrp2 and mClps expression under DD conditions (Fig. 4C), and the ratio of AdoMet to AdoHcy declined in ATRA-treated livers compared with control livers (Fig. 4D). These observations implied that the temporary changes of the ratio of AdoMet to AdoHcy in the livers were associated with hepatic mPlrp2 and mClps expression.
Fig. 4.
Effects of the administration of 5′-AMP and ATRA on the expression of the mPlrp2 and mClps genes in the liver. A: 5′-AMP (150 nmol/g) induced hepatic mPlrp2 and mClps gene expression in LD mice. Samples were collected 3.5 h after intraperitoneal injection. Results are expressed as double samples, representative of six animals in every group. Gapdh mRNA was monitored as an internal control. B: Quantification of AdoMet and AdoHcy in livers of LD mice treated by 5′-AMP compared with mice treated with saline. An elevated ratio of AdoMet to AdoHcy was observed in mice treated by 5′-AMP. Data are means ± SEM; n = 6; *P < 0.05. C: Injection of ATRA (2 nmol/g and 10 nmol/g) for 4 h inhibited hepatic mPlrp2 and mClps gene expression in DD mice. Data are means ± SEM; n = 6; *P < 0.05, **P < 0.01, compared with DD. D: Quantification of AdoMet and AdoHcy in the livers of DD mice treated with ATRA, compared with mice treated with oil. Lowered ratios of AdoMet to AdoHcy were observed in mice treated by ATRA. Data are means ± SEM; n = 6; *P < 0.05.
Mice fed a vitamin A-free diet activated hepatic mPlrp2 and mClps expression, along with related increases in the methylation of histone H3K4 associated with the mPlrp2 and mClps promoters
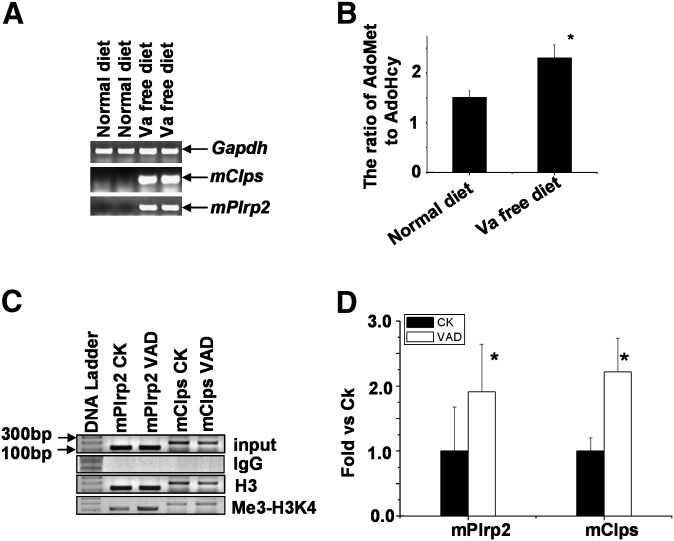
The gene products of mPlrp2 and mClps have been demonstrated to function in the hydrolysis of dietary retinyl esters in the gastrointestinal organs (19), implying the possibility of similar hydrolysis activities for hepatic retinyl esters. To determine whether hepatic mPlrp2 and mClps were related with the hepatic utilization of retinoids, mice were placed on a vitamin A-free diet for 7 days. Surprisingly, hepatic mPlrp2 and mClps mRNA were upregulated strongly in the livers of mice fed a vitamin A-free diet (Fig. 5A). Next, we detected the expression levels of other lipases in the liver, such as hepatic lipase, lipoprotein lipase, and carboxylester lipase, and found no obvious reproducible changes in these lipases in mice fed the vitamin A-free diet compared with that observed in mice fed a normal diet (data not shown). A subsequent analysis of AdoMet and AdoHcy showed that the ratio of AdoMet to AdoHcy in the liver is elevated in these animals compared with mice fed a normal diet. (Fig. 5B). These changes in AdoMet and AdoHcy levels resulted in an increase of total trimethylation of histone H3K4-related liver DNA in mice fed a vitamin A-free diet compared with control mice, and ChIP analysis indicated an increase in the binding of trimethylated H3K4 to mPlrp2 and mClps promoters in the liver DNA of mice fed vitamin A-free diets (Fig. 5C, D).
