Abstract
Id1 is frequently overexpressed in many cancer cells, but the functional significance of these findings is not known. To determine if Id1 could contribute to the development of hematopoietic malignancy, we reconstituted mice with hematopoietic cells overexpressing Id1. We showed for the first time that deregulated expression of Id1 leads to a myeloproliferative disease in mice, and immortalizes myeloid progenitors in vitro. In human cells, we demonstrate that Id genes are expressed in human acute myelogenous leukemia cells, and that knock down of Id1 expression inhibits leukemic cell line growth, suggesting that Id1 is required for leukemic cell proliferation. These findings established a causal relationship between Id1 overexpression and hematologic malignancy. Thus, deregulated expression of Id1 may contribute to the initiation of myeloid malignancy, and Id1 may represent a potential therapeutic target for early stage intervention in the treatment of hematopoietic malignancy.
Keywords: Id1, myeloproliferative disease, leukemia, therapeutic target, prevention
Introduction
Hematopoiesis is tightly regulated, in part, by transcription factors, which act alone or in combination to drive the expression of specific target genes that are essential for lineage commitment (Kondo et al., 1997; Akashi et al., 2000a; Rosmarin et al., 2005). Hematopoietic stem cell (HSC) express multiple transcription factors at low levels including PU.1, GATA-1, C/EBPα, E2A and others, which are selectively up- and downregulated to promote erythroid (GATA-1), myeloid (PU.1 and C/EBPα) and lymphoid (E2A) development (Hu et al., 1997; Akashi et al., 2003). Targeted deletion and overexpression of these transcription factors in mice leads to enhanced progenitor proliferation and impaired terminal differentiation. In humans, mutation or deregulated expression of transcription factors are frequently observed in patients with leukemia and lymphoma, which are characterized by increased HSC/progenitor cell proliferation and a block in hematopoietic differentiation (Tenen, 2003; Koschmieder et al., 2005; Rosenbauer et al., 2005). Thus, a complete understanding of the transcriptional network regulating this developmental process should lead to an improved understanding of the leukemogenic process, and treatment of leukemia.
Id proteins are members of the helix–loop–helix (HLH) family of transcription factors that are expressed in many tissues (Norton, 2000; Yokota and Mori, 2002; Ruzinova and Benezra, 2003; Wong et al., 2004). Id proteins competitively dimerize with other members of the HLH family (E2A, E2-2 and HEB), and inhibit their ability to bind DNA and transactivate specific target genes, which are required for the differentiation (Wilson et al., 1991; Jen et al., 1992). In addition to their ability to block differentiation, Id genes are expressed in actively proliferating cells, and are known to promote cell proliferation by inhibiting the expression of p16 and p21, and the function of Rb (Lasorella et al., 2000; Pagliuca et al., 2000; Alani et al., 2001; Ohtani et al., 2001). Thus, deregulation of Id gene expression could contribute to the development of cancer by disrupting these normal cellular processes. Human epithelial tumors derived from prostate, breast, ovarian, esophageal express high levels of Id1 compared to their normal cell counterparts, and inhibition of Id1 gene expression in breast cancer cell lines reduces their metastatic potential in vivo, and the highest levels of Id gene expression correlate with poor clinical outcomes (Lasorella et al., 2001; Ruzinova and Benezra, 2003; Sikder et al., 2003; Fong et al., 2004; Perk et al., 2005). Thus, deregulation of Id gene expression may be a common mechanism for cancer cells to escape the normal cues for differentiation and proliferation.
Id genes are expressed in several murine and human hematopoietic cell lines, as well as normal bone marrow cells (BMC) (Kreider et al., 1992; Ishiguro et al., 1995; Quesenberry et al., 1996; Cooper et al., 1997). High levels of Id1 mRNA are detected in purified primitive myeloid progenitors (common myeloid progenitor (CMP), granulocyte/macrophage progenitor (GMP)), whereas the levels are decreased during neutrophil differentiation (Cooper et al., 1997; Leeanansaksiri et al., 2005). Based on these data, we hypothesized that deregulated expression of Id1 in hematopoietic cells might contribute to the development of myeloid malignancies. In this report, we show that overexpression of Id1 immortalizes myeloid progenitors in vitro, leads to a myeloproliferative disease (MPD) in vivo. Furthermore, we demonstrate that Id genes are expressed in human acute myelogenous leukemia (AML) cell lines and AML patient samples, and that knock down of Id1 expression inhibits AML cell line growth, suggesting that Id1 is important for leukemic cell growth. Therefore, Id1 may represent potential therapeutic target to prevent initiation of MPD or treat AML.
