Abstract
The objective of this study was to determine whether 5-lipoxygenase (ALOX5) gene variants associated with cardiovascular disease affect eicosanoid production by monocytes. The study was a randomized, double-masked, parallel intervention trial with fish oil (5.0 g of fish oil daily, containing 2.0 g of eicosapentaenoic acid [EPA] and 1.0 g of docosahexaenoic acid [DHA]) or placebo oil (5.0 g of corn/soy mixture). A total of 116 subjects (68% female, 20–59 years old) of African American ancestry enrolled, and 98 subjects completed the study. Neither ALOX5 protein nor arachidonic acid-derived LTB4, LTD4, and LTE4 varied by genotype, but 5-hydroxyeicosatetraenoate (5-HETE), 6-trans-LTB4, 5-oxo-ETE, 15-HETE, and 5,15-diHETE levels were higher in subjects homozygous for the ALOX5 promoter allele containing five Sp1 element tandem repeats (“55” genotype) than in subjects with one deletion (d) (three or four repeats) and one common (“d5” genotype) allele or with two deletion (“dd”) alleles. The EPA-derived metabolites 5-HEPE and 15-HEPE and the DHA-derived metabolite 17-HDoHE had similar associations with genotype and increased with supplementation; 5-HEPE and 15-HEPE increased, and 5-oxo-ETE decreased to a greater degree in the 55 than in the other genotypes. This differential eicosanoid response is consistent with the previously observed interaction of these variants with dietary intake of omega-3 fatty acids in predicting cardiovascular disease risk.
Keywords: arachidonic acid, eicosanoids, leukotrienes, lipoxygenase, nutrition, omega-3 fatty acids
Eicosanoids, including leukotrienes (LT), are products of arachidonic acid (AA) metabolism that mediate inflammation during infectious, allergic, and other inflammatory diseases (1), including cardiovascular disease (CVD) (2). The ALOX5 gene encodes the 5-lipoxygenase (5-LO) enzyme, which acts with 5-lipoxygenase activating protein (FLAP) to catalyze the first two steps in LT synthesis, the production of 5-hydroperoxyeicosatetraenoate (5-HpETE), followed by its conversion to LTA4 or its glutathione peroxidase-dependent conversion to 5-hydroxyeicosatetraenoate (5-HETE) (3). 5-HETE is a bioactive semistable metabolite that reversibly forms a lactone (5-hydroxy-6δ-lactone) (4, 5) and is also converted by 5-hydroxyeicosanoid dehydrogenase (5-HEDH) to the active metabolite 5-oxo-ETE (6). LTA4 is unstable (7) and is metabolized by LTA4 hydrolase (LTA4H) to LTB4 (8) or by LTC4 synthase (LTC4S) to LTC4 (9), which is a precursor to the more stable metabolites LTD4 and LTE4 (10). These eicosanoids are produced by inflammatory cells such as macrophages and granulocytes, where they promote inflammation by binding to specific receptors on target cells (3, 11).
Inflammatory processes mediated by eicosanoid-producing macrophages in the vascular wall play important roles in the development of arterial plaque and the pathogenesis of CVD (12, 13). Consistent with this notion, variations in genes of the LT biosynthetic pathway have been associated with increased CVD risk (14–18), although some studies have reported equivocal results or no association (19–21). The promoter region of the ALOX5 gene has between three and eight tandem repeats of a consensus binding site for the transcription factors Sp1 and Egr1, with the most frequent allele having five such repeats (22). Variant alleles with three or four sites (or, rarely, more than five sites) are associated with greater intima-medial thickness of the carotid artery in healthy adults from Los Angeles, CA (23), and with occurrence of a first myocardial infarction in a case-control study of Costa Ricans (24). In both of those studies, the observed diet–gene interactions were more pronounced in subjects with high dietary AA intake, who carried the deletion (d) variants (i.e., the “3” or “4” type repeats, referred to jointly as “d” alleles), than in subjects with two common alleles (i.e., the “55” type genotype [containing five Sp1 element tandem repeats]). Conversely, in the Los Angeles study (23), this atherogenic effect was reversed in the subjects with d alleles who had high eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) intake. However, two subsequent studies, a case-control study of myocardial infarction in the United Kingdom (21) and a case-control study of CVD in California (25), did not report associations with ALOX5 promoter polymorphism, although they did not perform gene–diet interaction analyses.
The mechanism underlying this apparent difference in risk with ALOX5 promoter variants is not known. We propose that subjects with d alleles have greater cardiovascular risk because they have greater transcription from the ALOX5 gene due to lower levels of methylation of GC-rich promoter elements. In support of this hypothesis, our group has recently demonstrated higher ALOX5 mRNA in peripheral blood lymphocytes from healthy adults with one or two d alleles (the “dd” or “d5” genotype) versus two common alleles (the 55 genotype) but not in granulocytes or monocytes from the same subjects (26). Greater methylation was seen in subjects with the 3 versus the 4 allele. However, the opposite association has been reported in peripheral blood eosinophils isolated from symptomatic asthmatics, in whom higher levels of ALOX5 mRNA and LTC4 production were seen in the 55 genotype than in subjects with two non-5 alleles, who had primarily the dd genotype (27). Thus, the biology underlying the association of these genotypes with more severe CVD remains to be defined. This association may be primarily related to ALOX5 expression in monocytes rather than in other immune cell types, given the important role that macrophages (derived from monocytes) play in the pathogenesis of CVD (12, 13).
The apparent diet–gene interaction of ALOX5 variants with CVD risk (23, 24) makes intuitive sense in that intake of long-chain omega-3 fatty acids, particularly DHA and EPA, is thought to decrease risk of CVD, at least in part, by antagonizing the production of AA-derived proinflammatory eicosanoids (28). In addition to that, competition between EPA and AA as the substrate for ALOX5, which can reduce production of AA-derived 4-series LTs, the resulting EPA-derived LTs (e.g., LTB5) are significantly less inflammatory than the corresponding AA-derived molecules (29). We hypothesize that this anti-inflammatory effect of EPA would be of greater benefit in subjects with higher intrinsic production of proinflammatory eicosanoids, as may occur for subjects with the dd or d5 ALOX5 promoter genotype.
Thus, the aim of the present study was to conduct a placebo-controlled intervention trial of fish oil supplementation, randomized within six ALOX5 promoter genotypes, to determine whether monocytes from dd and d5 subjects would have higher ALOX5 protein and AA-derived, proinflammatory eicosanoid metabolite production than monocytes from the 55 genotype control subjects. A second goal was to determine whether fish oil supplements would cause a greater reduction in AA-derived eicosanoids in dd or d5 subjects than in 55 control subjects.
SUBJECTS AND METHODS
Study design
Subject recruiting and genotype analysis.
Healthy adults 20–59 years old who self-identified as African American, black, or of African ancestry were recruited into the study from three study sites: Davis, Sacramento, and Oakland, CA. Recruitment of study participants occurred at health fairs, community meetings, churches, and other public venues from May 2005 to March 2007. Potential study participants met with a study recruiter and completed a screening questionnaire at initial contact or via telephone in response to advertising. A buccal swab (DNA swab pack, SK-1; Isohelix, Kent, UK) was then taken for DNA isolation, and genotype analysis was performed as described previously (26). After genotyping, eligible subjects were contacted to schedule an appointment for a screening blood draw to be used for a complete blood count, lipid panel, and chemistry panel examination. Potential subjects were then invited for the baseline (week 0) blood draw, at which time they were randomized to treatment.
Ethical review and trial registration.
The institutional review boards of the University of California Davis and Alta Bates Summit Medical Center reviewed and approved this protocol. Written informed consent was obtained from all study participants.
Inclusion and exclusion criteria.
Eligible subjects had one of the six ALOX5 genotype groups of interest to the study. Potential subjects who reported a physician-diagnosed chronic inflammation-related disease, including CVD, hypertension, diabetes, or a lipid disorder that required regular use of anti-inflammatory or lipid-lowering medication, were excluded. Subjects with an abnormal result on a standard chemistry panel, lipid panel, or complete blood count that suggested underlying disease were also excluded and referred to their physicians for further evaluation. Other exclusions are listed in the supplementary online material.
Randomization to treatment groups within genotypes.
Subjects within each of the six ALOX5 genotypes (dd genotypes “33,” “34,” and “44”; d5 genotypes “35” and “45”; and control genotype 55) were allocated to receive treatment or placebo, using six randomization lists and a randomized block design with a block size of two.
Fish oil and placebo treatments.
Fish oil concentrate and placebo capsules (1.0 g/capsule) were provided in bulk by Ocean Nutrition Canada (Dartmouth, Nova Scotia, Canada). The fish oil was 40/20 ethyl ester (lot nos. 10524 and 8980), and the placebo was corn/soybean placebo (lot nos. 10525 and 8981). Additional information is provided in the supplementary online material.
Sample size.
Our sample size was based on our principal outcome, to detect differences in ALOX5 mRNA expression levels at baseline between variant and control (55) genotypes, and on expected reductions in ALOX5-derived LT metabolites. Our goal was to recruit 30 subjects in order to retain 24 in all genotype groups, except for the homozygous 44 genotype because of the lower prevalence of that genotype (23), which had a frequency of only 2.5% (9/354 subjects) in this population.
Laboratory methods
Blood draws.
Blood (80 ml) was drawn from the antecubital vein into sodium-heparin tubes at week 0 (baseline) and at week 6 (final or visit 2) after an overnight fast of 12 h duration. The blood samples from all sites were processed at the Western Human Nutrition Research Center in Davis. Blood was mixed well and stored at room temperature until it was processed within 4 h of collection.
Plasma preparation and monocyte isolation.
Blood was centrifuged at 1,500 g for 10 min at 25°C. Plasma was pooled from all tubes for each subject and centrifuged at 2,500 g for 20 min at 20°C to remove platelets. Plasma was stored at −80°C. The buffy coat was removed and used for monocyte isolation. Ficoll step gradients were used to separate the peripheral blood mononuclear cells (PBMCs) and granulocytes (using Histopaque 1119 and 1077; Sigma, St. Louis, MO). Monocytes were separated from the PBMCs by positive selection using anti-CD14 antibody on magnetic beads (Miltenyi Biotec, Auburn, CA). Purity of monocytes was assessed by flow cytometry analysis using phycoerythrin-labeled anti-CD14 (Miltenyi) and isotype control antibody and by FACSCalibur flow cytometry (Beckton Dickenson, San Jose, CA). The lowest measured percent purity was 50%, with only 9% being <80% pure. The mean (±standard deviation [SD]) percent purity was 90.9 ± 8.4%. Percent purity did not correlate with ALOX5 metabolite concentrations (data not shown). Viability was assessed by Trypan blue exclusion and always exceeded 84%. The mean viability was 97.4 ± 2.6%.