Fig. 5.
The mPlrp2 and mClps genes are activated in the livers of mice fed a vitamin A-free diet. A: RT-PCR analysis of mPlrp2 and mClps mRNA in mice fed a vitamin A-free diet for 7 days. Results are expressed as double samples, representative of six animals in every group. Gapdh mRNA was monitored as an internal control. B: Quantification of AdoMet and AdoHcy in the livers of mice fed a vitamin A-free diet compared with mice fed a normal diet. Increased ratios of AdoMet and AdoHcy were observed in vitamin A-free-diet mice. C: Representative PCR profiles, and D, quantitative real-time RT-PCR analysis of the binding of histone H3K4-3Me to the mPlrp2 and mClps promoter sequences in vitamin A-free and control mice, respectively. Data are means ± SEM; n = 6; *P < 0.05.
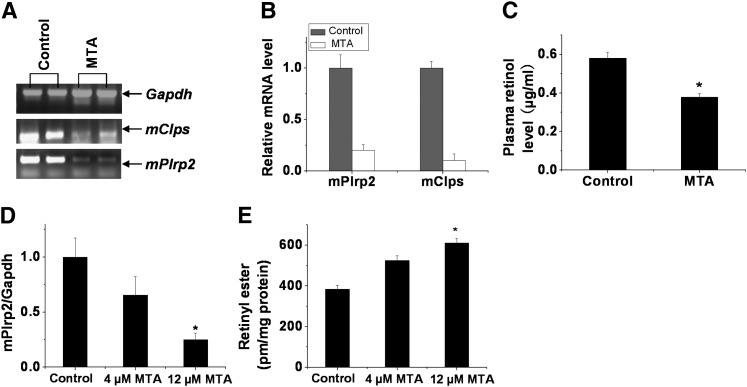
A methylase inhibitor blocked hepatic mPlrp2 and mClps expression and markedly lowered the levels of free retinol in the plasma and impaired hydrolysis of retinyl ester in the HSC-T6 cell line
AdoMet serves as a methyl donor in a series of biomethylation reactions, and the by-product, AdoHcy, is a potent feedback inhibitor of AdoMet-dependent methyltransferase. Therefore, the ratio of AdoMet to AdoHcy reflects the methylating capacity of tissues (27, 28). To clarify whether the changes of the ratio of AdoMet to AdoHcy are correlated with mPlrp2 and mClps expression by the regulation of methylase, we examined the effects of a methylase inhibitor on mPlrp2 and mClps gene expression in the liver. MTA, an effective methylation blocker, was used to treat mice fed the vitamin A-free diet. Figure 6A, B showed that severe attenuation of hepatic mPlrp2 and mClps gene expression was observed in MTA-treated animals. Subsequent HPLC analysis showed that the peak of retinol from plasma extraction markedly declined in MTA-treated mice compared with control animals treated with saline. Quantitative analysis of peak levels revealed that MTA-treated mice exhibited significantly decreased levels of free retinol in the plasma compared with control animals (Fig. 6C, n = 5). MTA inhibited mPlrp2 expression and hydrolysis of retinyl ester in a dose-dependent manner in the HSC-T6 cell line (Fig. 6D, E), and a linear correlation was found between the expression of mPlrp2 and the levels of retinyl ester. These data indicated that mPlrp2 and mClps are involved in the hydrolysis of retinyl esters in HSCs in the liver and that their expression is regulated by DNA methylation.
Fig. 6.