Results
Id1 enhances primitive myeloid progenitor cell proliferation and immortalizes bone marrow cells in vitro
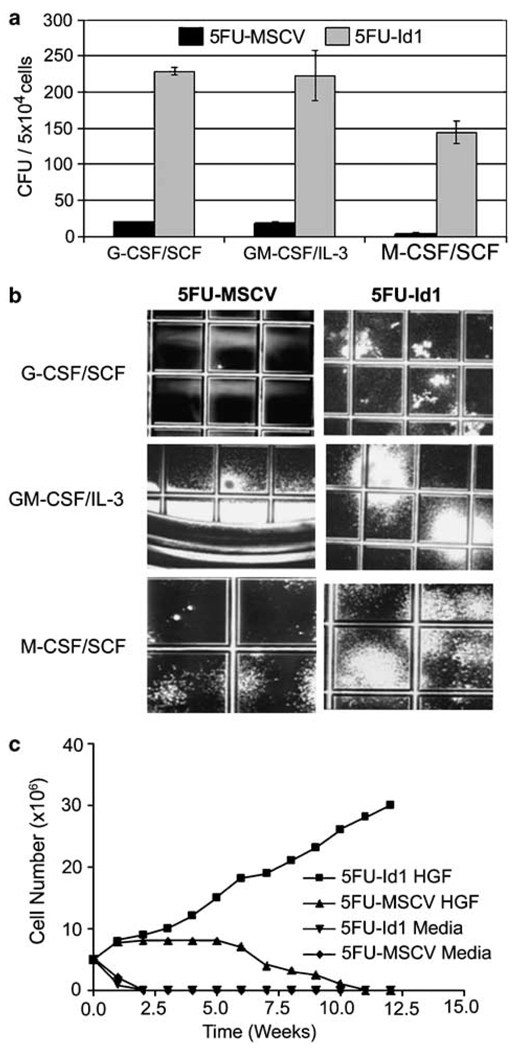
We previously reported that Id1 expression increases during the early stages of myelopoiesis and overexpression of Id1 enhances proliferation of the EML progenitor cells (Leeanansaksiri et al., 2005), suggesting that Id1 might promote primitive myeloid progenitor proliferation. To test this hypothesis, BMC were transduced with retroviral vectors that express Id1 and green fluorescent protein (GFP; MSCV-Id1-GFP) or GFP alone (MSCV-GFP). Transduced cells (GFP+) were sorted and then plated in myeloid colony assays. Id1-transduced (5FU-Id1) BMC showed enhanced colony formation in the presence of all hematopoietic growth factors (cytokines) tested (SCF/G-CSF, SCF/M-CSF and GM-CSF/IL-3) compared to control BMC transduced with MSCV-GFP (5FU-MSCV) (5FU-MSCV: 20.5 ± 0.5, 5FU-Id1: 229 ± 5 with G-CSF/SCF; 5FU-MSCV: 19 ± 1, 5FU-Id1: 223 ± 35 with GM-CSF/IL-3; 5FU-MSCV: 3.5 ± 1.5, 5FU-Id1: 144 ± 16 with M-CSF/SCF; Figure 1a). Furthermore, 5FU-Id1 BMC showed increased colony size in the presence of all growth factors tested (Figures 1a and b). Morphological analysis of 5FU-Id1 BMC from individual G-CSF/SCF- and GM-CSF/IL-3-induced colonies demonstrated increased numbers of immature myeloid cells including myeloblasts, promyelocytes and terminally differentiated granulocytes and macrophages (data not shown). Collectively, these data suggest that Id1 promotes increased proliferation and expansion of primitive progenitors and immature myeloid cells.
Figure 1.
Effect of Id1 overexpression on bone marrow cells (BMC) proliferation in suspension and semisolid media. Mouse BMC were harvested 3 days after 5-FU treatment, and then transduced with MSCV-Id1-GFP (5FU-Id1) or MSCV-GFP (5FU-MSCV). (a) Sorted green fluorescent protein (GFP) positive cells were placed in semisolid agar media supplemented with cytokine combination as indicated (50 ng/ml hG-CSF, 100 ng/ml hM-CSF, 100 ng/ml mSCF, 20 ng/ml mGM-CSF, 30 ng/ml mIL-3). Colony forming units (CFU) from GFP positive cells were scored after 10 days. Data are presented as CFU per 5 × 104 BMC ± s.e.m. of duplicate plates. (b) Photographs demonstrating size and frequency of CFU; grid = 2 mm. (c) Proliferation of sorted GFP positive cells in suspension culture with or without cytokines (100 ng/ml mSCF, 50 ng/ml hIL-6, 100 ng/ml hTPO, 100 ng/ml hFlt3L) in complete Iscove’s modified Dulbecco’s medium (cIMDM). Results are representative of two separate experiments.
To determine the long-term proliferative potential of Id1-transduced BMC in vitro, transduced BMC were cultured in complete Iscove’s modified Dulbecco’s medium (cIMDM) containing four cytokines. 5FU-MSCV BMC and 5FU-Id1 BMC proliferated to the same extent over the first 2 weeks in cultures, whereas cells cultured in the absence of cytokine did not survive beyond 1 week (Figure 1c). However, by the third and fourth week of culture, 5FU-Id1 BMC showed a significant increase in the number of viable cells compared to 5FU-MSCV BMC, which contained both proliferating and differentiating cells. Furthermore, 5FU-Id1 BMC escaped proliferative senescence and continued to divide for over 1 year in culture, whereas 5FU-MSCV BMC did not proliferate beyond 10 weeks. In addition, the 5FU-Id1 BMC were dependent on the presence of stem cell factor (SCF) for survival as > 95% of the cells did not survive after 72 h when cultured in medium without mSCF. Collectively, Id1 promoted primitive hematopoietic progenitor proliferation, and combined with SCF, Id1 was sufficient to immortalize primitive BMC in vitro.