Monocyte cultures.
The calcium ionophore A23187 is commonly used to stimulate 5-LO activity in monocytes and granulocytes (30, 31) and was found to produce an equivalent response to ionomycin in preliminary studies (data not shown). Purified monocytes (1 × 106 cells/ml) were cultured at 37°C in 5% CO2 using RPM1 1640 complete medium supplemented with 10% heat-inactivated autologous plasma. Supernatant and cell pellets were collected at 15, 30, and 60 min following stimulation with the calcium ionophore A23187 (Sigma, St. Louis, MO) diluted in cell culture-grade dimethyl sulfoxide (DMSO; Sigma, St. Louis, MO) at a final concentration of 10 μM (0.1% DMSO) in the cell culture medium. A 60 min DMSO control culture was also included. Preliminary analysis of all time points from a subset of subjects revealed that the 60 min A23187-stimulated culture induced the highest production of ALOX5 metabolites. Therefore, the 60 min A23187 and DMSO time points were analyzed for the remainder of the subjects.
Oxylipid analysis.
Cell culture supernatants (0.2 ml) were spiked with 5 µl of antioxidant solution (0.2 mg/ml BHT/EDTA solution in a methanol:water ratio of 1:1) and a suite of deuterated oxylipid surrogates including d4-prostaglandin E2, d4-LTB4, and d4-5-HETE (Cayman Chemical, Ann Arbor, MI). Supernatants were then extracted using 60 mg of OasisTM HLB matrix in solid phase extraction cartridges (Waters Corp., Milford, MA). Analytes were eluted from the solid phase extraction cartridges with 0.5 ml of methanol followed by 2.0 ml of ethyl acetate, into tubes containing 2 µl of glycerol. Samples were brought to near dryness under a vacuum and reconstituted with 100 µl of an internal standard solution consisting of 400 nM cyclohexyl-urido-dodecanoic acid (Cayman Chemical, Ann Arbor, MI) in methanol. Analytes were chromatographically separated using an ultra-performance liquid chromatography system equipped with a 2.1 × 150 mm Acquity BEH C18 reversed-phase column and quantified by negative mode electrospray ionization with a Quattro Micro tandem mass spectrometer run in multireaction monitoring mode (Waters Corp.). Oxylipids and deuterated surrogates were quantified using internal standard ratio response methodologies measured against a minimum 5-point calibration curve bracketing all reported concentrations. Samples (1 in 20) were analyzed in replicate to assess analytical precision.
Genotyping.
Genotyping of the ALOX5 promoter repeat polymorphism was performed as described previously (26).
RNA analysis.
Total RNA was isolated from purified monocytes prior to culturing for gene expression analysis as described previously (26). Briefly, total RNA was isolated and reverse transcribed into cDNA. Real-time PCR was carried out in triplicate, and expression levels were normalized to that of β-glucuronidase as an endogenous control. Data are presented as relative units (ru).
Western blots.
Cells to be used for Western blot analysis were frozen in whole-cell lysis buffer and processed at a later time (QIAshredder kit; Qiagen, Valencia, CA). Western blotting was performed, and bands for 5-LO and β-actin proteins were quantified using standard methods, as described in the online supplementary materials.
Ancestry informative markers.
Using genomic DNA purified from PBMC as template, a set of 93 validated single-nucleotide polymorphism ancestry informative markers (AIMs) was analyzed as described previously (32). Ancestry information was determined using the Bayesian clustering algorithm performed by Structure version 2.3.3 software (33). The analysis was performed using European (EUR), sub-Saharan African (AFR), East Asian (EAS), and Amerindian (AMI) reference population samples under the conditions previously described (26).
Statistical analysis
Descriptive analysis.
Western blot data were available for 95 subjects at baseline to analyze genotype effects and for 68 subjects at both baseline and 6 weeks to analyze treatment effects. RNA data were available for 83 subjects at baseline and 6 weeks to analyze treatment effects. ALOX5 protein data were normally distributed. RNA data were normally distributed (LTA4H) or were normalized by log10 (FLAP) or square root (ALOX5) transformation.
Oxylipid data for 114 paired samples (A23187 and DMSO cultures at 60 min) were available for analysis of the effect of A23187 on oxylipid production and from 115 A23187-stimulated cultures for analysis of genotype effects at baseline. Oxylipid data for 97 subjects were available at baseline and 6 weeks to analyze treatment effects. At baseline, zero values were not seen in either the DMSO or the A23187 supernatants for 5-HETE, 5-HEPE, 15-HEPE, 5-oxo-ETE, and 6-trans-LTB4 in either culture (n = 228 samples) and were uncommon for 15-HETE (n = 1), 9-HETE (n = 4), 6-trans-LTB4 (n = 4), and LTB4 (n = 3) but were more common for LTD4 (n = 37), LTE4 (n = 54), and LTB5 (n = 67), particularly in the DMSO cultures. To allow transformations when zero values were present, a small positive value approximately equal to the lowest detected value was added to all values for a particular metabolite. The AA metabolite 9-HETE is used as a marker of generalized lipid peroxidation, as indicated by its appearance following myeloperoxidase-mediated hypochlorous acid production (34). Correlation plots of each metabolite concentration to 9-HETE concentration were individually examined to visually identify high outliers that could have skewed the analysis by increasing variance. Such exclusions were made for LTB4 (n = 1), LTD4 (n = 3), and LTE4 (n = 2) during a comparison of the A23187 to DMSO cultures. In such cases, the same statistical analysis was also performed using rank data to confirm the possibility that exclusion of these outliers did not unduly affect the results of the statistical analysis. For A23187 cultures, only one sample was considered an outlier (genotype 33, placebo), and this value was excluded only for LTB4, LTD4, and LTE4 analysis. These metabolites were all quantified relative to the same deuterated surrogate (d4-LTB4), and an unusually low recovery of the surrogate in this sample was noted, suggesting a laboratory error.
Statistical methods.
ALOX5 protein and metabolite data were compared among genotypes at baseline by one-factor ANOVA. Data that were not normally distributed were analyzed using values from the best transformation (i.e., either log10 or square root) to most closely approximate a normal distribution. Untransformed data are presented in tables and graphs. When the overall P value was <0.05, post hoc multiple comparisons versus the 55 control group were made using the Holm-Sidak method when comparing all subjects (dd, d5, and 55 genotypes), subjects with 3 and 5 alleles only (genotypes 33, 35, and 55), and subjects with 4 and 5 alleles only (genotypes 44, 45, and 55). When subjects with only 3 and 4 alleles were examined (genotypes 33, 34, and 44), all pair-wise comparisons were made. Two group comparisons (35 vs. 45) were made using the Student t-test.
The effect of the fish oil intervention on RNA, protein, and oxylipids at baseline and the end of the study was analyzed using two-way repeated measures ANOVA. The interaction of genotype with intervention was analyzed using two-factor ANOVA to compare the differences in oxylipid concentrations between baseline and the end of the study by treatment group and genotype. Differences were often not normally distributed, and the same approach to normalization was used as described above for baseline oxylipid concentrations. No outliers were identified in oxylipid data at the second visit. Statistical analysis was performed using SigmaStat version 3.10 software (Systat, San Jose, CA).
RESULTS
Baseline data
Subject characteristics.
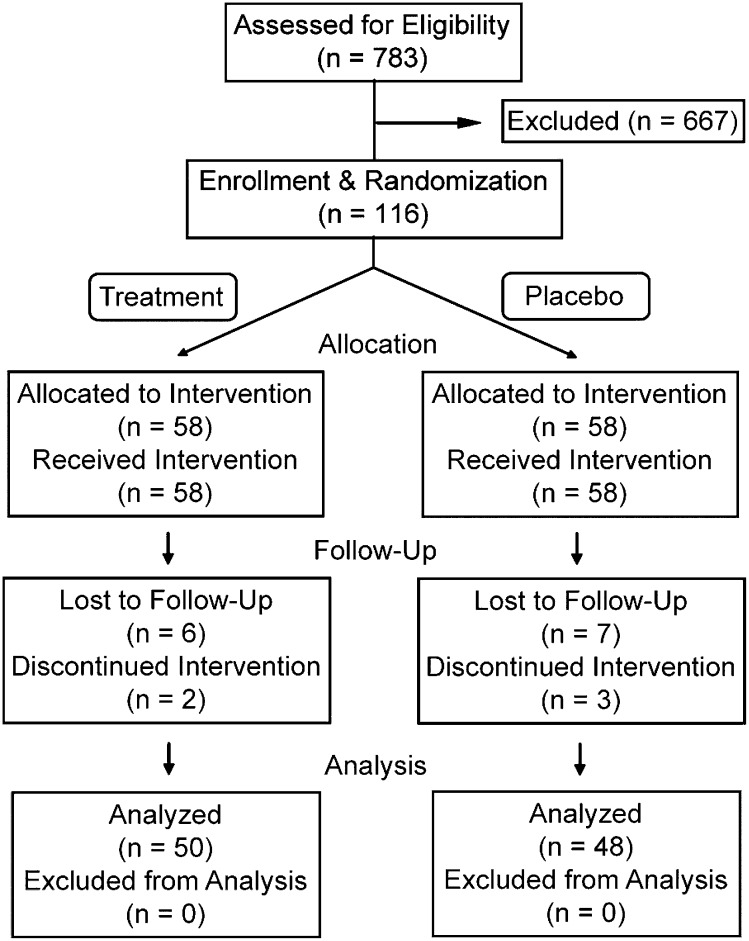
A total of 783 individuals were screened for entry into the study (Fig. 1). Of these, 116 subjects were entered and randomized among the six ALOX5 genotype groups to receive either fish oil or placebo treatment. Subjects who were entered into the study were younger and had a lower mean body mass index (BMI) than subjects who were screened but not enrolled (see supplementary Table I). Of the 116 subjects who entered the study, 98 subjects completed the 6 week intervention (Fig. 1 and Table 1). Baseline characteristics for the 116 enrolled subjects were compared by treatment and genotype, using the six genotypes used for randomization as well as the three pooled genotype groups (dd, d5, and 55 [Table 1). Of the total, 68% (79/116) of subjects were female, the median age (25th/75th percentiles) was 35.4 years old (25.0/46.5 years, respectively), and the mean BMI (±SD) was 27.6 ± 4.6 kg/m2. Significant differences were not seen among the subjects grouped by sex or genotype, with the exception of a modest difference in BMI among the six genotype groups, as described in online supplementary materials. The 18 subjects who did not complete the study did not differ from the 98 subjects who completed the study, with regard to age, sex, and BMI (see supplementary Tables II and III). However, the genotype distribution did differ between these groups: the noncompletion rate was 27% (9/33) for the dd group, 8% (4/53) for the d5 group, and 17% (5/30) for the 55 group (P = 0.048, chi-square test).