MTA impairs the expression of mPlrp2 and mClps in the livers of mice fed a vitamin A-free diet and hydrolysis of retinyl ester in the HSC-T6 cell line. A: RT-PCR analysis of mPlrp2 and mClps mRNA in vitamin A-free-diet mice treated with MTA. Mice were placed on a vitamin A-free diet for 7 days and injected with MTA (100 nmol/g/day) on the last two days. Results are expressed as double samples, representative of six animals in every group. Gapdh mRNA was monitored as an internal control. B: Quantitative real-time RT-PCR analysis of mPlrp2 and mClps mRNA in vitamin A-free-diet mice treated with MTA. C: Quantification of retinol levels in plasma extracted from vitamin A-free-diet mice treated with MTA compared with mice treated with saline. Decreased plasma retinol levels were observed in MTA-treated mice. D: Quantitative real-time RT-PCR analysis of mPlrp2 in HSC-T6 cells treated with the indicated dosages of MTA. E: Quantification of retinyl esters from HSC-T6 cells treated with the indicated dosages of MTA compared with the control. HSC-T6 cells were incubated for 12 h with palmitic acid (100 μM), after which the cells were treated with 5 μM retinol for an additional 12 h, and then the indicated dosages of MTA were introduced into the cells. Retinyl ester concentrations were determined 12 h later. Data are means ± SEM; n = 6; *P < 0.05.
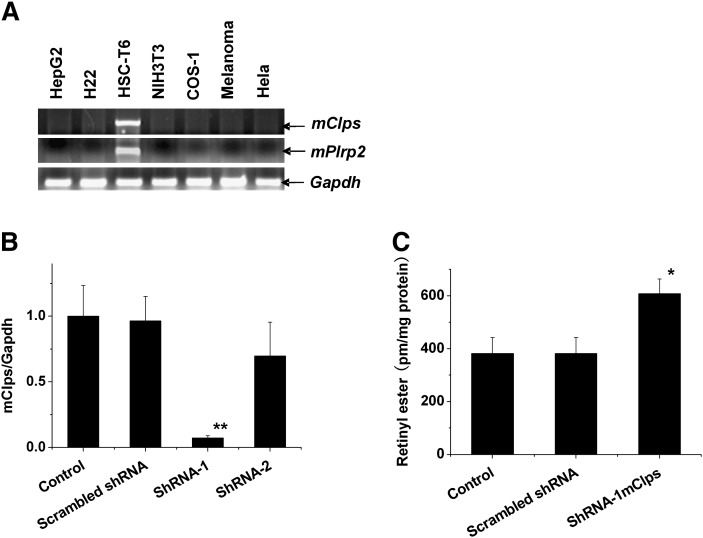
HSCs specifically expressed mClps and mPlrp2
HSCs are primarily involved in the storage of retinol esters. Interestingly, cultured HSC-T6 cells in the activated state specifically expressed the mClps and mPlrp2 genes (Fig. 7A), whereas other cell lines, including some hepatic cell lines, failed to exhibit detectable expression of mClps and mPlrp2. To examine the role of mPlrp2 and mClps in the retinoid metabolism of HSCs, we performed gene-silencing studies in the rat HSC-T6 cell line. As shown in Fig. 7B, C, the knockdown of mClps by 80% in HSC-T6 cells led to the retention of more retinyl esters compared with control and scrambled shRNA-treated cells, indicating a role for this gene in the hydrolysis of retinyl esters associated with the metabolism of retinoids in HSCs. Then we performed PLRP2 immunohistochemistry for establishing the intracellular site of mPlrp2 expression in the liver. As shown in supplementary Fig. II, mPlrp2 is especially expressed in portal tracts (rich HSCs) in the liver.
Fig. 7.