Id1 immortalizes SCF-dependent CMP-like cells that retain the ability to differentiate in response to G-CSF and GM-CSF in vitro
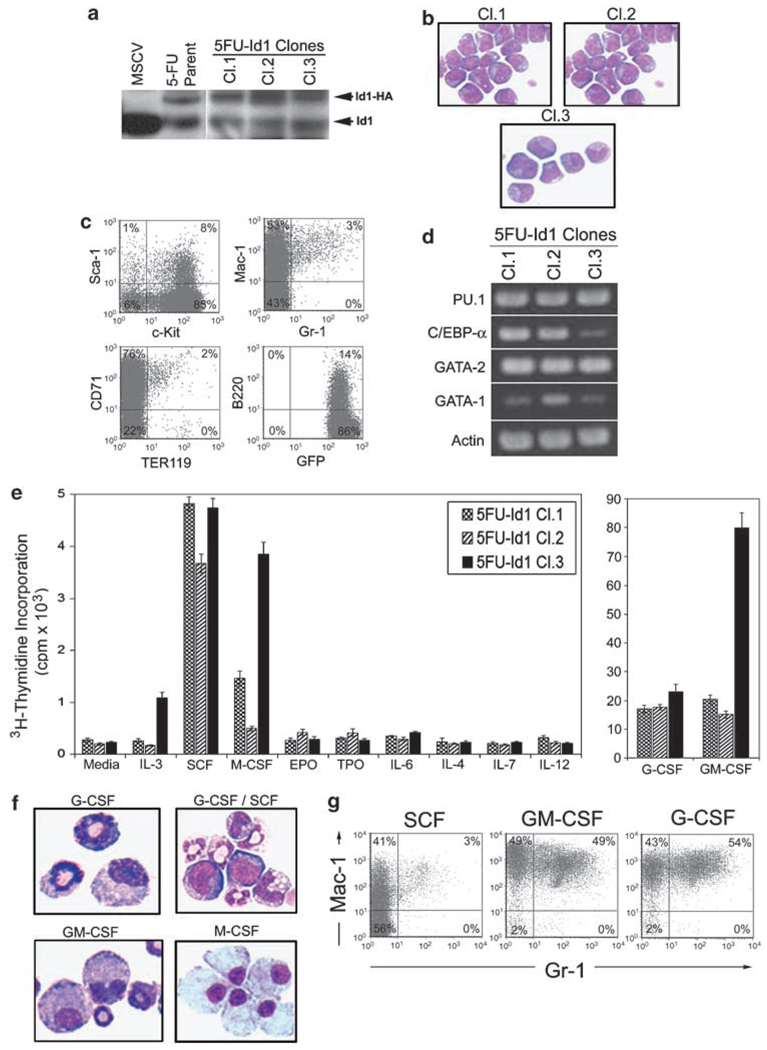
To determine if the Id1-immortalized progenitors were blocked in differentiation at a particular stage of development, single-cell clones were derived by limiting dilution analysis (5FU-Id1 Cl.1, Cl.2 and Cl.3) and their morphology, immunophenotype and their ability to proliferate and differentiate in response to cytokine were evaluated. As expected, the 5FU-Id1 clones were GFP+ by flow cytometry and expressed additional levels of exogenous Id1-HA by western blot analysis (Figure 2a). In morphological analysis, the majority of cells in the 5FU-Id1 clones were myeloblast-like cells with some promyelocytes and myelocytes (Figure 2b). Analysis of cell surface marker expression by flow cytometry demonstrated that the 5FU-Id1-cloned cells were c-Kit+, Sca-1lo/−, Gr-1lo, Mac-1+, TER119− and B220lo/− (Figure 2c), indicating that these progenitors were blocked at an early stage of myeloid development with an immunophenotype similar to CMP/GMP (Akashi et al., 2000b). In addition, semiquantitative RT–PCR analysis of the 5FU-Id1 clones demonstrated that all clones express Gata-1, Gata-2, Pu.1 and Cebpα, which is most consistent with the expression pattern observed in CMP (Figure 2d). Chromosomal analysis of 5FU-Id1 clones showed normal karyotype for the 5FU-Id1 Cl.2 and Cl.3, whereas 5FU-Id1 Cl.1 showed a deletion on one of the X chromosomes (data not shown). Furthermore, we analysed the 5FU-Id1 clones for their proliferative response to multiple cytokines in 3H-thymidine incorporation assay and found that all clones proliferate in response to granulocyte colony-stimulating factor (G-CSF), granulocyte monocyte colony-stimulating factor (GM-CSF) and SCF and to a much lesser extent to monocyte colony-stimulating factor (M-CSF) and interleukin-3 (IL-3; 5FU-Id1 Cl.3), but not to EPO, TPO, IL-2, IL-4, IL-6 and IL-12, suggesting that the 5FU-Id1 clones respond to cytokine that support myeloid development (Figure 2e). We also evaluated the differentiation ability of the 5FU-Id clones in response to G-CSF, GM-CSF and M-CSF. We found that all 5FU-Id1 clones (5FU-Id1 Cl.2 shown) could be induced to differentiate into morphologically identifiable granulocytes and macrophages (Figure 2f), and more than 97% of these cells expressed high levels of Gr-1+/Mac-1+ (granulocytes), or Gr-1+/Mac-1+ (macrophages) in cultures with G-CSF and GM-CSF in comparison to cell lines maintained in SCF (Figure 2g), which only express moderate levels of Mac-1. Thus, the Id1 overexpressing BMC are CMP-like hematopoietic progenitors that are blocked in differentiation when cultured in SCF, however, these cells can be induced to differentiate into granulocytes and macrophages in cultures supplemented with G-CSF, GM-CSF and M-CSF.
Figure 2.
