Abstract
13C-Cholesterol was produced with high efficiency by a genetically engineered yeast strain. The method produces ∼1 mg of cholesterol per gram of glucose using 100 ml of culture medium. Uniform 94% enrichment where the most abundant product is the fully enriched isotopomer (u-13C27) is obtained using (u-13C6, 99%) glucose medium. High enrichment is very important for relaxation experiments, but for NMR applications where carbon-carbon couplings are measured, this is problematic. A good compromise between sensitivity and cost consists in diluting (u-13C6, 25%) with natural-abundance glucose. With a 2:3 ratio, the maximal amount of singlets can be obtained in 1 dimensional (D) carbon and 2D heteronuclear single-quantum correlation (HSQC) spectra with 6× intensity increase relative to natural-abundance samples. The use of (1-13C1-glucose, 99%) or (2-13C1-glucose, 99%) as isotope sources allows the labeling of the cholesterol in multiple mostly nonvicinal positions and reach 45× intensity increase. As an alternative, the dilution of (u-13C6, 99%) glucose can be used to simultaneously enrich eleven pairs of 13C up to ∼1,000× natural-abundance probability, which should be very beneficial to double-quantum NMR experiments including the INADEQUATE and related pulse sequences. The flexibility of the method and the potential to adapt the culture protocol to specific needs should find many applications in chemistry and biology and in different fields of NMR and MS.
Keywords: isotope enrichment, mass spectrometry, yeast
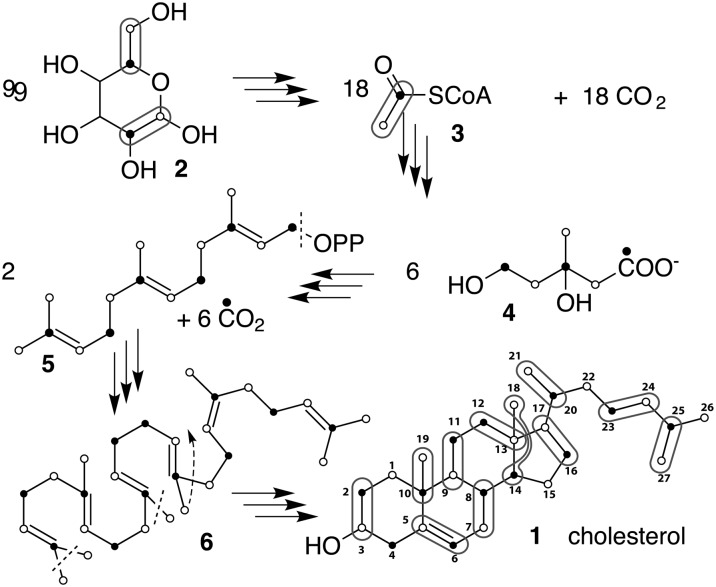
Sterols are important lipids in most eukaryotes. In particular, cholesterol has attracted a lot of attention because of its involvement in cardiovascular diseases in humans and because it has been suggested to play an important role in the formation of membrane microdomains (1). Until now, only very limited and expensive sources of labeled cholesterol were available. Cholesterol enriched at carbons 3 and 4 or 23–27 are commercially available, but other positions require de novo biosynthesis. Small amounts of low-enrichment cholesterol have been obtained in vivo by skin injection of enriched mevalonate in rat (2) or feeding mammals and humans with 13C enriched precursors (3, 4). An in vitro alternative based on human hepatoma Hep G2 cells cultures (5) produces small amounts of cholesterol with higher enrichment levels, but if these methods can produce samples for MS analysis and 14C labeling, they are not efficient enough for NMR applications. We have previously engineered a Saccharomyces cerevisiae strain by deleting the ERG5 and ERG6 genes and introducing plasmids that express the DHCR7 and DHCR24 genes from Danio rerio, leading to the synthesis of cholesterol (6). Subsequently, we integrated cassettes expressing DHCR7 and DHCR24 into the ERG5 and ERG6 loci, creating deletions of the latter genes, to create a more stable strain that efficiently produces cholesterol as its major (<95%) sterol (7). The metabolically engineered yeast use the same biosynthesis pathway as animals (Fig. 1) and efficiently produce 13C-enriched cholesterol with different labeling patterns with a yield of ∼1 mg of cholesterol per gram of glucose in 100 ml of culture medium.
Fig. 1.