Fig. 1.
A consort diagram describes subject recruitment and retention. Of the 667 subjects excluded, 338 subjects were excluded based on criteria evaluated during initial screening; 28 subjects were excluded due to ineligible genotype; 100 subjects with eligible genotypes were excluded because these genotype groups were full; 38 subjects were excluded with abnormal blood test results at a second screening step; and 113 subjects declined to participate. Subjects were randomized within six ALOX5 genotypes, as shown in Table 1.
TABLE 1.
Allocation of subjects to treatment by ALOX5 genotype
| ALOX5 Sp1 promoter variant genotypea |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| dd |
d5 |
||||||||
| Subject group | 33 | 34 | 44 | Subtotal | 35 | 45 | Subtotal | 55 | All subjects |
| Fish oil groupb | |||||||||
| Allocated to treatment | 7 | 7 | 2 | 16 | 15 | 11 | 26 | 16 | 58 |
| Discontinued treatmentc | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 2 |
| Lost to follow-up | 2 | 0 | 0 | 2 | 0 | 1 | 1 | 3 | 6 |
| Completed study | 4 | 7 | 2 | 13 | 14 | 10 | 24 | 13 | 50 |
| Placebo groupb | |||||||||
| Allocated to treatment | 7 | 8 | 2 | 17 | 15 | 12 | 27 | 14 | 58 |
| Discontinued treatment | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 3 |
| Lost to follow-up | 0 | 4 | 0 | 4 | 2 | 0 | 2 | 1 | 7 |
| Completed study | 5 | 4 | 2 | 11 | 13 | 12 | 25 | 12 | 48 |
| All subjects | |||||||||
| Allocated to treatment | 14 | 15 | 4 | 33 | 30 | 23 | 53 | 30 | 116 |
| Discontinued treatment | 3 | 0 | 0 | 3 | 1 | 0 | 1 | 1 | 5 |
| Lost to follow-up | 2 | 4 | 0 | 6 | 2 | 1 | 3 | 4 | 13 |
| Completed study | 9 | 11 | 4 | 24 | 27 | 22 | 49 | 25 | 98 |
These genotypes indicate the number of tandem Sp1 binding-sites (3, 4, or 5) in ALOX5 promoter regions 33, 34, 44, 35, 45, and 55. Some analyses were done using subjects homozygous for deletion alleles (dd genotypes 33, 34, and 44), subjects heterozygous for deletion alleles (d5 genotypes 35 and 45), and subjects homozygous for the most common allele (genotype 55).
Subjects were randomly allocated to the fish oil or placebo treatment group (Fig. 1) within six genotype groups.
Reasons for discontinuing treatment in the fish oil group included difficulty with blood draw (genotype 33) and personal reasons not related to the study (genotype 35), whereas reasons for discontinuing treatment in the placebo group included pregnancy (genotype 33), acne exacerbation (genotype 55), and personal reasons not related to the study (genotype 33).
Ancestry informative markers.
Since the frequency of the ALOX5 promoter deletion alleles varies by ancestry group (25) AIMs were used to determine if the percentage of AFR ancestry varied by ALOX5 genotype. The overall AFR mean for all subjects with available DNA (n = 114) was 72.7% ± 16.0%. No significant differences were seen among the six ALOX5 genotype groups (P = 0.14), but means differed among the three ALOX5 genotype groups (P = 0.020). The AFR means for the dd, d5, and 55 groups were 75.7% ± 15.0%, 74.7% ± 15.2%, and 65.6% ± 16.7%, respectively. Both the dd and d5 groups differed from the 55 group (P < 0.05, adjusting for multiple comparisons) but not from one another. The overall mean for EUR ancestry was 24.2% ± 14.7% for all subjects. Significant differences were seen among the six ALOX5 genotype groups (P = 0.038), although no groups differed from one another when all multiple comparisons were used. EUR means differed among the three ALOX genotype groups (P = 0.003). The EUR means for the dd, d5, and 55 groups were 20.1% ± 11.1%, 22.5% ± 14.8%, and 31.9% ± 15.7%, respectively. Both the dd and the d5 means differed from those of the 55 group (P < 0.05, adjusting for multiple comparisons) but not from one another. The percentages of AMI and EAS markers were low and did not differ by genotype. The AMI median (25th/75th percentiles) for all subjects was 1.0% (0.5%/2.2%, respectively), and the EAS median was 0.5% (0.3%/1.0%, respectively).
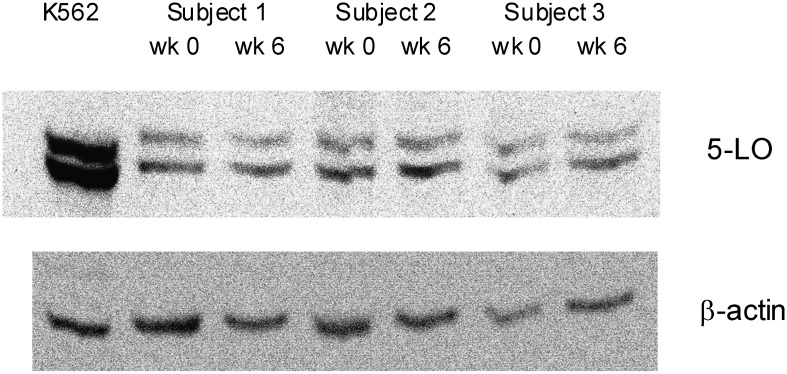
ALOX5 protein.
We measured ALOX5 protein levels by Western blot analysis of unstimulated monocytes. Two bands corresponding to ALOX5 protein were identified in all samples and were quantified relative to β-actin expression (Fig. 2). The different mobility of the two bands presumably results from phosphorylation (22). Protein expression levels at baseline representing the sum of the two band densities (mean ratio ± SD to β-actin) in the dd, d5, and 55 genotype groups were 1.54 ± 0.38 (n = 25), 1.63 ± 0.71 (n = 45), and 1.38 ± 0.39 (n = 25), respectively, and were not significantly different (P = 0.23, by one-factor ANOVA). Differences were not seen by ANOVA for either ALOX5 band independently, for the ratio between the two bands, or when ALOX5 band densities were analyzed without adjustment for β-actin, and neither were significant differences seen when protein levels were compared among all six genotypes defined using the 3 and 4 allele groups separately (data not shown).
Fig. 2.
Western blot analyses of arachidonate 5-LO protein and β-actin are shown. These representative blots show K562 control cells and monocyte samples (0.5 × 106 cells per lane) from three subjects at baseline (week 0) and follow-up (week 6). The 5-LO bands are approximately 80 and 78 kDa.
Metabolite profiles.
We next determined the metabolite profiles in supernatants of monocyte cultures by quantifying three AA- and EPA-derived metabolites produced directly by ALOX5/FLAP activity: 5-HETE, 5-HEPE (from EPA), and 6-trans-LTB4. In addition, we measured 5-oxo-ETE (which additionally requires 5-HEDH activity), LTB4 and LTB5 (LTB5 is EPA-derived; LTB4 and LTB5 production additionally requires LTA4H activity), and LTC4 and LTD4 (which additionally require LTC4S activity). The LTB4 degradation products 20-hydroxy LTB4 and 20-carboxy LTB4 were also measured and found at trace levels in all analyzed samples. The 15-lipoxygenase (15-LO) metabolites 15-HETE, 15-HEPE (EPA-derived), lipoxin A4 (LXA4), 8,15-diHETE, and 5,15-diHETE also were measured (35), as were 17-hydroxy docosahexaenoate (17-HDoHE), an intermediate in the synthesis of anti-inflammatory, DHA-derived resolvins (36), and 9-HETE, generally considered a nonenzymatic peroxidation product (37). Concentrations of most ALOX5 metabolites increased significantly in response to A23187 stimulation (Table 2). The greatest fold increases were seen for LTB4, 6-trans-LTB4, and LTE4. Smaller increases were seen for 15-LO metabolites and also for the nonenzymatic metabolite 9-HETE, indicating generally increased oxidative metabolism in these monocytes following A23187 stimulation. Relatively high levels of some metabolites (e.g., 5-HETE) in the control cultures suggest that the process of isolating and culturing the monocytes (e.g., positive selection, plastic adherence) caused some activation of these cells.
TABLE 2.
Oxylipid concentrations from A23187 and DMSO treatments at baseline
| Metabolite | Mean A23187 concentration ± SEM (nmol/l) | Mean DMSO ± SEM (nmol/l) | Mean difference ± SEM (nmol/l) | Fold changea | P valueb |
|---|---|---|---|---|---|
| Group 1c | |||||
| LTB4d | 6.52 ± 0.34 | 0.71 ± 0.12 | 5.81 ± 0.38 | 14.80 | <0.001 |
| LTB5d | 0.21 ± 0.06 | 0.24 ± 0.04 | −0.02 ± 0.06 | 0.85 | 0.18 |
| LTD4 | 1.30 ± 0.09 | 0.85 ± 0.07 | 0.46 ± 0.11 | 1.42 | <0.001 |
| LTE4d | 3.75 ± 0.36 | 1.42 ± 0.19 | 2.26 ± 0.42 | 2.82 | <0.001 |
| Group 2 | |||||
| 6-trans-LTB4 | 11.8 ± 1.40 | 5.90 ± 0.87 | 5.88 ± 1.05 | 2.28 | <0.001 |
| 5-oxo-ETE | 2.07 ± 0.20 | 1.69 ± 0.19 | 0.38 ± 0.22 | 1.34 | 0.004 |
| 5-HETE | 397 ± 28.8 | 286 ± 24.2 | 111 ± 20.8 | 1.57 | <0.001 |
| 5-HEPE | 25.9 ± 2.63 | 18.8 ± 1.83 | 7.07 ± 1.55 | 1.36 | <0.001 |
| 15-HETE | 10.98 ± 1.08 | 9.25 ± 0.78 | 1.73 ± 0.74 | 1.21 | <0.001 |
| 15-HEPE | 1.41 ± 0.13 | 1.56 ± 0.21 | −0.15 ± 0.23 | 1.14 | 0.16 |
| 5,15-DiHETE | 4.00 ± 0.50 | 2.86 ± 0.40 | 1.14 ± 0.35 | 1.20 | <0.001 |
| 9-HETE | 11.0 ± 1.17 | 9.37 ± 0.92 | 1.62 ± 0.76 | 1.17 | <0.001 |
| Lipoxin A4 | 1.75 ± 0.14 | 1.30 ± 0.10 | 0.46 ± 0.13 | 1.38 | 0.001 |
| 17-HDoHE | 6.16 ± 0.79 | 4.47 ± 0.48 | 1.70 ± 0.56 | 1.33 | <0.001 |
| Group 3 | |||||
| 8,15-DiHETE | 0.84 ± 0.12 | 0.66 ± 0.07 | 0.17 ± 0.14 | 1.06 | 0.27 |
Data show mean ± SEM (nmol/l) results from paired monocyte culture supernatants treated with 10 μM A23187 and 0.1% DMSO from 114 subjects at baseline.