Hematopoietic phenotype of cloned Id1 overexpressing bone marrow cells (BMC). BMC infected with retroviral vectors that express pMSCV-Id1-GFP were sorted and maintained in complete Iscove’s modified Dulbecco’s medium (cIMDM) with 10% fetal calf serum (FCS), P/S, 100 ng/ml mSCF, 100 ng/ml thrombopoietin (hTpo), 100 ng/ml hFlt-3L and 50 ng/ml hIL-6, then cloned by single cell culture. The stem cell factor (SCF)-dependent Id1 overexpressing green fluorescent protein (GFP) positive cells were maintained in cIMDM supplemented 100 ng/ml mSCF. (a) 5FU-Id1 immortalized BMC cloned cell lines (Cl.1, Cl.2, Cl3) and parent cells were examined for Id1 protein expression by western blot. BMC transfected with MSCV-GFP were used as control. Exogenous Id1 protein, tagged with HA, and endogenous Id1 are indicated, respectively. (b) Morphology of Wright–Giemsa stained 5FU-Id1 cloned cells; × 1000 magnification. (c) Cloned cells were analysed by flow cytometry for the expression of hematopoietic developmental surface markers. Isotype control antibodies were used as negative controls to exclude nonspecific background staining. Percentages of positive cells are indicated in each quadrant. (d) Reverse transcriptase (RT)-PCR products for hematopoietic transcriptional regulators (PU.1, C/EBPα, GATA-2, GATA-1) in 5FU-Id1-cloned cell lines were shown. (e) Proliferation of 5FU-Id1 cell lines indicated by 3H-thymidine incorporation in the presence of the indicated cytokine. Bars indicate average 3H-thymidine incorporation in c.p.m. × 103 over 16 h as measured from triplicate wells ± s.e.m. (f, g) For induction of myeloid differentiation, cloned cells were cultured with hG-CSF (100 ng/ml), mGM-CSF (100 ng/ml) or hM-CSF (100 ng/ml) for 5 days. Morphology and flow cytometry analysis of myeloid cell surface markers (Gr-1, Mac-1) on 5FU-Id1 Cl.2 cells after culture for 5 days in the indicated cyto were shown (Wright–Giemsa, × 1000 magnification).
Overexpression of Id1 in bone marrow cells induces a myeloproliferative disease in mice
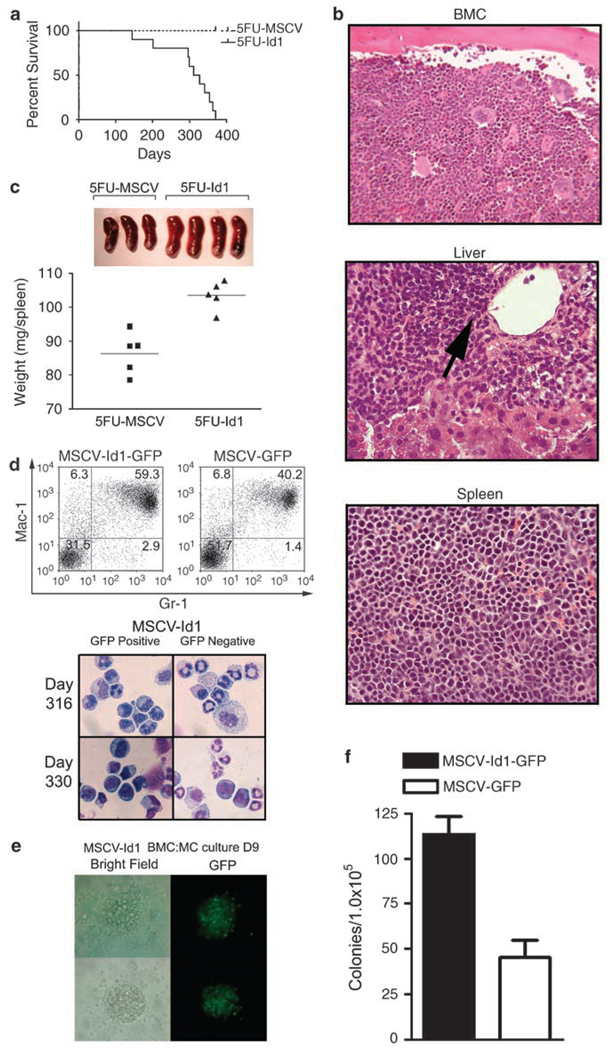
Our results demonstrate that overexpression of Id1 in normal hematopoietic progenitors leads to immortalization of CMP-like cells in vitro, which suggests that overexpression of Id1 in vivo could result in a myeloproliferative disorder. Though we and another group had previously observed an increased myeloid development in the bone marrow (BM) and peripheral blood (PB) of mice transplanted with Id1-transduced BMC (Buitenhuis et al., 2005; Leeanansaksiri et al., 2005), these mice were not further monitored to determine if they might have increased myeloid proliferation and progress to an MPD or an AML. Therefore, we transplanted mice with 5FU-Id1 and 5FU-MSCV BMC to evaluate their survival over the course of 1 year. All mice transplanted with 5FU-Id1 BMC became moribund and died in 1 year, whereas mice transplanted with mouse stem cell virus (MSCV)-transduced BMC survived (Figure 3a). Histopathological analysis of sick or moribund mice showed myeloid and erythroid hyperplasia in BM and erythroid hyperplasia in spleen (Figure 3b). In addition, myeloid cell infiltration was observed in the liver of sick animals. Furthermore, spleens of the 5FU-Id1 BMC transplant recipients were significantly enlarged (5FU-MSCV: 86.4 ± 2.8, 5FU-Id1: 103.6 ± 1.9 mg, P < 0.01; Figure 3c) and there was evidence of extramedullary hematopoiesis including increased numbers of Gr-1+/Mac-1+ granulocytes, and Gr-1−/Mac-1+ macrophages in the spleens of these animals (data not shown). Complete blood counts of PB cells from sick mice showed consistent monocytosis and low hemoglobin and hematocrit levels. Furthermore, one mouse showed leukocytosis with blasts in the PB, a feature of chronic myelogenous leukemia in blast crisis (Supplementary Table S1). Increased numbers of immature myeloid cells were detected in BMC of sick mice transplanted with 5FU-Id1 BMC after 316 and 330 days by flow cytometry and cytocentrifuge preparations (Figure 3d), and these BMC contained increased numbers of progenitors that could give rise to GFP-positive colonies in vitro (Figures 3e and f). Based on observations from recipient mice transplanted with 5FU-Id1 BMC, and the diagnostic criteria of hematologic diseased mouse models (Kogan et al., 2002), these mice were diagnosed with an MPD. In addition, cell lines with a CMP-like phenotype, similar to those derived from direct infection of BM, could be readily established by subculturing BMC from these sick mice in the media containing SCF (data not shown). Mice transplanted with BM or spleen cells from 5FU-Id1 BMC sick mice did not develop acute myeloid disease in secondary recipients, suggesting that Id1 overexpression alone is not sufficient to give rise to a transplantable leukemia in vivo. Taken together, these data demonstrated that overexpression of Id1 drives the expansion of early myeloid progenitors that differentiate in vivo, which results in the development of an MPD.