Biosynthetic pathway of cholesterol produced by yeast grown on a glucose medium. Rounded frames indicate the 11 pairs of carbons stemming from glucose through acetyl CoA (3). Filled and empty dots correspond to the carbonyl and the Me of (3) respectively. Of the 54 carbons of glucose needed to make cholesterol, 18 are lost during entry into the citric acid cycle and six when mevalonate (4) is decarboxylated. Finally, three methyl groups are further removed after the cyclization of squalene (6) while Me(18) migrates from C(14) to C(13).
EXPERIMENTAL PROCEDURES
Materials
The Saccharomyces cervisiae strain used was RH6829 (MATa ura3 leu2 his3 trp1 bar1 erg5Δ::HIS5-GPD-DHCR24 erg6Δ::TRP1-GPD-DHCR7 (7). Yeast nitrogen base and yeast extracts were obtained from US Biological and Difco, respectively. D-glucose (99%), leucine (95%), and uracil (99%) were obtained from Aristar, Fluka, and Sigma, respectively. The pyrogallol and petroleum ether with high boiling point (b.p. 40–60) come from Sigma and potassium hydroxide (85%) and methanol were obtained from Acros. All enriched sources of glucose and chloroform-D (99.8%) were purchased from Cambridge Isotope Laboratory.
Preparation of media
Three samples of cholesterol were prepared. The 94% enriched cholesterol was prepared in a 450 ml medium containing 0.7% yeast nitrogen base, 0.5% yeast extracts, and 1.5% (6.75 g) (u-13C6, 99%) glucose. It was mixed under magnetic stirring and filter sterilized. A 4.5 ml solution of leucine and uracil was added from a 10× stock solution at 0.4 mg/l. The production medium was inoculated with 45 μl (1:10000) RH6829 saturated yeast culture and incubated at 30°C in a 3-l flask for two days with shaking at 230 rpm. We obtained ∼5.5 mg of cholesterol after lipid extraction (see below). The 10% enriched cholesterol was prepared from 2.5 l medium where 9.90 g of (u-13C6, 24.0%) was diluted with 15.0 g of natural-abundance glucose (1%). For growth the medium was split into three flasks. We obtained ∼46 mg of cholesterol. The partially labeled cholesterol was prepared in 300 ml medium (0.7% yeast nitrogen base, 0.5% yeast extract) at 1% glucose concentration. A total of 0.98 g of (1-13C1, 98.1%) was diluted with 2 g of natural-abundance glucose and resulted in 1.7 mg of cholesterol enriched at the position of the empty circles in Fig. 1.
Lipid extraction
Cholesterol extraction and purification was performed as previously described (7). Briefly, the saturated culture media were centrifuged at 3,000 rpm for 5 min and the cells harvested and washed with water in glass tubes. The cells were resuspended with 1 ml 60% potassium hydroxide, 1 ml of 0.5% pyrogallol in methanol, and 1.5 ml methanol in screw-cap glass tubes and incubated for 2 h at 85°C, tightly closed. Sterols were extracted three times with 2 ml fractions of petroleum ether. The petroleum ether phases were evaporated under nitrogen flow and the samples stored at −80°C.
The cholesterol was then purified using a C18 reversed-phase column by HPLC. The extracts were dissolved in acetone at 45°C, centrifuged, and the soluble extract was applied to an Uptisphere 120A 5 μm ODB column (Laubscher Labs, Switzerland) and eluted isocratically with 70% acetonitrile/30% ethanol (v/v). The cholesterol peak was identified by spotting 5 μl onto a TLC plate. The TLC plate was immersed in staining solution (50 mg FeCl3:6H2O, 90 ml H2O, 5 ml glacial acetic acid, 5 ml sulfuric acid) for 1 min, then heated for 3 min at 110°C and scanned. The cholesterol peak fractions were pooled and dried under nitrogen.
NMR analysis
All the NMR measurements were performed at 37°C on a Bruker Avance-III 500 MHz spectrometer equipped with a TCI low-temperature probe. The 5.5 mg of 94% labeled cholesterol, 5 mg of 10% labeled cholesterol, and the 1.7 mg of “1-C” labeled cholesterol were dissolved in 0.6 ml CDCl3 separately. The proton-decoupled 13C experiment was recorded in 512 scans with a 236 ppm window, 3 s recycling delay, and 10 μs hard pulses.