The mClps gene is specifically expressed in the HSC-T6 cell line. A: RT-PCR analysis of mPlrp2 and mClps mRNA in multiple cell lines. B: Total RNA from mClps knockdown HSCs was subjected to realtime RT-PCR analysis. C: Quantifi cation of retinyl esters from knockdown HSCs compared with control and scrambled shRNA. HSC-T6 cells were incubated for 12 h with palmitic acid (100 μM), after which the cells were treated with 5 μM retinol for an additional 12 h, and then shRNAs were introduced into the cells. Retinyl ester concentrations were determined 12 h later. Results represent means ± SEM from five HSC preparations. * P < 0.05; ** P < 0.01.
DISCUSSION
We are endeavoring to understand the basic biochemistry of retinoid metabolism, not only because of its fundamental scientific interest, but also for a better understanding of the role that vitamin A nutrition and retinoids play in health and disease. Under normal nutritional conditions, the main site of vitamin A storage is the liver, where over 95% of total retinoids are found as retinyl esters (29, 30). The hydrolysis of retinyl esters is especially crucial in the digestion and intestinal absorption of dietary vitamin A and in the hepatic uptake, storage, and mobilization of vitamin A. Although PTL, PLRP2, and their partner, CLPS, were found to be the main hydrolases of retinyl esters in intestinal juice (19), the enzymes physiologically involved in hepatic retinyl ester hydrolysis remain unclear. Much early work showed that retinyl palmitate hydrolyzing activities are present in rat liver homogenates and that the activity varied markedly, with over a 50-fold range among the livers of individual rats (31), raising the question of whether these REH activities detected in liver homogenates actually contributed to retinoids released in physiological livers. A few types of lipases, such as hepatic lipase, carboxylesterases, and lipoprotein lipase, are found in hepatocytes. All of these known enzymes hydrolyze a variety of substrates, including retinoids, and no enzyme has been shown to be absolutely specific for retinyl esters (11).
In our previous work, a reduction in light illumination activated the expression of the pancreas-specific lipases mPlrp2 and mClps in the liver (20), and light-modulated retinoic acid signals regulated circulatory 5′-AMP levels by modulating CD73 activity, switching on mPlrp2 and mClps gene expression in the liver (21). This study showed that temporary changes in the ratio of AdoMet to AdoHcy in specific tissues were associated with mPlrp2 and mClps expression. That correlative relationship between this ratio and mPlrp2 and mClps gene expression was supported by several findings: i) a higher ratio of AdoMet to AdoHcy in the pancreas compared with the liver; ii) an elevated hepatic ratio of AdoMet to AdoHcy in darkness or weak light illumination; and iii) an increasing ratio of AdoMet to AdoHcy after 5′-AMP administration. In these independent studies on the elevation of the ratio of AdoMet to AdoHcy, the mPlrp2 and mClps genes were found to be activated in liver tissues. This sensitive switch to both genes by temporary changes of the ratio of AdoMet to AdoHcy implied specific physiological functions. Homeostasis of free retinol in the plasma was controlled by vitamin A uptake from the diet and release of liver retinoid stores. A stable hydrolase activity in the degradation of liver retinoid stores was unreasonable for maintaining a relative constant circulatory level of retinol. The specific genes for hydrolyzing hepatic retinyl esters should be regulated by the status of the utilization of vitamin A under physiological conditions. Thus, both lipase genes mPlrp2 and mClps could be optimal candidate genes involved in hepatic hydrolysis of retinol esters.