Fold change indicates the median A23187:DMSO ratio. Paired samples with zero values for either treatment were excluded from the ratio calculation.
P values indicate difference between A23187 and DMSO by paired Wilcoxon signed-rank test or t-test.
Groups 1, 2, and 3 are based on correlations among metabolites as seen in Table 3.
N = 113 subjects for LTB4, 111 subjects for LTB5, and 112 subjects for LTE4, as described in Subjects and Methods.
Correlation among metabolites.
We next constructed a correlation matrix using the oxylipid data from the A23187-stimulated supernatants to determine if metabolite concentrations from the same or closely linked enzymes were correlated (Table 3). Modest but statistically significant correlations were seen between LTB4 and two cysteinyl LTs, LTC4 and LTD4. The synthesis of these metabolites, referred to here as group 1, requires both ALOX5 and FLAP activity as well as that of LTA4H for LTB4 synthesis and LTC4S for synthesis of LTD4 and LTE4. By comparison, much stronger correlations (with R values of ∼0.7 or greater [Table 3) were seen among metabolites produced directly by ALOX5 prior to the committed LT synthesis steps, such as 5-HETE, 5-HEPE, 6-trans-LTB4, and 5-oxo-ETE. These metabolites also correlated strongly with 15-LO-derived products (15-HETE; 15-HEPE; 5,15-DiHETE; and lipoxin A4), 9-HETE, and 17-HDoHE (and are collectively referred to as group 2). Last, the 15-LO/12-LO metabolite 8,15-DiHETE did not correlate consistently with metabolites in group 1 or 2 and was thus considered by itself as group 3 (Table 3).
TABLE 3.
Oxylipid metabolites from monocyte cultures stimulated with A23187 at baseline
| Metabolite | LTD4a | LTE4a | 6-trans-LTB4b | 5-HETEb | 5-HEPEb | 15-HETEb | 15-HEPEb | 9-HETEb | 5,15-DiHETEb | 17-HDoHEb | Lipoxin A4b | 8,15-DiHETEc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LTB4a | 0.243d | 0.552 | 0.387 | 0.214 | 0.204 | 0.118 | 0.211 | 0.162 | 0.182 | 0.129 | 0.060 | −0.001 |
| 0.0093e | <0.001 | <0.001 | 0.022 | 0.029 | 0.212 | 0.024 | 0.084 | 0.053 | 0.170 | 0.528 | 0.988 | |
| LTD4 | 0.149 | −0.168 | −0.204 | −0.177 | −0.321 | −0.147 | −0.236 | −0.208 | −0.300 | −0.065 | −0.158 | |
| 0.113 | 0.074 | 0.030 | 0.060 | <0.001 | 0.118 | 0.012 | 0.027 | 0.001 | 0.494 | 0.094 | ||
| LTE4 | 0.257 | 0.090 | −0.037 | 0.041 | 0.109 | 0.044 | 0.006 | 0.057 | −0.052 | −0.202 | ||
| 0.006 | 0.338 | 0.695 | 0.665 | 0.248 | 0.645 | 0.946 | 0.550 | 0.581 | 0.031 | |||
| 6-trans-LTB4 | 0.871 | 0.689 | 0.842 | 0.683 | 0.778 | 0.804 | 0.714 | 0.668 | 0.271 | |||
| <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.004 | ||||
| 5-HETE | 0.810 | 0.887 | 0.755 | 0.838 | 0.862 | 0.749 | 0.696 | 0.296 | ||||
| <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.001 | |||||
| 5-HEPE | 0.736 | 0.678 | 0.808 | 0.858 | 0.723 | 0.675 | 0.282 | |||||
| <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.00 | ||||||
| 15-HETE | 0.737 | 0.893 | 0.842 | 0.848 | 0.705 | 0.201 | ||||||
| <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.031 | |||||||
| 15-HEPE | 0.706 | 0.709 | 0.690 | 0.577 | 0.122 | |||||||
| <0.001 | <0.001 | <0.001 | <0.001 | 0.192 | ||||||||
| 9-HETE | 0.874 | 0.848 | 0.708 | 0.231 | ||||||||
| <0.001 | <0.001 | <0.001 | 0.013 | |||||||||
| 5,15-DiHETE | 0.791 | 0.783 | 0.335 | |||||||||
| <0.001 | <0.001 | <0.001 | ||||||||||
| 17-HDoHE | 0.613 | 0.239 | ||||||||||
| <0.001 | 0.010 | |||||||||||
| Lipoxin A4 | 0.275 | |||||||||||
| 0.003 |
Data show a correlation matrix of oxylipid metabolites for 115 subjects (n = 114 for LTB4, LTD4, and LTE4) from monocyte cultures stimulated with A23187 for 60 min at the baseline visit.
These metabolites are considered together as Group 1, based on correlations with LTB4, shown in boldface type.
These metabolites are considered together as Group 2, based on correlations with one another, shown in italicized type.
This metabolite did not correlate consistently with Group 1 or 2 metabolites and is considered alone as Group 3.
Spearman rank order correlation coefficient.
P value for correlation.
Correlation of ALOX5 mRNA and protein with metabolites.
We next examined the association among ALOX5 mRNA, protein, and oxylipid levels from A23187-stimulated monocytes. Protein and RNA levels were measured in monocytes prior to stimulation. ALOX5 mRNA levels were negatively correlated with all 10 group 2 metabolites (P < 0.05, n = 99) (see supplementary Fig. I) but not with the other metabolites. For group 2 metabolites derived from AA, the Spearman correlation coefficients ranged from −0.263 (P = 0.009) for 9-HETE to −0.306 (P = 0.002) for 5-HETE. The correlations were somewhat weaker for the two EPA metabolites 5-HEPE (R = −0.213, P = 0.034) and 15-HEPE (R = −0.231, P = 0.022) and for the DHA metabolite 17-HDoHE (R = −0.230, P = 0.022) from group 2. Analysis of FLAP and LTA4H mRNA levels did not reveal significant correlations with any metabolites (data not shown). ALOX5 protein levels also did not correlate with ALOX5 mRNA levels (Spearman R = 0.143, P = 0.20, n = 83) or any metabolite (P > 0.05), with the exception of 15-HEPE, for which a modest negative correlation was seen (Spearman R = −0.221, P = 0.032, n = 95).
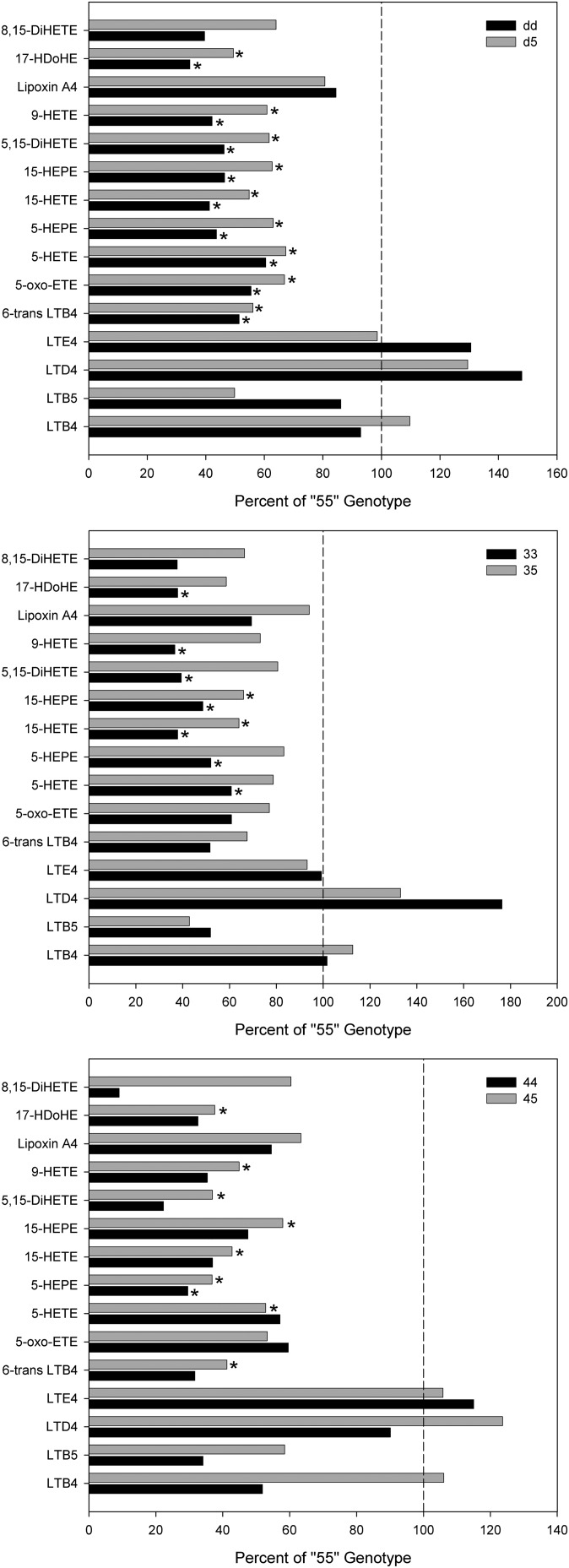
Comparison of metabolite levels among dd, d5, and 55 genotypes.