GC-MS analysis
Samples were dissolved in chloroform:methanol (1:1) and injected into a VARIAN CP-3800 Gas Chromatograph equipped with a Factor Four Capillary Column VF-5ms 15 m × 0.32 mm ID DF = 0.10 and analyzed by a Varian 320 MS triple quadrupole with electron energy set to −70 Ev at 200°C. Sterols were eluted with a linear gradient from 195–230°C at 4°C per min (7).
RESULTS AND DISCUSSION
The efficiency of the conversion of 13C glucose into cholesterol was tested using (u-13C6, 99%) glucose. The medium, including 6.75 g of enriched glucose as a carbon source, produced about 5.5 mg of cholesterol with 92% 13C enrichment. The most abundant isotopomer was the fully-enriched (u-13C27) cholesterol, and a simulation of the mass distribution (see supplementary Fig. I) indicates that the presence of 2.25 g of yeast extract with the 6.75 g of enriched glucose in the medium corresponded only to about 8% dilution of the isotope source. Only 2% of mass corresponds to the light isotopomers of natural-abundance cholesterol (386 and 387). The rest shows the predicted distribution of heavy isotopomers decreasing from 413 mass units. The nearly total independence of the metabolism of cholesterol relative to the presence of yeast extract explains this small contribution and makes it unnecessary to use more sophisticated culture protocols (8, 9) used for the biosynthesis of 13C or 15N enriched proteins or metabolic studies. This contamination would, however, be a problem for use as an internal standard for MS applications and could be circumvented by preparation of a small preculture in 13C medium to use for inoculation.
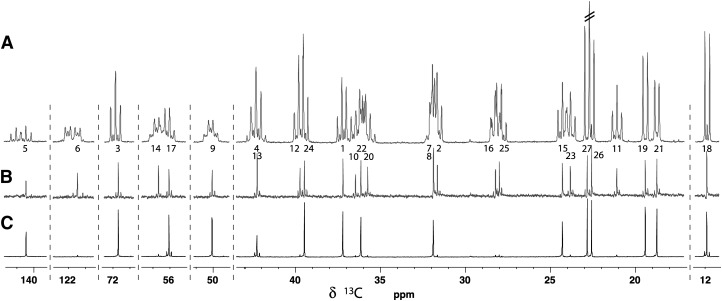
The proton-decoupled carbon spectrum is shown in Fig. 2A where the 13C-13C coupling patterns are clearly visible. When recording carbon spectra at low resolution, signals have up to four one-bond coupling partners resulting in quintet-like structures because the resolution is not high enough to resolve the slightly different values of the 1JCC couplings. In high-resolution spectra, whether it is a 1D {1H} 13C spectrum or high-resolution HSQC, 40 additional couplings larger than 1 Hz further increase the multiplicities or broaden signals. (See DFT-GIAO calculations of JCC in supplementary Table I.) The most extreme situations arise for carbons 6, 8, 13, and 14 that are expected to have eight coupling partners with JCC > 1 Hz resulting in transitions down to 1/256th the amplitude they would have as singlets. In terms of sensitivity, the paradox is that for some carbons, the fully-enriched material is expected to show less intense signals than natural-abundance cholesterol. In order to observe intense singlets, one should either use NMR methods able to refocus or decouple 13C-13C couplings or reduce the level of enrichment. The first being outside the scope this paper, we concentrated on the second option.
Fig. 2.
{1H} 13C spectra of cholesterol produced using (A) (u-13C6, 99%) glucose, (B) 2:3 (u-13C6-, 24%)/(u-13C6, 1.07%) glucose, and (C) 1:2 (1-13C1, 99%)/(u-13C6, 1.07%) glucose. In A, all carbons exhibit a multiplet pattern due to the n 1JCC of the fully enriched isotopomer (for example, the doublet of carbon 18) overlapping with minor structures corresponding to n-1 couplings (the singlet between the lines of the doublet of carbon 18) with a relative integral of c.a. 6%. The structure of the carbon 6 corresponds to a ABKX spin system where A and B are the strongly coupled carbons 7 and 8. Carbons 9 and 14 are also affected by these nearly degenerated resonances. Carbon pairs 20/22, and 1/10 also show some second-order effects but they are not strong enough to significantly affect remote coupling partners. All carbons in B have singlet structures but satellite signals due to 1JCC are observed for the carbon pairs originating from the same glucose (rounded frames of Fig. 1). Carbons 7 and 8 are so strongly coupled that they come out as a singlet. In C, only carbons 13, 17, and 18 show satellite structures because they are directly bound to enriched positions (empty circles in Fig. 1).