ATRA has been shown to play a negative role in retinol metabolism in the ferret liver (32). Our current study demonstrates that in mice exposed to darkness, the ratio of AdoMet to AdoHcy decreased, inhibiting both mPlrp2 and mClps gene expression in the liver, and that ATRA feedback inhibited hepatic mPlrp2 and mClps gene expression by decreasing the ratio of AdoMet to AdoHcy. Moreover, mice fed a vitamin A-free diet also exhibited an elevated ratio of AdoMet to AdoHcy, strongly activating hepatic mPlrp2 and mClps expression. We observed an increase in the binding of trimethylated H3K4 to the promoter sequences of mPlrp2 and mClps in the liver of vitamin A-free mice. H3K4 trimethylation is a primary signal that is sufficient for initiating a gene repression pathway in vivo (33). In vitamin A-free mice, increased H3K4 trimethylation relaxed this repression of related gene transcription. Moreover, a methylase inhibitor impaired both hepatic mPlrp2 and mClps gene expression, resulting in the decrease of retinol levels in the plasma. Importantly, both of these genes were expressed specifically in an activated HSC-T6 cell line, and were only detected in portal tracts in the liver. Knockdown of the expression of these genes decreased retinol loss in vitro. Together, these observations strongly support the conclusion that hepatic mPlrp2 and mClps were the primary genes involved in the hydrolysis of hepatic retinyl esters.
With the regulation of the light-modulated retinoic acid signal, liver tissues displayed a reverse alteration in the ratio of AdoMet to AdoHcy, which was an important metabolic indicator for cellular methylation status, also known as methylation potential (27). An altered ratio of AdoMet to AdoHcy resulted in protein carboxymethylation (34, 35), inhibiting or activating the expression of specific genes. In conclusion, the results of this study demonstrate that temporary changes in the ratio of AdoMet to AdoHcy switch hepatic mPlrp2 and mClps gene expression and are associated with the hydrolysis of hepatic retinyl esters for the utilization of vitamin A in the mouse liver.
Supplementary Material
Footnotes
Abbreviations:
- AdoHcy
- S-adenosylhomocysteine
- AdoMet
- S-adenosylmethionine
- ATRA
- all-trans retinoic acid
- CD73
- ecto-5′-nucleotidase
- ChIP
- chromatin immunoprecipitation
- DD
- dark/dark
- LD
- light/dark
- HSC
- hepatic stellate cell
- LED
- light-emitting diode
- mClps
- procolipase
- mPlrp2
- pancreatic-related protein 2
- MTA
- methylthioadenosine
- PLRP2
- pancreatic lipase-related protein 2
- PTL
- pancreatic triglyceride lipase
- REH
- retinyl ester hydrolase
This work was supported by the National Science Foundation of China (30730030, 31071130) and a Nanjing University of Science & Technology fund (2010ZDJH14).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
REFERENCES
- 1.Theodosiou M., Laudet V., Schubert M. 2010. From carrot to clinic: an overview of the retinoic acid signaling pathway. Cell. Mol. Life Sci. 67: 1423–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chambon P. 2005. The nuclear receptor superfamily: a personal retrospect on the first two decades. Mol. Endocrinol. 19: 1418–1428. [DOI] [PubMed] [Google Scholar]
- 3.Buczyłko J., Saari J. C., Crouch R. K., Palczewski K. 1996. Mechanisms of opsin activation. J. Biol. Chem. 271: 20621–20630. [DOI] [PubMed] [Google Scholar]
- 4.Wald G. 1968. The molecular basis of visual excitation. Nature. 219: 800–807. [DOI] [PubMed] [Google Scholar]