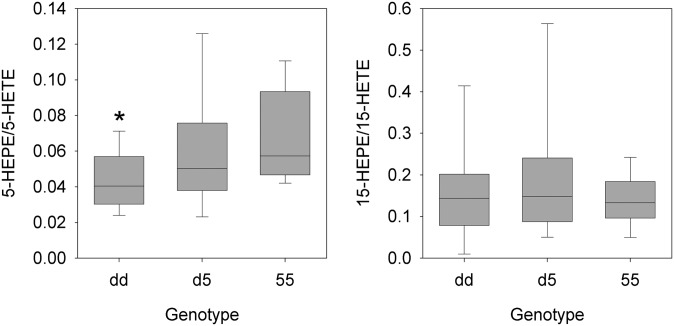
To assess potential functional differences among ALOX5 promoter alleles, we next compared A23187-stimulated metabolite levels among the different genotype groups. Group 1 LT concentrations (LTB4, LTB5, LTD4, and LTE4) did not vary by genotype, although LTD4 concentration tended to be higher in the dd group (Table 4 and Fig. 3). However, all group 2 metabolites, with the exception of lipoxin A4, were significantly higher in the 55 group than in either of the other two genotype groups. This was observed for both AA- and EPA-derived metabolites, as well as for the DHA-derived metabolite 17-HDoHE (Table 4 and Fig. 3). The EPA-derived 5-HEPE to AA-derived 5-HETE ratio differed by genotype (P = 0.003) (Fig. 4), with the dd genotype being lowest, the d5 genotype being intermediate, and the 55 genotype highest, although only the dd versus 55 comparison was significant (P < 0.05 by post hoc comparison of ANOVA on ranks). Such differences were not seen for the 15-HEPE-to-15-HETE ratio (Fig. 4) or LTB5-to-LTB4 ratio (data not shown).
TABLE 4.
Mean (± SEM) oxylipid concentrations (nmol/l) from A23187-stimulated monocyte culture supernatants at baseline by genotype group for 115 subjects
| Mean ± SEM oxylipid concentrations (nmol/l) for the genotype shown |
||||
|---|---|---|---|---|
| Metabolite | dd (n = 32) | d5 (n = 53) | 55 (n = 30) | Pa |
| Group 1b | ||||
| LTB4c | 5.94 ± 0.64 | 7.01 ± 0.55 | 6.39 ± 0.55 | 0.460 |
| LTB5 | 0.25 ± 0.12 | 0.14 ± 0.018 | 0.29 ± 0.19 | 0.565 |
| LTD4c | 1.52 ± 0.16 | 1.33 ± 0.15 | 1.03 ± 0.16 | 0.095 |
| LTE4c | 4.54 ± 0.74 | 3.43 ± 0.45 | 3.48 ± 0.75 | 0.402 |
| Group 2 | ||||
| 6-trans-LTB4 | 9.15 ± 1.67d | 9.99 ± 1.29d | 17.80 ± 4.33 | 0.039 |
| 5-oxo-ETE | 1.59 ± 0.21d | 1.92 ± 0.22d | 2.87 ± 0.58 | 0.038 |
| 5-HETE | 326 ± 44 | 363 ± 42d | 539 ± 60 | 0.004 |
| 5-HEPE | 16.9 ± 4.04d | 24.4 ± 3.27d | 38.7 ± 6.47 | <0.001 |
| 15-HETE | 7.23 ± 1.16 | 9.61 ± 1.35d | 17.53 ± 2.80 | <0.001 |
| 15-HEPE | 0.97 ± 0.16d | 1.31 ± 0.16d | 2.09 ± 0.36 | 0.001 |
| 5,15-DiHETE | 2.75 ± 0.69d | 3.67 ± 0.66d | 5.95 ± 1.24 | 0.006 |
| 9-HETE | 7.02 ± 1.38d | 10.14 ± 1.62d | 16.65 ± 2.85 | <0.001 |
| Lipoxin A4 | 1.71 ± 0.30 | 1.63 ± 0.18 | 2.02 ± 0.32 | 0.451 |
| 17-HDoHE | 3.65 ± 1.01d | 5.22 ± 0.77d | 10.56 ± 2.30 | <0.001 |
| Group 3 | ||||
| 8,15-DiHETE | 0.50 ± 0.084 | 0.80 ± 0.12 | 1.25 ± 0.38 | 0.091 |
Data are mean (± SEM) oxylipid concentrations (nmol/l) from A23187-stimulated monocyte culture supernatants at baseline by genotype group for 115 subjects.
One-factor ANOVA.
Groups 1, 2, and 3 are based on correlations among metabolites as seen in Table 3.
One of these 32 dd subjects was excluded as an outlier for LTB4, LTC4, and LTE4 analysis.
These results showed significant differences from the 55 genotype group by post hoc comparison.
Fig. 3.
Relative mean concentration values of oxylipid metabolites are shown 60 min after A23187 stimulation of monocyte cultures from subjects with different ALOX5 promoter genotypes at baseline. Bars show percentages relative to the mean levels for the 55 control group and dd and d5 subjects (top panel); 33 and 35 subjects (middle panel); and 44 and 45 subjects (bottom panel). *, Significant differences from the 55 group mean by one-factor ANOVA. Concentrations, standard errors and numbers of subjects are given in Table 4 (also see supplementary Tables IV and V).
Fig. 4.
Ratios of EPA-derived (5-HEPE and 15-HEPE) to AA-derived (5-HETE and 15-HETE) 5-LO and 15-LO metabolites are shown at 60 min after A23187 stimulation of monocyte cultures from subjects with different ALOX5 promoter genotypes at baseline. Box plots show median, 10th, 25th, 50th, 75th, and 90th percentiles for subjects with the three ALOX5 genotypes of interest. *, Significant difference from the corresponding 55 genotype group. Numbers of subjects in the dd, d5, and 55 genotype groups were 32, 53, and 30, respectively.
Comparison of metabolite levels by individual promoter alleles.
To determine if these differences were consistent across the individual ALOX5 alleles, the same analysis was performed for the 33, 35, and 55 groups, the 44, 45, and 55 groups, and the 33, 34, and 44 groups (see supplementary Tables IV, V, and VI, respectively). Essentially the same results were seen, namely, A23187-stimulated metabolite levels were highest in the 55 control group. While the 35 group had intermediate levels, only the comparison between the 33 and 55 groups was significant for most metabolites (Fig. 3; also see supplementary Table IV). We next compared subjects with 4 and 5 alleles and found the same pattern, that is, the 55 control subjects had the highest group 2 metabolite concentrations, and the 44 homozygotes had the lowest (Fig. 3; also see supplementary Table V). However, only the 45-versus-55 comparisons were significantly different, which may have resulted from the small number of 44 subjects in the study (n = 4). Homozygous dd subjects were next subdivided into the 33, 34, and 44 genotypes in order to compare differences in metabolite concentrations between the d alleles. No significant differences were found among these groups (see supplementary Table VI). However, when subjects with one deletion allele and one common allele (i.e., the 35 and 45 genotypes) were compared, subjects with one 3 allele had significantly higher concentrations of several group 2 metabolites than did subjects with one 4 allele (see supplementary Table VII).
Association of ancestry with metabolite level.
Because subjects in the 55 genotype group had a higher percentage of EUR ancestry and higher group 2 metabolite levels, we analyzed the association of both EUR ancestry with group 2 metabolite levels at baseline. Percent EUR ancestry correlated positively with most of the metabolites (6-trans-LTB4; lipoxin A4; 5-oxo-ETE; 15-HETE; 5,15-diHETE; 9-HETE; 17-HDoHE; 5-HEPE; and 15-HEPE) but Spearman correlation coefficients were low (0.19–0.22), and P values were marginal (0.048–0.019). Correlations were not significant when subjects in the 55 group were excluded from analysis. Analysis of covariance was used to determine if percent EUR altered the association of metabolite levels with ALOX5 genotypes. The association between the 55 genotype and higher group 2 metabolite levels remained consistent with the data shown in Table 4, although statistical significance was lost for 6-trans-LTB4 and 5-oxo-ETE, which had P values near 0.05 in the unadjusted analysis (Table 4). Thus, ALOX5 genotypes were associated with these metabolite levels independent of EUR ancestry. Essentially the same results were seen when we adjusted for AFR ancestry.
Effect of intervention
Subject characteristics.
Among the 116 participants who entered the study, 98 subjects completed the 6 week intervention, and complete data were available for the analysis of 97 subjects. The genotype and treatment group distributions of these subjects are shown in supplementary Table III.
ALOX5 protein and mRNA.
Using repeated measures two-factor ANOVA, we analyzed changes in ALOX5 protein and mRNA (in unstimulated monocytes) levels between baseline and the 6 week visit to identify treatment effects. Total ALOX5 protein levels increased between baseline and the 6 week follow-up visit in both groups (P = 0.017), but there was no effect observed for the fish oil treatment (P = 0.66 for treatment and P = 0.11 for interaction). Nor did the treatment affect the individual ALOX5 bands seen by Western blotting or the ratio between the two. The mean ALOX5:β-actin protein ratios for the placebo group at baseline and at 6 weeks were 1.53 and 1.60, respectively (standard error of the mean [SEM] = 0.089; n = 31), while the corresponding values for the fish oil group were 1.43 and 1.78, respectively (SEM = 0.081; n = 37). Similarly, ALOX5 mRNA levels did not change between visits (P = 0.76), and there was no effect of the fish oil treatment (P = 0.84 for treatment and P = 0.45 for interaction). The mean ALOX5 RNA levels for the placebo group at baseline and 6 weeks were 0.360 and 0.350 ru, respectively (SEM = 0.00798; n = 41), while the corresponding values for the fish oil group were 0.348 and 0.350 ru, respectively (SEM = 0.00789; n = 42).
Oxylipid metabolites: treatment effects.
We next assessed changes in A23187-stimulated oxylipid concentrations in monocyte supernatants between baseline and the end of the study, differences between the fish oil and placebo groups at both time points, and changes between the groups over time. No differences were seen between the fish oil and placebo groups at baseline for any of the 15 oxylipid metabolites (Table 5). Five of the 11 AA-derived oxylipid metabolites (as indicated by a P value of <0.05 under the Visit column in Table 5) decreased from baseline to the end of the study in both the fish oil and the control groups. By comparison, the three EPA-derived metabolites, LTB5, 5-HEPE, and 15-HEPE, all increased significantly in the fish oil group and either showed no change in the placebo group or, in the case of 5-HEPE, decreased significantly. In the case of the one DHA-derived metabolite, 17-HDoHE, levels decreased in the placebo group but not in the fish oil group. Thus, concentrations of all four omega-3 fatty acid-derived metabolites were higher in the fish oil group than in the placebo group at the end of the study.
TABLE 5.