Figure 3.
Long-term survival of recipient mice and characterization of transplanted Id1 overexpressing bone marrow cells (BMC). Lethally irradiated 8- to 12-week-old female C57BL/6 (CD45.1) mice (n = 12 for each group) were transplanted with 1 × 106 5FU-Id1 or 5FU-MSCV BMC and monitored. (a) Percent survival over time was shown with survival curve. Day 0 is the time of transplantation initiation. All mice with Id1 overexpressing hematopoietic cells (5FU-Id1) died within 12 months after injection, whereas control (5FU-MSCV) recipient mice survived. (b) Moribund animals were euthanized and BM, liver and spleen were prepared for histopathological examination. Arrow indicates extramedullary myeloid proliferation in recipient liver (H&E stain, × 400). (c) Gross morphology of transplant recipient spleens (upper panel) and wet weight of spleens 6 months after transplantation (P < 0.05). (d) BMC that express pMSCV-Id1-GFP or pMSCV-GFP from recipient mice were analysed for Gr-1/Mac-1 expression and prepared for morphologic examination. Giemsa stained cytospins from bone marrow sorted for green fluorescent protein (GFP)-positive (5FU-Id1; donor) and GFP-negative (host) cells, 316–330 days after transplantation; × 1000 magnification. (e) GFP-positive donor cells (5FU-Id1) were cultured in methylcellulose to demonstrate colony forming units (CFU) potential. For MSCV-Id1-GFP BMC methylcellulose colony assays, cells were plated in 1.1% methylcellulose medium supplemented with 10% fetal calf serum (FCS), mSCF, mGM-CSF, mIL-3 at same concentrations as described in Materials and methods. Shown are two colonies representative of multiple colonies in bright field (left panels) and green fluorescence (right panels). (f) CFU were scored and data are presented as CFU per 1 × 105 BMC ± s.e.m. of triplicate plates.
Id1 expression is upregulated during human myeloid differentiation and is expressed in human AML cell lines and AML patient bone marrow
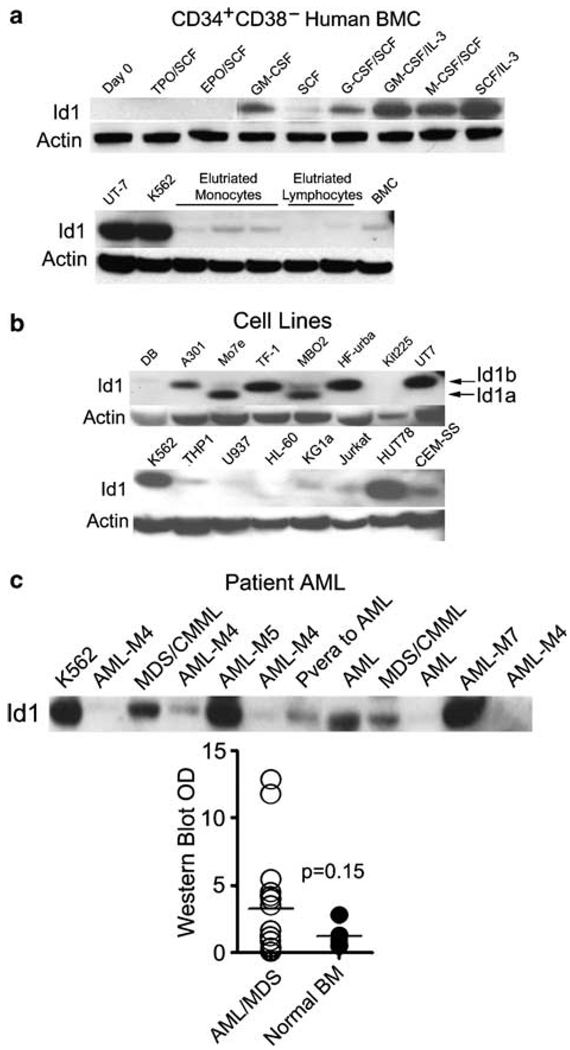
Human MPD or myelodysplastic syndrome (MDS) often precedes AML. Therefore, to evaluate if Id1 overexpression correlates with development of human leukemia, we examined Id1 expression in AML cell lines and primary AML or MDS patient samples. We previously found that Id1 expression is upregulated in murine BMC by cytokines that promote myeloid development (Leeanansaksiri et al., 2005). To evaluate if Id1 expression is also upregulated during human myeloid development, we purified human CD34+CD38− BMC from healthy BM, and cultured these cells in various cytokines. Id1 protein is expressed at low levels in unfractionated human BMC, and not detected in freshly isolated (day 0) CD34+CD38− cells, or CD34+CD38− cells cultured for 6 days in SCF/EPO or SCF/TPO as measured by western blotting, suggesting that Id1 is not induced during the early stages of erythroid or megakaryocyte development in these cultures (Figure 4a, upper panel). In comparison, Id1 protein expression was upregulated in CD34+CD38− cells that were cultured for 6 days in cytokines that promote myeloid development including GM-CSF, G-CSF/SCF, GM-CSF/IL-3, M-CSF/SCF and SCF/IL-3. These cultures contained many immature myeloid cells as well as granulocytes and macrophages at all stages of development by examination of Wright–Giemsa stained cytocentrifuge preparations (data not shown). Thus, Id1 protein expression increases during the early stages of human myeloid progenitor development, as it does in mouse hematopoietic cells.