- 5.Blomhoff R., Green M. H., Green J. B., Berg T., Norum K. R. 1991. Vitamin A metabolism: new perspectives on absorption, transport, and storage. Physiol. Rev. 71: 951–990. [DOI] [PubMed] [Google Scholar]
- 6.Futterman S., Andrews J. S. 1964. The composition of liver vitamin A ester and the synthesis of vitamin A ester by liver microsomes. J. Biol. Chem. 239: 4077–4080. [PubMed] [Google Scholar]
- 7.Helgerud P., Petersen L. B., Norum K. R. 1982. Acyl CoA:retinol acyltransferase in rat small intestine: its activity and some properties of the enzymic reaction. J. Lipid Res. 23: 609–618. [PubMed] [Google Scholar]
- 8.MacDonald P. N., Ong D. E. 1988. Evidence for a lecithin-retinol acyltransferase activity in the rat small intestine. J. Biol. Chem. 263: 12478–12482. [PubMed] [Google Scholar]
- 9.Liu L., Gudas L. J. 2005. Disruption of the lecithin:retinol acyltransferase gene makes mice more susceptible to vitamin A deficiency. J. Biol. Chem. 280: 40226–40234. [DOI] [PubMed] [Google Scholar]
- 10.Blomhoff R., Norum K. R., Berg T. 1985. Hepatic uptake of [3H]retinol bound to the serum retinol binding protein involves both parenchymal and perisinusoidal stellate cells. J. Biol. Chem. 260: 13571–13575. [PubMed] [Google Scholar]
- 11.Harrison E. H. 1998. Lipases and carboxylesterases: possible roles in the hepatic metabolism of retinol. Annu. Rev. Nutr. 18: 259–276. [DOI] [PubMed] [Google Scholar]
- 12.Mello T., Nakatsuka A., Fears S., Davis W., Tsukamoto H., Bosron W. F., Sanghani S. P. 2008. Expression of carboxylesterase and lipase genes in rat liver cell-types. Biochem. Biophys. Res. Commun. 374: 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schindler R., Mentlein R., Feldheim W. 1998. Purification and characterization of retinyl ester hydrolase as a member of the non-specific carboxylesterase supergene family. Eur. J. Biochem. 251: 863–873. [DOI] [PubMed] [Google Scholar]
- 14.Weng W., Li L., van Bennekum A. M., Potter S. H., Harrison E. H., Blaner W. S., Breslow J. L., Fisher E. A. 1999. Intestinal absorption of dietary cholesteryl ester is decreased but retinyl ester absorption is normal in carboxyl ester lipase knockout mice. Biochemistry. 38: 4143–4149. [DOI] [PubMed] [Google Scholar]
- 15.Thirstrup K., Verger R., Carriere F. 1994. Evidence for a pancreatic lipase subfamily with new kinetic properties. Biochemistry. 33: 2748–2756. [DOI] [PubMed] [Google Scholar]
- 16.Jennens M. L., Lowe M. E. 1995. Rat GP-3 is a pancreatic lipase with kinetic properties that differ from colipase-dependent pancreatic lipase. J. Lipid Res. 36: 2374–2381. [PubMed] [Google Scholar]
- 17.Blaner W. S., Obunike J. C., Kulandsky S. B., Al-Haideri M., Piantedosi R., Deck-elbaum R. M., Goldberg I. J. 1994. Lipoprotein lipase hydrolysis of retinyl ester. Possible implications for retinoid uptake by cells. J. Biol. Chem. 269: 16559–16565. [PubMed] [Google Scholar]
- 18.D'Agostino D., Lowe M. E. 2004. Pancreatic lipase-related protein 2 is the major colipase-dependent pancreatic lipase in suckling mice. J. Nutr. 134: 132–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reboul E., Berton A., Moussa M., Kreuzer C., Crenon I., Borel P. 2006. Pancreatic lipase and pancreatic lipase-related protein 2, but not pancreatic lipase-related protein 1, hydrolyze retinyl palmitate in physiological conditions. Biochim. Biophys. Acta. 1761: 4–10. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J., Kaasik K., Blackburn M. R., Lee C. C. 2006. Constant darkness is a circadian metabolic signal in mammals. Nature. 439: 340–343. [DOI] [PubMed] [Google Scholar]