Oxylipid concentrations from A23187-stimulated monocyte supernatants by treatment group at baseline and final visits
| Mean ± SEM oxylipid concentrations (nmol/l) |
|||||||
|---|---|---|---|---|---|---|---|
| Baseline visit |
Final visit |
Two-factor ANOVA P values |
|||||
| Metabolite | Fish oil | Placebo | Fish oil | Placebo | Treatment | Visit | Treatment X visit |
| Group 1a | |||||||
| LTB4 | 6.03 ± 0.51 | 6.88 ± 0.52 | 6.04 ± 0.51 | 5.49 ± 0.52 | 0.24 | 0.11 | 0.080 |
| LTB5 | 0.326 ± 0.270 | 0.099 ± 0.273 | 0.954 ± 0.270 | 0.104 ± 0.273 | <0.001 | 0.082 | 0.019 |
| LTD4 | 1.31 ± 0.12 | 2.61 ± 0.62 | 0.98 ± 0.12 | 2.62 ± 0.62 | 0.089 | 0.19 | 0.088 |
| LTE4 | 3.55 ± 0.48 | 3.50 ± 0.50 | 2.92 ± 0.48 | 3.07 ± 0.50 | 0.96 | 0.56 | 0.54 |
| Group 2 | |||||||
| 6-trans-LTB4 | 12.03 ± 1.60 | 12.23 ± 1.62 | 9.16 ± 1.60 | 8.41 ± 1.62 | 0.65 | 0.023 | 0.11 |
| 5-oxo-ETE | 1.95 ± 0.26 | 2.28 ± 0.26 | 1.70 ± 0.26 | 1.63 ± 0.26 | 0.98 | 0.067 | 0.43 |
| 5-HETE | 412 ± 43 | 410 ± 44 | 343 ± 43 | 337 ± 44 | 0.76 | 0.013 | 0.84 |
| 5-HEPE | 25.7 ± 10.7 | 27.9 ± 10.8 | 124.4 ± 10.7bc | 20.9 ± 10.8 | <0.001 | <0.001 | <0.001 |
| 15-HETE | 11.14 ± 1.43 | 11.53 ± 1.45 | 8.98 ± 1.43 | 6.68 ± 1.45 | 0.48 | 0.003 | 0.11 |
| 15-HEPE | 1.50 ± 0.37 | 1.43 ± 0.37 | 3.65 ± 0.37bc | 0.97 ± 0.37 | 0.008 | 0.059 | <0.001 |
| 5,15-DiHETE | 4.14 ± 0.68 | 4.16 ± 0.68 | 3.17 ± 0.68 | 2.84 ± 0.68 | 0.38 | 0.009 | 0.29 |
| 9-HETE | 10.16 ± 1.59 | 12.64 ± 1.61 | 8.86 ± 1.59 | 7.62 ± 1.61 | 0.86 | 0.001 | 0.12 |
| Lipoxin A4 | 1.87 ± 0.21 | 1.69 ± 0.21 | 1.56 ± 0.21 | 1.56 ± 0.21 | 0.51 | 0.25 | 0.94 |
| 17-HDoHE | 6.09 ± 1.06 | 6.74 ± 1.07 | 7.05 ± 1.06c | 3.67 ± 1.07 | 0.14 | 0.15 | 0.004 |
| Group 3 | |||||||
| 8,15-DiHETE | 0.69 ± 0.16 | 1.05 ± 0.16 | 0.61 ± 0.16 | 0.61 ± 0.16 | 0.63 | 0.26 | 0.54 |
Data show means ± SEM oxylipid concentrations (nmol/l) from A23187-stimulated monocyte supernatants by treatment groups (n = 49 fish oil, n = 48 placebo) at the baseline and final (after 6 weeks supplementation) visits.
Groups 1, 2, and 3 are based on correlations among metabolites, as seen in Table 3.
Significant increase from baseline.
Significantly different from placebo at final visit.
Oxylipid metabolites: treatment X genotype interactions.
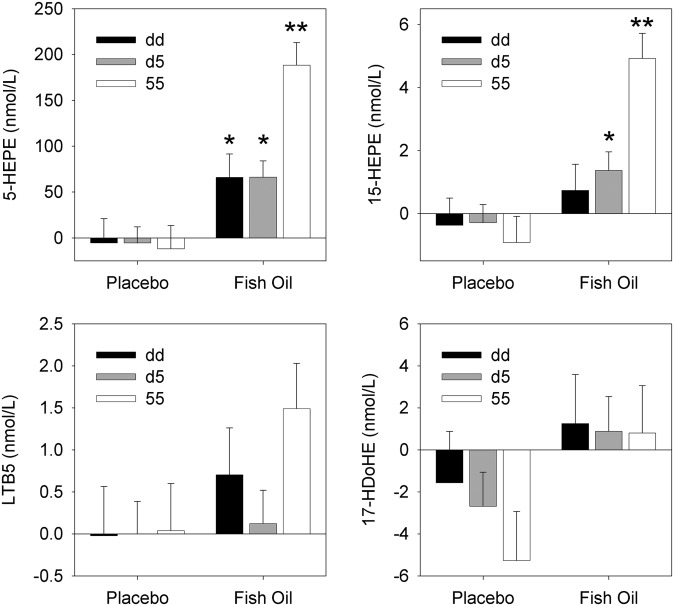
We next assessed whether genotype modified the effect of treatment on oxylipid metabolite levels in supernatants from A23187-stimulated monocytes by using repeated measures two-factor ANOVA. For 5-HEPE, genotype alone was not significant (P = 0.10), but both the treatment effect (P < 0.001) and the genotype X treatment interaction (P = 0.008) were significant, demonstrating that 5-HEPE increased significantly between visit 1 and visit 2 for subjects receiving fish oil in all three genotype groups, dd, d5, and 55 (P ≤ 0.025). In addition, the increase in the 55 group was significantly greater than the increases seen in the dd and d5 groups (P ≤ 0.003), which did not differ from one another (P = 0.92) (Fig. 5). When the change in 5-HEPE was expressed as a percent increase over baseline, the increases in all three groups were similar (563%, 495%, and 482% in the dd, d5 and 55 groups, respectively) and did not differ significantly. Similarly, for 15-HEPE, the effect of genotype alone was not significant (P = 0.27), but both treatment effect (P < 0.001) and genotype X treatment interaction (P = 0.007) were significant, indicating significant increases in 15-HEPE in the d5 and 55 (but not dd) subjects receiving fish oil, relative to that of placebo (P ≤ 0.046). Similar to the results with 5-HEPE, the fish oil-induced increase in 15-HEPE was significantly greater in the 55 group than in the other two genotype groups (P ≤ 0.001), which did not differ from one another (P = 0.53), and this difference among genotypes was not seen when change was expressed as a percentage of baseline. By comparison, gene X treatment interactions were not significant for LTB5 or 17-HDoHE (Fig. 5).
Fig. 5.
Mean (±SEM) changes in oxylipid concentrations are shown from baseline to follow-up among the three ALOX5 genotype groups of interest in response to placebo and fish oil interventions. Significant increases were seen in all of these EPA-derived (5-HEPE, 15-HEPE, and LTB5) and DHA-derived (17-HDoHE) metabolites in the fish oil treatment compared with the placebo group (P < 0.05). Genotype X treatment interactions were seen for 5-HEPE and 15-HEPE. For these metabolites, either a single (*) or double (**) asterisk indicates a within-genotype difference between the treatment and placebo groups, while the double asterisk additionally indicates that the 55 genotype, within the fish oil treatment group, had a significantly greater increase than the other genotype groups. The numbers of fish oil/placebo subjects in the dd, d5, and 55 genotype groups were 12/13 subjects, 24/25 subjects, and 13/12 subjects, respectively.
Additionally, a gene by treatment interaction was not seen for the 5-HEPE:5-HETE ratio. Although the differences among genotypes seen with this ratio at baseline were not significant at follow-up, the trends were similar (data not shown). The 5-HEPE:5-HETE ratio increased substantially with fish oil treatment, as expected (P < 0.001). The median (25th/75th percentiles) 5-HEPE:5-HETE ratio for the fish oil group at follow-up was 0.28 (0.14/0.56, respectively; n = 49) and 0.044 for the placebo group (0.032/0.068, respectively; n = 48).
No significant genotype X treatment interactions were seen for 10 of the 11 AA-derived metabolites. However, a significant interaction was seen for 5-oxo-ETE (P = 0.033) by repeated measures two-factor ANOVA of rank data. For the 55 subjects, 5-oxo-ETE decreased in the treatment group and increased in the placebo group. The median (25th/75th percentiles) change in the 55 group was −0.800 nmol/l (−1.968/0.0753 nmol/l, respectively) for the treatment group and 0.496 nmol/l (−0.563/0.995 nmol/l, respectively) nmol/l for the placebo group. In the other genotypes, slight decreases were seen in both the treatment and the placebo groups. The change in 5-oxo-ETE levels in the d5 group was −0.0579 nmol/l (−0.549/0.473 nmol/l, respectively) for the treatment group and −0.400 nmol/l (−1.168/0.133 nmol/l, respectively) for the placebo group. In the dd group, the change was −0.463 nmol/l (−0.849/1.346 nmol/l, respectively) in the treatment group and −0.611 nmol/l (−1.289/−0.156 nmol/l, respectively) nmol/l in the placebo group.
DISCUSSION
Our a priori hypothesis predicted that ALOX5 protein levels and production of LTB4 and other ALOX5-derived metabolites by monocytes would be higher in subjects with the dd promoter genotype than in subjects with the 55 genotype. However, ALOX5 protein and LTB4 levels did not vary by genotype. This finding did not support our a priori hypothesis but was consistent with our recent report that ALOX5 mRNA levels did not vary by genotype in monocytes from the subjects in the present study, although higher levels were seen in dd and d5 subjects than in 55 subjects, as predicted, when mRNA levels were examined in lymphocytes (26). While other studies have not examined the association between ALOX5 genotype and LT production in monocytes, one study did report that both ALOX5 mRNA and LTC4 production were higher in eosinophils isolated from asthmatic subjects with the 55 genotype than in those from non-5 homozygous subjects (27). These results indicate that the effect of these promoter variants on ALOX5 expression vary by cell type.