Figure 4.
Evaluation of Id1 protein expression in human bone marrow cells (BMC), human leukemic cell lines and primary acute myelogenous leukemia (AML). (a) Human BMC were thawed and stained with directly conjugated fluorescent antibodies to human CD34-FITC (Clone 581), human CD38-PE (Clone HIT2; BD Biosciences) and then sorted for CD34+/CD38−. Purified CD34+CD38− human BMC were stimulated in complete Iscove’s modified Dulbecco’s medium (cIMDM) with or without cytokines for 6 days prior to harvest and lysis for western blotting (upper panel). Human monocyte and lymphocyte populations were elutriated and analysed for Id1 protein expression. UT-7 and K562 cells were used as positive controls (lower panel). (b) Human hematopoietic cell line lysates were prepared to measure the expression level of Id1 protein. Total cell lysates were separated by SDS–polyacrylamide gel electrophoresis (PAGE) and western blotted to detect Id1. Subsequently, the blots were stripped and stained with anti-actin antibody to demonstrate equal loading. (c) Protein (30 µg) extracted from thawed BM mononuclear cells (MNC) of AML patients and K562 cells were analysed by western blotting for the presence of Id1 protein. Id1 protein levels were evaluated semiquantitatively by measuring the optical density (OD) of Id1 bands compared to that present in K562 (arbitrary value = 10). OD was measured by Image J image analysis software (Image J 1.37V, National Institute of Health, MD, USA; http://rsb.info.nih.gov/ij). The graph shows the distribution of Id1 protein expression levels in 20 patients with AML/MDS and 4 with normal BM MNC. Data are presented as individual values (circles) and medians (horizontal bars; P = 0.15; Student’s t-test).
Next, we measured the levels of Id1 expression in human leukemic cell lines by western blot analysis. Id1 was expressed in a number of myeloid cell lines including K562, UT-7, TF-1, MBO2, KG1a, THP-1, but not in the U937 or HL-60 leukemia cell lines, demonstrating that Id1 is constitutively expressed in some, though not all, myeloid leukemia cell lines (Figure 4b). In comparison, Id1 is expressed at low levels in unfractionated BMC and purified populations of PB mononuclear cells (monocytes and lymphocytes; Figure 4a, lower panel), indicating that relatively high levels of constitutive Id1 expression are maintained in some myeloid cell lines. Furthermore, we detected both forms of Id1, Id1a (149 amino acids) and Id1b (156 amino acids), in some of leukemic cell lysates, however, to date no functional difference has been reported for Id1a and Id1b isoforms. In addition, Id1 was expressed in some of the T-cell lines (Jurkat, HUT78, CEM-SS and A301), and in one B-cell line (HF-Urba; Figure 4b). Collectively, these results suggest that Id1 may be expressed in leukemic patients’ samples and involved in the pathogenesis of leukemia. Therefore, we examined Id1 expression in AML or MDS patients’ samples by western blot analysis, and measured Id1 protein expression with optical densitometry (OD). There were two distinct populations in patients group based on Id1 expression. Id1 was expressed at higher levels in 12 of 18 AML (patients diagnosed with AML-M1, -M2, -M4, -M5 and -M7) and 2 out of 2 MDS BMC compared to normal BMC (Figure 4c), suggesting that Id1 protein is overexpressed in some, but not all myeloid malignancies.