- 21.Pang W., Li C., Zhao Y., Wang S., Dong W., Jiang P., Zhang J. 2008. The environmental light influences the circulatory levels of retinoic acid and associates with hepatic lipid metabolism. Endocrinology. 149: 6336–6342. [DOI] [PubMed] [Google Scholar]
- 22.She Q. B., Nagao I., Hayakawa T., Tsuge H. 1994. A simple HPLC method for the determination of S-adenosylmethionine and S-adenosylhomocysteine in rat tissues: the effect of vitamin B6 deficiency on these concentrations in rat liver. Biochem. Biophys. Res. Commun. 205: 1748–1754. [DOI] [PubMed] [Google Scholar]
- 23.Teerlink T., Copper M. P., Klaassen I., Braakhuis B. J. 1997. Simultaneous analysis of retinol, all-trans- and 13-cis-retinoic acid and 13-cis-4-oxoretinoic acid in plasma by liquid chromatography using on-column concentration after single-phase fluid extraction. J. Chromatogr. B Biomed. Sci. Appl. 694: 83–92. [DOI] [PubMed] [Google Scholar]
- 24.Braunstein M., Rose A. B., Holmes S. G., Allis C. D., Broach J. R. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7: 592–604. [DOI] [PubMed] [Google Scholar]
- 25.Spencer V. A., Sun J. M., Li L., Davie J. R. 2003. Chromatin immunoprecipitation: a tool for studying histone acetylation and transcription factor binding. Methods. 31: 67–75. [DOI] [PubMed] [Google Scholar]
- 26.Lowe M. E. 1994. Pancreatic triglyceride lipase and colipase: insights into dietary fat digestion. Gastroenterology. 107: 1524–1536. [DOI] [PubMed] [Google Scholar]
- 27.Hermes M., Osswald H., Kloor D. 2007. Adenosine metabolism and its effect on methylation potential in cultured cells: methodological considerations. Cell Mol. Biol. (Noisy-le-grand). 52 (Suppl.): OL874–OL881. [PubMed] [Google Scholar]
- 28.Hoffman D. R., Cornatzer W. E., Duerre J. A. 1979. Relationship between tissue levels of S-adenosylmethionine, S-adenylhomocysteine, and transmethylation reactions. Can. J. Biochem. 57: 56–65. [DOI] [PubMed] [Google Scholar]
- 29.Linder M. C., Anderson G. H., Ascarelli I. 1971. Quantitative distribution of vitamin A in Kupffer cell and hepatocyte populations of rat liver. J. Biol. Chem. 246: 5538–5540. [PubMed] [Google Scholar]
- 30.Harrison E. H., Blaner W. S., Goodman D. S., Ross A. C. 1987. Subcellular localization of retinoids, retinoid-binding proteins, and acyl-CoA:retinol acyltransferase in rat liver. J. Lipid Res. 28: 973–981. [PubMed] [Google Scholar]
- 31.Blaner W. S., Prystowsky J. H., Smith J. E., Goodman D. S. 1984. Rat liver retinyl palmitate hydrolase activity. Relationship to cholesteryl oleate and triolein hydrolase activities. Biochim. Biophys. Acta. 794: 419–427. [DOI] [PubMed] [Google Scholar]
- 32.Wang X. D., Krinsky N. I., Russell R. M. 1993. Retinoic acid regulates retinol metabolism via feedback inhibition of retinol oxidation and stimulation of retinol esterification in ferret liver. J. Nutr. 123: 1277–1285. [DOI] [PubMed] [Google Scholar]
- 33.Santos-Rosa H., Schneider R., Bannister A. J., Sherriff J., Bernstein B. E., Emre N. C., Schreiber S. L., Mellor J., Kouzarides T. 2002. Active genes are tri-methylated at K4 of histone H3. Nature. 419: 407–411. [DOI] [PubMed] [Google Scholar]
- 34.Backlund P. S., Jr, Carotti D., Cantoni G. L. 1986. Effects of the S-adenosylhomocysteine hydrolase inhibitors 3-deazaadenosine and 3-deazaaristeromycin on RNA methylation and synthesis. Eur. J. Biochem. 160: 245–251. [DOI] [PubMed] [Google Scholar]
- 35.Bartel R. L., Borchardt R. T. 1984. Effects of adenosine dialdehyde on S-adenosylhomocysteine hydrolase and S-adenosylmethionine-dependent transmethylations in mouse L929 cells. Mol. Pharmacol. 25: 418–424. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.