Having a method with nearly quantitative enrichment efficiency in hand, we could envisage to produce samples with different isotope distributions listed in Table 1. For HSQC and related experiments (HSQC-NOESY, etc.) one usually wants to obtain singlets with maximal intensities and minimize additional doublet or triplet due to 13C coupling partners. For these experiments the maximal proportion of singlet (7–15%) can be obtained with 20–30% average enrichment using diluted (u-13C6, 25%) glucose. (See the discussion in the supplementary data and supplementary Figs. II–IV for more details about the distribution of multiplicities as a function of the level of enrichment and dilution of labeled glucose.) We produced 10% uniformly enriched cholesterol by mixing (u-13C6, 24.0%) with 1.5 equivalents of natural-abundance (u-13C6, 1.07%) glucose. On average, about 6.8% of the carbons come as singlets (see the distribution in supplementary Fig. V) whereas the proportion of doublets due to 1JCC should be close to 2.45%, which is not disturbing as these doublets spread their intensities over pairs of peaks (Fig. 2B).
TABLE 1.
Conditions for the production of enriched cholesterol and applications
| Growth Medium |
Cholesterol |
Av.% of 13C Multiplicity |
|||||
|---|---|---|---|---|---|---|---|
| Dilution Lab./Nat. (g) | Enrichment in % (g) | (lJCC) |
(nJCC)a |
||||
| Labeled glucose | s | d | s | d | Applications and Remarks | ||
| (u-13C6, 99%) | 1:0 (6.75 g) | 94b (5.4 mg) | 0.5c | 20.6d | 0.0c | 1.1 | Experiments not affected by dense 13C-13C coupling networks |
| (u-13C6, 99%) | 1:0.5 | 65.9 | 3.0 | 25.1 | 0.7 | 5.2 | DQ experimentse |
| (u-13C6, 99%) | 1:2 | 32.2 | 4.5 | 18.3 | 2.1 | 8.7 | DQ experimentse |
| (u-13C6, 99%) | 1:8.5 | 10.09 | 2.5 | 6.4 | 2.2 | 5.3 | DQ experimentse |
| (u-13C6, 24.0%) | 2:3 (9.9:15 g) | 9.49 (46 mg) | 6.8 | 2.5 | 6.6 | 3.3 | For all C for SQ experimentsf |
| (1-C-13C6, 99%) | 1:0 | 27.25 | 45.0 | 2.7 | 35.5 | 8.9 | For C 1, 3, 5, 7, 9, 15, 19, 21, 22, 24, 26, 27 at low res. SQg |
| (1-C-13C6, 99%) | 1:2(1 g/2 g) | 8.94 (1.7 mg) | 15.1 | 0.3 | 13.9 | 1.8 | For C 1, 3, 5, 7, 9, 15, 19, 21, 22, 24, 26, 27 at low-res SQg |
| (2-C-13C6, 99%) | 1:0 | 22.11% | 47.6 | 1.3 | 25.8 | 15.0 | For C 2, 4, 6, 10, 16, 20, 23, 25 at low-res SQg |
| (2-C-13C6, 99%) | 1:2 | 7.34% | 13.2 | 0.9 | 10.3 | 14.7 | For C 2, 4, 6, 8, 10, 11, 13, 14, 16, 20, 23, 25 at low-res SQg |
The criterion for split is JCC > 1 Hz.
Based on MS data. The yeast extract decreased slightly the enrichment level.
High-multiplicity spectra, negligible amount of singlets.
This is due to the 5 methyls coupling with unit probabilities with their directly-bound carbons.
“DQ” stands for experiments based on 13C-13C double quantum coherences (ADEQUATES, INADEQUATES, etc.).
“SQ” stands for experiments based on isolated 13C (HSQC, 13C spectra, DEPT, etc.).
JCC > 25 Hz.
The alternation of carbons stemming from the methyl and carbonyl of acetyl CoA in the precursor 5 (see Fig. 1) and the availability of (1-13C1, 99%) and (2-13C1, 99%) glucose makes it possible to significantly increase the proportion of singlets by selectively labeling either the filled or the empty carbons of Fig. 1. With singly-labeled glucose, the maximum level of enrichment can reach c.a. 50% (see supplementary Figs. VI and VII) because carbons 5 and 6 of glucose accounting for half of the AcCoA are not enriched. We produced cholesterol with ∼9% average enrichment diluting (1-13C1, 99%) glucose with natural-abundance glucose 1:2. The amplitudes of the singlets were about 11–15% of the maximum for all carbons, except for carbon 13 where 6% of the maximal amplitude was predicted by calculations (see Fig. 2C).