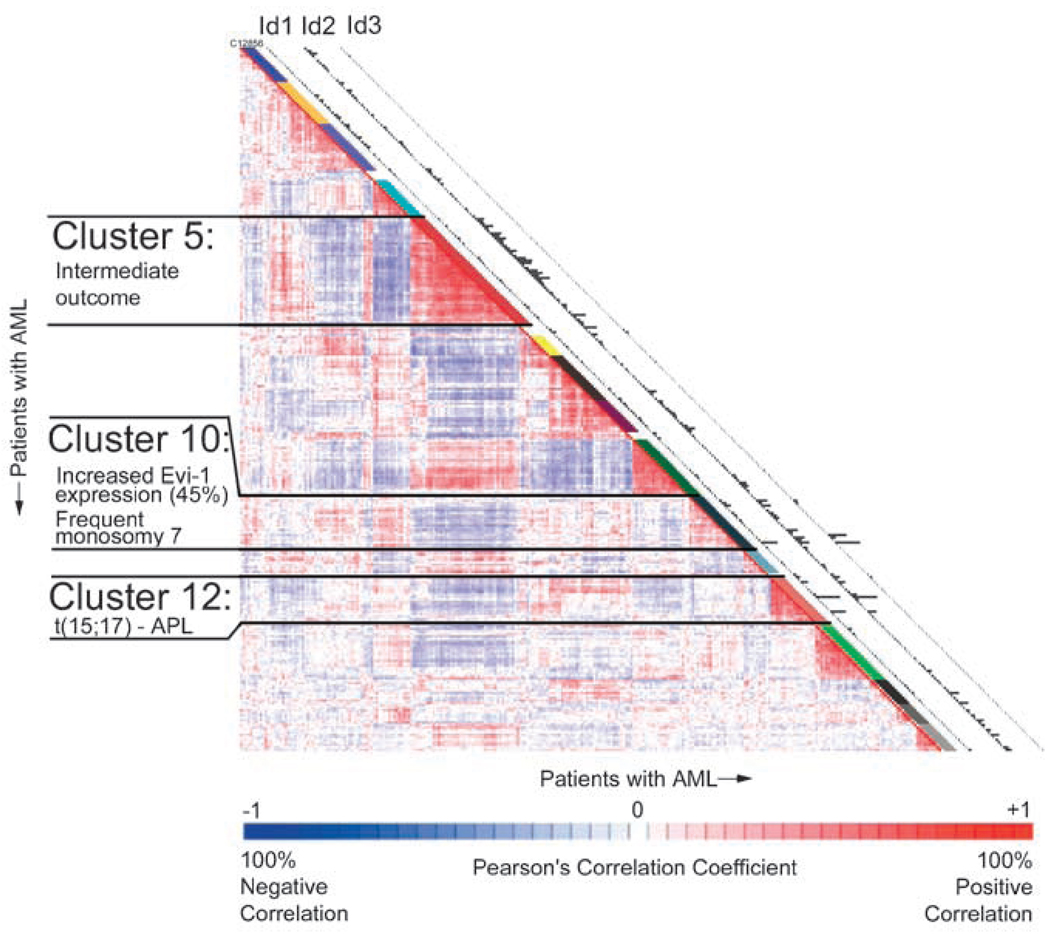
To further study the relationship between human leukemic cells and Id1 expression, we analysed a cohort of 285 cases of primary AML for Id1, Id2 and Id3 gene expression that had been previously profiled for gene expression (Valk et al., 2004; Bullinger and Valk, 2005). We found that within the 285 patient set, Id1 was expressed in 50 and Id2 was expressed in 55 of the patient AML samples, whereas the expression of Id3 was rare. Among 50 patients samples expressing Id1, 19 samples also expressed Id2 (Supplementary Table S2). Therefore, Id3 was not further analysed. Increased Id1 and Id2 expression levels were evenly distributed in French–American–British subtypes M1–M6 in this AML cohort (Table 1). However, 61 and 50% of patients with 5q and/or 7q deletion, and 28 and 22% of patients with t(15;17) showed increased expression of Id1 and Id2, respectively, suggesting that elevated Id1 expression may contribute to a block in granulocyte differentiation (Table 1). In addition, a significant number of AML patients with a normal karyotype showed increased Id1 (17%) and/or Id2 (14%) expressions. Id1 expression was not correlated with any specific cluster, whereas Id2 expression was highly represented in clusters 5, 10 and 12 among the 16 pre-established AML clusters in this cohort (Figure 5). Among 78 patients, 23 AML patients having FLT3-ITD (29%) and 8 AML patients among 23 patients having EVI1 (35%) showed increased level of Id1 expression, whereas increased Id2 expression level correlated with NRAS (30%), KRAS (44%), CEBPA (6%) and EVI1(30%) expression, suggesting that Id gene expression may be induced by downstream signal transduction pathways of activated oncogenes in these AML. Additional genes (32) were identified as being positively correlated with the expression of Id1 and Id2, and 4 genes were negatively correlated (Supplementary Table S3). Also, no CEBPA mutations were found in cases of AML with increased Id1. Collectively, Id1 is constitutively expressed in some AML or MDS patients’ samples, and may contribute to the pathogenesis of AML or MDS in human.
Table 1.
Characteristics of AML patients expressing Id
| No. | No. of Id1 positive (%) |
No. of Id2 positive (%) |
No. of Id3 positive (%) |
|
|---|---|---|---|---|
| Sex | ||||
| Male | 137 | 23 (17) | 21 (15) | 3 (2) |
| Female | 148 | 27 (18) | 34 (23) | 1 (1) |
| Age (year) | ||||
| < 35 | 76 | 19 (25) | 12 (16) | 2 (3) |
| 35–60 | 177 | 28 (16) | 36 (20) | 1 (1) |
| > 60 | 32 | 3 (9) | 7 (21) | 1 (3) |
| WBC count (× 109 per liter) | ||||
| ≤ 20 | 113 | 18 (16) | 29 (26) | 4 (4)* |
| > 20 | 157 | 32 (20) | 26 (17) | 0 (0) |
| FAB | ||||
| M0 | 6 | 1 (17) | — | — |
| M1 | 57 | 11 (19) | 15 (26) | — |
| M2 | 59 | 10 (17) | 9 (15) | — |
| M3 | 18 | 3 (17) | 2 (11) | — |
| M4 | 46 | 8 (17) | 9 (19) | 2 (3) |
| M5 | 64 | 7 (11) | 15 (23) | 1 (2) |
| M6 | 3 | 1 (33) | 1 (33) | — |
| Not determined | 32 | 9 (28) | 4 (12) | 1 (3) |
| Cytogenetic abnormalities | ||||
| t(11q23) | 3 | — | — | — |
| −5/7(q) | 18 | 11 (61)* | 9 (50)** | 3 (17)* |
| +8 | 16 | 3 (19) | 4 (25) | — |
| −9q | 3 | — | 1 (33) | — |
| Inv(16)/t(16:16) | 19 | 2(11) | 2 (11) | — |
| t(15;17) | 18 | 5 (28) | 4 (22) | — |
| t(6;9) | 4 | 1 (25) | — | — |
| t(8;21) | 22 | 2 (9) | 1 (4.5) | — |
| t(9;22) | 1 | 1 (100) | 1 (100) | — |
| Normal karyotype | 121 | 20 (17) | 17 (14) | — |
| Complex (> 3 chromosome aberrations) | 7 | 1 (14) | 3 (43) | — |
| Other abnormal karyotypes | 34 | 2 (6) | 9 (27) | — |
| Failure | 4 | 1(25) | 1 (25) | — |
| Not determined | 10 | 1 (10) | 1 (10) | 1 (10) |
| Molecular abnormalities | ||||
| FLT3-ITD | 78 | 23 (29)** | 7 (9)** | — |
| FLT3-TKD | 33 | 7 (21) | 7 (21) | — |
| NRAS | 26 | 4 (15) | 8 (30)* | — |
| KRAS | 9 | — | 4 (44) | — |
| CEBPA | 17 | — | 1 (6) | — |
| EVI1 | 23 | 8 (35)* | 7 (30) | 3 (13)*** |
Abbreviation: FAB, French–American–British subtypes. P-values calculated using χ2-test,
P = 0.05–0.01,
P = 0.01–0.001,
P < 0.001.