Another principal finding of this study was that production of several ALOX5 metabolites, including 5-HETE, 5-oxo-ETE, and 6-trans-LTB4, was greater in monocytes from subjects with the 55 genotype than in subjects with the d genotypes. This result is somewhat surprising given the fact that ALOX5 protein (and RNA) levels did not vary with genotype in this cell population. The lack of association between ALOX5 protein and metabolite levels is not entirely unexpected as protein levels are not the sole determinant of enzyme activity. For example, ALOX5 activity can be modulated by many factors, including serine phosphorylation, translocation from the cytosol to the nuclear envelope, and the availability of substrate, which is determined by cytoplasmic phospholipase A2 activity (22). Because these factors would not be reflected in Western blot analyses, total ALOX5 protein levels may thus not correlate with metabolite levels. However, ALOX5 metabolite levels were associated with genotype, and it is difficult to imagine how the promoter deletion genotypes of interest would affect metabolite levels without affecting ALOX5 gene expression. Again, differences in activity may provide an explanation. For example, it is possible that the promoter alleles are in linkage disequilibrium with functional coding variants in the ALOX5 gene that affect activity of the enzyme, rather than its expression. Such variants could potentially explain both the lack of correlation of ALOX5 protein levels with metabolite levels and the different metabolite levels seen among the ALOX5 genotypes. In-depth sequence analysis of the ALOX5 structural gene may help to resolve this question.
Our data also show that concentrations of the 15-LO metabolites and 9-HETE were higher in the 55 genotype than in the deletion genotypes, as seen for the ALOX5 metabolites. The concentrations of these metabolites correlate strongly with one another, so the consistent genotype association is not surprising. It is not clear, however, why these metabolites correlate with one another. Coordinate regulation of enzyme activity related to the redox status of the cell might explain this observation. For example, the first catalytic step of ALOX5 is to produce 5-HpETE, which may be released from the catalytic site, although such leakiness is typically not seen (22). The high concentrations of 5-HETE in the present study suggest that such release is occurring in these samples, as previously reported for monocytes (30, 31) and other cells (38). Lipid hydroperoxides such as 5-HpETE increase ALOX5 enzymatic activity by oxidizing the catalytic iron center of the enzyme from Fe2+ to Fe3+ (22) and could also stimulate 15-LO activity by the same mechanism (35). This activity could explain the correlation among the group 2 metabolites, although it would not necessarily explain the higher levels of these metabolites in the 55 genotype.
In addition, another finding of our study was that these group 2 metabolites correlated negatively with ALOX5 mRNA levels. This modest but consistent association suggests that the cellular redox status of a monocyte may also affect ALOX5 mRNA transcription and/or stability but not translation, as protein levels were not correlated with these metabolite levels. Moreover, this association appears to be independent of the promoter variant genotypes because mRNA expression did not vary by genotype (26). Similarly, a negative correlation was also seen between several group 2 metabolites and LTD4 concentrations. This association could result from greater LTD4 degradation via ω- and β-oxidation (3) under conditions of higher lipid peroxide production. However, this activity might also be expected to affect LTB4 and LTE4 levels as well, which was not observed, so such a phenomenon would appear to be LTD4-specific but remains speculative.
Our original hypothesis predicted that ALOX5 metabolite levels would be higher in the deletion genotype subjects than in the 55 genotype subjects, which was based on the observation that these deletion alleles are associated with an increased risk of CVD (23, 24). We inferred from this association that elevated production of LTs might exacerbate inflammation in the arterial wall and would be reflected in ALOX5 mRNA/protein expression and/or eicosanoid production in peripheral blood monocytes. However, peripheral blood monocytes may not be reliable proxy cells for macrophages, which have a more activated phenotype in vascular lesions (13) and in other tissues than monocytes. Macrophages isolated from the lung, for example, have significantly higher ALOX5 expression than monocytes isolated from peripheral blood (31, 39). It would thus be interesting to determine whether the promoter variants affect ALOX5 expression under inflammatory conditions that may be more representative of a vascular lesion than the experimental conditions used here. It is also possible that production of eicosanoids by other cell types might account for the association of deletion alleles with increased risk of CVD, and examination of activated endothelial cells or fibroblasts might yield different results than we have seen with monocytes.
We hypothesized that fish oil consumption would decrease production of AA-derived oxylipid metabolites in the present study. While an overall decrease in several AA-derived group 2 metabolites was seen in both groups over the study period, the decrease in the fish oil group was no greater than that in the placebo group. The reason for the decrease in both groups is unclear. As ALOX5 mRNA levels did not change over this period, and ALOX5 protein levels actually increased, changes in ALOX5 gene expression are not a likely cause. It also seems unlikely that consuming linoleic acid-rich corn/soy placebo oil would have had such an effect, even if increased intake of fish oil might. However, both placebo and fish oil contained the antioxidant vitamin E, which could have decreased HETE formation in both groups if the vitamin E content of monocytes was correspondingly increased. Perhaps this decreased oxidative stress and ALOX5 activation in monocytes also allowed greater stability of the enzyme itself, thus accounting for the increase in ALOX5 protein as well, although this is speculative.
Many fish oil intervention trials have demonstrated decreased production of proinflammatory AA-derived eicosanoids and increased production of less-inflammatory EPA-derived eicosanoids (29, 40). The failure to see decreased production of AA-derived eicosanoids in the fish oil group above that seen in the placebo group may have been due to high dietary AA intake and/or the relatively short duration of the intervention. By comparison, the concentrations of EPA-derived metabolites, including LTB5, 5-HEPE, and 15-HEPE, did increase with supplementation, as expected, as did the concentration of the DHA-derived metabolite 17-HDoHE. Thus, our results are consistent with those of previously reported clinical interventions with EPA supplementation (40, 41).
A unique feature of this study was its within-genotype randomization, which allowed testing of the hypothesis that the genotype of interest would have a significant effect on response to fish oil supplementation. Two such gene treatment interactions were found for the EPA-derived metabolites 5-HEPE and 15-HEPE. The concentration of 5-HEPE increased in all genotypes, but the increase was significantly greater in the 55 group than in either the dd or d5 group, which did not differ from one another. In the case of 15-HEPE, significant increases were seen only for the d5 and 55 genotypes receiving the fish oil supplement. These interactions are consistent with the higher levels of group 2 metabolites also seen in the 55 genotype. The only decrease in an AA-derived eicosanoid (5-oxo-ETE) as a result of the fish oil intervention was also seen in the 55 genotype group. The 55 genotype is clearly associated with higher concentrations of most group 2 eicosanoid metabolites, relative to the other genotypes and with greater increases in the EPA-derived metabolites after the level of intake increased as a result of fish oil supplementation. Analyses of the individual promoter alleles also support this notion, as significant differences in metabolite levels were also observed between the 55 and the 35 and 45 genotypes but not between the 33 and 34 genotypes.
This intervention trial was based on observational data indicating that subjects with deletion alleles had a higher risk of CVD than subjects with the 55 genotype but that this risk decreased when such subjects increased their level of omega-3 intake (23, 24). We thus hypothesized that subjects in the dd and perhaps d5 genotypes would have a better response to fish oil supplementation than would the 55 subjects. When we examine the production of EPA-derived metabolites, it is evident the 55 subjects had a quantitatively greater increase in some EPA-derived metabolites than the dd and d5 subjects did. This finding was not predicted by our a priori hypothesis and does not explain how increasing omega-3 fatty acid intake would be more beneficial to the deletion genotypes. However, the dd subjects at baseline had a significantly lower 5-HEPE:5-HETE ratio than the other genotypes, indicating a relatively lower production of EPA-derived eicosanoids in the dd group. This ratio was increased significantly by the intervention irrespective of genotype. It is thus plausible to postulate that there may be a greater benefit to the dd subjects as a result of this intervention because it shifts the balance of eicosanoid production away from AA and toward EPA as a substrate in the dd group, which may have anti-inflammatory benefits.
In summary, this placebo-controlled intervention trial, randomized within genotypes, identified a significant gene–diet interaction, which demonstrates that genotype-determined differences in response to omega-3 intake are evident either when baseline metabolite concentrations are examined or in response to this fish oil intervention. However, these suggestive data do not define a mechanism that accounts for these associations. Defining such a mechanism will require future studies to examine more immune (or other) cell types under conditions designed to more closely approximate conditions found in diseased tissue such as arterial plaque.
Supplementary Material
Acknowledgments
The authors thank the volunteer subjects for participating in this study, the research staff from the Western Human Nutrition Research Center and Ethnic Health Institute who helped carry out this project, Jan Peerson from the UC Davis Department of Nutrition for help with statistical analysis, and Russell Shigeta from the Department of Biochemistry and Molecular Medicine for technical support.
Footnotes
Abbreviations:
- 17-HDoHE
- 17-hydroxy docosahexaenoate
- 5-HEDH
- 5-hydroxyeicosanoid dehydrogenase
- 5-HETE
- 5-hydroxyeicosatetraenoate
- 5-HpETE
- 5-hydroperoxyeicosatetraenoate
- 5-LO
- 5-lipoxygenase
- AA
- arachidonic acid
- AFR
- sub-Saharan African
- AIM
- ancestry informative marker
- AMI
- Amerindian
- BMI
- body mass index
- CVD
- cardiovascular disease
- d
- deletion
- DHA
- docosahexaenoic acid
- EAS
- East Asian
- EPA
- eicosapentaenoic acid
- EUR
- European
- FLAP
- 5-lipoxygenase activating protein
- LT
- leukotrienes
- PBMC
- peripheral blood mononuclear cells
- ru
- relative units (ru)
This work was supported by National Institutes of Health Grants HL-079353 (to H.A.), AT-003411 (to C.B.S.), and P-60MD0222 (to UC Davis Center of Excellence in Nutritional Genomics). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies. Support was also received from US Department of Agriculture Grant CRIS Project 5306-51530-006-00D (to C.B.S.). P.A. was supported through a fellowship award from the Gustavus and Louise Pfeiffer Research Foundation. A portion of this work was conducted in a facility constructed with support from the National Institutes of Health Research Facilities Improvement Program (RR-10600-01, CA-62528-01, and RR-14514-01) from the National Center for Research Resources. Reference to a company or product name does not imply approval or recommendation of the product by the US Department of Agriculture to the exclusion of others that may be suitable.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of materials and methods (Study Design, Laboratory Methods, and Results), seven tables, and one figure.