An alternative and possibly more effective approach is to take advantage of the tendency of the biosynthetic pathway to generate pairs of enriched positions and chose NMR experiments based on 13C-13C double-quantum magnetization. The dilution of (u-13C6, 99%) glucose allows reaching a proportion of doublet of about 25% at 65.87% enrichment, which is comparable with 0.01% at natural abundance. (See the supplementary discussion and supplementary Figs. IV and III).
The availability of enriched cholesterol for NMR experiments should find many applications in liquid and solid-state NMR but also in the more specialized methods studying membranes and liquid crystals. The dynamics of cholesterol, the observation of changes in its chemical environment, and the structure determination of complexes should all benefit from enrichment and contribute to elucidate some of the unresolved questions concerning cholesterol. The protocol can be extended to other sterols produced in yeast, for example, ergosterol, the natural sterol, and campesterol, which can also be efficiently produced in genetically modified yeast.
Supplementary Material
Acknowledgments
Rupali Shivapurkar thanks J. Thomas Hannich for help with sterol isolation, Mohammadali Foroozandeh for the NMR experiment set-up help, and André Pinto for assistance on the NMR spectrometer.
Footnotes
This work was supported by the State of Geneva, the Swiss NSF funds 200021-116171, 200020-126650, 206021_1288746, 3100A0-128670, partial funding from the NCCR Chemical Biology funded by the Swiss National Science Foundation, the ESF (Euromembranes grant to H.R.) and SystemsX.ch evaluated by the SNSF.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of seven figures, one table, and discussion.
REFERENCES
- 1.Simons K., Ikonen E. 1997. Functional rafts in cell membranes. Nature. 387: 569–572. [DOI] [PubMed] [Google Scholar]
- 2.Popják G., Edmond J., Anet F. A. L., Easton N. R., Jr 1977. Carbon-13 NMR studies on cholesterol biosynthesized from 13C mevalonates. J. Am. Chem. Soc. 99: 931–935. [DOI] [PubMed] [Google Scholar]
- 3.Ouguerram K., Nguyen P., Krempf M., Pouteau E., Briand F., Bailhache E., Magot T. 2004. Selective uptake of high density lipoproteins cholesteryl ester in the dog, a species lacking in cholesteryl ester transfer protein activity: An in vivo approach using stable isotopes. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 138: 339–345. [DOI] [PubMed] [Google Scholar]
- 4.Ouguerram K., Krempf M., Maugeais C., Maugre P., Darmaun D., Magot T. 2002. A new labeling approach using stable isotopes to study in vivo plasma cholesterol metabolism in humans. Metabolism. 51: 5–11. [DOI] [PubMed] [Google Scholar]
- 5.Kelleher J. K., Kharroubi A. T., Aldaghlas T. A., Shambat I. B., Kennedy K. A., Holleran A. L., Masterson T. M. 1994. Isotopomer spectral analysis of cholesterol synthesis: applications in human hepatoma cells. Am. J. Physiol. Endocrinol. Metab. 266: E384–E395. [DOI] [PubMed] [Google Scholar]
- 6.te Welscher Y. M., ten Napel H. H., Balagué M. M., Souza C. M., Riezman H., de Kruijff B., Breukink E. 2008. Natamycin blocks fungal growth by binding specifically to ergosterol without permeabilizing the membrane. J. Biol. Chem. 283: 6393–6401. [DOI] [PubMed] [Google Scholar]
- 7.Guan X. L., Riezman I., Wenk M. R., Riezman H. 2010. Yeast lipid analysis and quantification by mass spectrometry. Methods Enzymol. 470: 369–391. [DOI] [PubMed] [Google Scholar]
- 8.Rembacz K. P., Faber K. N., Stellaard F. 2007. Selective conversion of plasma glucose into CO2 by Saccharomyces cerevisiae for the measurement of 13C abundance by isotope ratio mass spectrometry: proof of principle. Rapid Commun. Mass Spectrom. 21: 3169–3174. [DOI] [PubMed] [Google Scholar]
- 9.Kohno T., Kusunoki H., Sato K., Wakamatsu K. 1998. A new general method for the biosynthesis of stable isotope-enriched peptides using a decahistidine-tagged ubiquitin fusion system: An application to the production of mastoparan-X uniformly enriched with 15N and 15N/13C. J. Biomol. NMR. 12: 109–121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.