REFERENCES
- 1.Boyce J. A. 2008. Eicosanoids in asthma, allergic inflammation, and host defense. Curr. Mol. Med. 8: 335–349. [DOI] [PubMed] [Google Scholar]
- 2.Allayee H., Roth N., Hodis H. N. 2009. Polyunsaturated fatty acids and cardiovascular disease: implications for nutrigenetics. J. Nutrigenet. Nutrigenomics. 2: 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy R. C., Gijon M. A. 2007. Biosynthesis and metabolism of leukotrienes. Biochem. J. 405: 379–395. [DOI] [PubMed] [Google Scholar]
- 4.Teiber J. F., Draganov D. I., La Du B. N. 2003. Lactonase and lactonizing activities of human serum paraoxonase (PON1) and rabbit serum PON3. Biochem. Pharmacol. 66: 887–896. [DOI] [PubMed] [Google Scholar]
- 5.Connelly P. W., Picardo C. M., Potter P. M., Teiber J. F., Maguire G. F., Ng D. S. 2011. Mouse serum paraoxonase-1 lactonase activity is specific for medium-chain length fatty acid lactones. Biochim. Biophys. Acta. 1811: 39–45. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y., Styhler A., Powell W. S. 1996. Synthesis of 5-oxo-6,8,11,14-eicosatetraenoic acid by human monocytes and lymphocytes. J. Leukoc. Biol. 59: 847–854. [DOI] [PubMed] [Google Scholar]
- 7.Fiore S., Serhan C. N. 1989. Phospholipid bilayers enhance the stability of leukotriene A4 and epoxytetraenes: stabilization of eicosanoids by liposomes. Biochem. Biophys. Res. Commun. 159: 477–481. [DOI] [PubMed] [Google Scholar]
- 8.Newman J. W., Morisseau C., Hammock B. D. 2005. Epoxide hydrolases: their roles and interactions with lipid metabolism. Prog. Lipid Res. 44: 1–51. [DOI] [PubMed] [Google Scholar]
- 9.Maclouf J. A., Murphy R. C. 1988. Transcellular metabolism of neutrophil-derived leukotriene A4 by human platelets. A potential cellular source of leukotriene C4. J. Biol. Chem. 263: 174–181. [PubMed] [Google Scholar]
- 10.Ezra D., Foster A., Cirino M., Rokach J., Letts L. G. 1987. Biliary and urinary excretion of peptide leukotrienes in the domestic pig. Prostaglandins. 33: 717–725. [DOI] [PubMed] [Google Scholar]
- 11.Powell W. S., Rokach J. 2005. Biochemistry, biology and chemistry of the 5-lipoxygenase product 5-oxo-ETE. Prog. Lipid Res. 44: 154–183. [DOI] [PubMed] [Google Scholar]
- 12.Lotzer K., Funk C. D., Habenicht A. J. 2005. The 5-lipoxygenase pathway in arterial wall biology and atherosclerosis. Biochim. Biophys. Acta. 1736: 30–37. [DOI] [PubMed] [Google Scholar]
- 13.Mehrabian M., Allayee H. 2003. 5-lipoxygenase and atherosclerosis. Curr. Opin. Lipidol. 14: 447–457. [DOI] [PubMed] [Google Scholar]
- 14.Helgadottir A., Manolescu A., Thorleifsson G., Gretarsdottir S., Jonsdottir H., Thorsteinsdottir U., Samani N. J., Gudmundsson G., Grant S. F., Thorgeirsson G., et al. 2004. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat. Genet. 36: 233–239. [DOI] [PubMed] [Google Scholar]
- 15.Helgadottir A., Gretarsdottir S., St. Clair D., Manolescu A., Cheung J., Thorleifsson G., Pasdar A., Grant S. F., Whalley L. J., Hakonarson H., et al. 2005. Association between the gene encoding 5-lipoxygenase-activating protein and stroke replicated in a Scottish population. Am. J. Hum. Genet. 76: 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helgadottir A., Manolescu A., Helgason A., Thorleifsson G., Thorsteinsdottir U., Gudbjartsson D. F., Gretarsdottir S., Magnusson K. P., Gudmundsson G., Hicks A., et al. 2006. A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction. Nat. Genet. 38: 68–74. [DOI] [PubMed] [Google Scholar]
- 17.Girelli D., Martinelli N., Trabetti E., Olivieri O., Cavallari U., Malerba G., Busti F., Friso S., Pizzolo F., Pignatti P. F., et al. 2007. ALOX5AP gene variants and risk of coronary artery disease: an angiography-based study. Eur. J. Hum. Genet. 15: 959–966. [DOI] [PubMed] [Google Scholar]
- 18.Shah S. H., Hauser E. R., Crosslin D., Wang L., Haynes C., Connelly J., Nelson S., Johnson J., Gadson S., Nelson C. L., et al. 2008. ALOX5AP variants are associated with in-stent restenosis after percutaneous coronary intervention. Atherosclerosis. 201: 148–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemaitre R. N., Rice K., Marciante K., Bis J. C., Lumley T. S., Wiggins K. L., Smith N. L., Heckbert S. R., Psaty B. M. 2009. Variation in eicosanoid genes, non-fatal myocardial infarction and ischemic stroke. Atherosclerosis. 204: e58–e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zee R. Y., Cheng S., Hegener H. H., Erlich H. A., Ridker P. M. 2006. Genetic variants of arachidonate 5-lipoxygenase-activating protein, and risk of incident myocardial infarction and ischemic stroke: a nested case-control approach. Stroke. 37: 2007–2011. [DOI] [PubMed] [Google Scholar]
- 21.Maznyczka A., Braund P., Mangino M., Samani N. J. 2008. Arachidonate 5-lipoxygenase (5-LO) promoter genotype and risk of myocardial infarction: a case-control study. Atherosclerosis. 199: 328–332. [DOI] [PubMed] [Google Scholar]
- 22.Radmark O., Werz O., Steinhilber D., Samuelsson B. 2007. 5-Lipoxygenase: regulation of expression and enzyme activity. Trends Biochem. Sci. 32: 332–341. [DOI] [PubMed] [Google Scholar]
- 23.Dwyer J. H., Allayee H., Dwyer K. M., Fan J., Wu H., Mar R., Lusis A. J., Mehrabian M. 2004. Arachidonate 5-lipoxygenase promoter genotype, dietary arachidonic acid, and atherosclerosis. N. Engl. J. Med. 350: 29–37. [DOI] [PubMed] [Google Scholar]
- 24.Allayee H., Baylin A., Hartiala J., Wijesuriya H., Mehrabian M., Lusis A. J., Campos H. 2008. Nutrigenetic association of the 5-lipoxygenase gene with myocardial infarction. Am. J. Clin. Nutr. 88: 934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Assimes T. L., Knowles J. W., Priest J. R., Basu A., Volcik K. A., Southwick A., Tabor H. K., Hartiala J., Allayee H., Grove M. L., et al. 2008. Common polymorphisms of ALOX5 and ALOX5AP and risk of coronary artery disease. Hum. Genet. 123: 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vikman S., Brena R. M., Armstrong P., Hartiala J., Stephensen C. B., Allayee H. 2009. Functional analysis of 5-lipoxygenase promoter repeat variants. Hum. Mol. Genet. 18: 4521–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalayci O., Birben E., Sackesen C., Keskin O., Tahan F., Wechsler M. E., Civelek E., Soyer O. U., Adalioglu G., Tuncer A., et al. 2006. ALOX5 promoter genotype, asthma severity and LTC production by eosinophils. Allergy. 61: 97–103. [DOI] [PubMed] [Google Scholar]
- 28.Calder P. C. 2004. n-3 Fatty acids and cardiovascular disease: evidence explained and mechanisms explored. Clin. Sci. (Lond.). 107: 1–11. [DOI] [PubMed] [Google Scholar]
- 29.Grimm H., Mayer K., Mayser P., Eigenbrodt E. 2002. Regulatory potential of n-3 fatty acids in immunological and inflammatory processes. Br. J. Nutr. 87(Suppl 1): S59–S67. [DOI] [PubMed] [Google Scholar]
- 30.Goldyne M. E., Burrish G. F., Poubelle P., Borgeat P. 1984. Arachidonic acid metabolism among human mononuclear leukocytes. Lipoxygenase-related pathways. J. Biol. Chem. 259: 8815–8819. [PubMed] [Google Scholar]
- 31.Bigby T. D., Holtzman M. J. 1987. Enhanced 5-lipoxygenase activity in lung macrophages compared to monocytes from normal subjects. J. Immunol. 138: 1546–1550. [PubMed] [Google Scholar]
- 32.Nassir R., Kosoy R., Tian C., White P. A., Butler L. M., Silva G., Kittles R., Alarcon-Riquelme M. E., Gregersen P. K., Belmont J. W., et al. 2009. An ancestry informative marker set for determining continental origin: validation and extension using human genome diversity panels. BMC Genet. 10: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pritchard J. K., Stephens M., Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics. 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang R., Shen Z., Nauseef W. M., Hazen S. L. 2002. Defects in leukocyte-mediated initiation of lipid peroxidation in plasma as studied in myeloperoxidase-deficient subjects: systematic identification of multiple endogenous diffusible substrates for myeloperoxidase in plasma. Blood. 99: 1802–1810. [PubMed] [Google Scholar]
- 35.Kuhn H., O'Donnell V. B. 2006. Inflammation and immune regulation by 12/15-lipoxygenases. Prog. Lipid Res. 45: 334–356. [DOI] [PubMed] [Google Scholar]
- 36.Serhan C. N. 2007. Resolution phase of inflammation: novel endogenous anti-inflammatory and proresolving lipid mediators and pathways. Annu. Rev. Immunol. 25: 101–137. [DOI] [PubMed] [Google Scholar]
- 37.Guido D. M., McKenna R., Mathews W. R. 1993. Quantitation of hydroperoxy-eicosatetraenoic acids and hydroxy-eicosatetraenoic acids as indicators of lipid peroxidation using gas chromatography-mass spectrometry. Anal. Biochem. 209: 123–129. [DOI] [PubMed] [Google Scholar]
- 38.Kiss L., Schutte H., Mayer K., Grimm H., Padberg W., Seeger W., Grimminger F. 2000. Synthesis of arachidonic acid-derived lipoxygenase and cytochrome P450 products in the intact human lung vasculature. Am. J. Respir. Crit. Care Med. 161: 1917–1923. [DOI] [PubMed] [Google Scholar]
- 39.Pueringer R. J., Bahns C. C., Hunninghake G. W. 1992. Alveolar macrophages have greater amounts of the enzyme 5-lipoxygenase than do monocytes. J. Appl. Physiol. 73: 781–786. [DOI] [PubMed] [Google Scholar]
- 40.Calder P. C. 2007. Immunomodulation by omega-3 fatty acids. Prostaglandins Leukot. Essent. Fatty Acids. 77: 327–335. [DOI] [PubMed] [Google Scholar]
- 41.Miller C. C., Tang W., Ziboh V. A., Fletcher M. P. 1991. Dietary supplementation with ethyl ester concentrates of fish oil (n-3) and borage oil (n-6) polyunsaturated fatty acids induces epidermal generation of local putative anti-inflammatory metabolites. J. Invest. Dermatol. 96: 98–103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.