Figure 5.
Microarray analysis from 285 acute myelogenous leukemia (AML) patients. The correlation view displays pairwise correlations between AML patients. The 16 clusters identified in the cohort of 285 AML patients using 2856 probe sets on the basis of the correlation view. Clinical and molecular data are depicted in the columns along the original diagonal of the correlation view. The expression levels of Id1, Id2, Id3 (probe set: 208937_s_at, 201565_s_at, 207826_s_at 209543_s_at for each) in the 285 AML patients are plotted in the column (bars are proportional to the level of mRNA expression).
Id1 expression is required for Mo7e cell growth in vitro
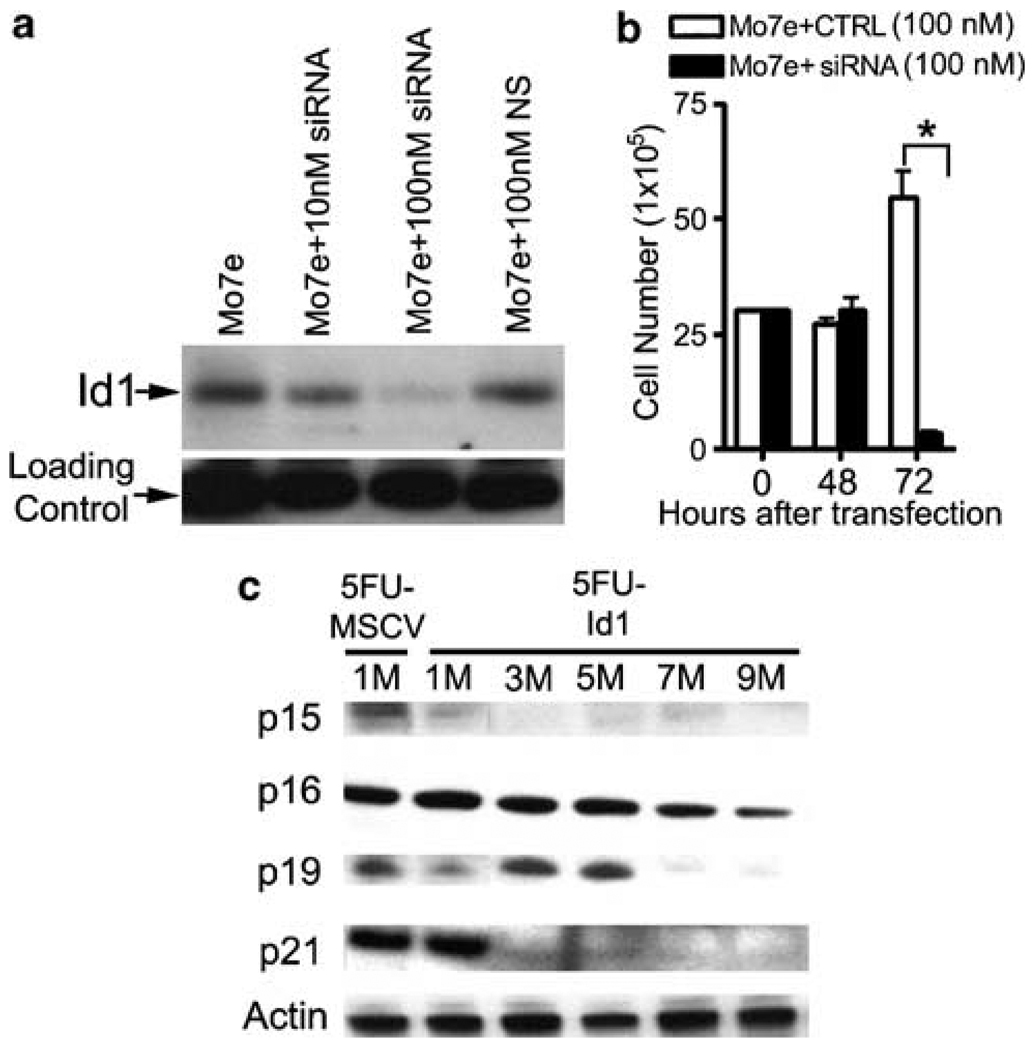
Based on the data that some human AML cell lines and AML patients samples show constitutively high levels of Id1 expression, we hypothesized that Id1 might contribute to the deregulated growth of human AML cells. Therefore, we knocked down Id1 expression in Mo7e cells using siRNA, and then measured cell growth in vitro by determining viable cell number with trypan blue exclusion method. At 48 h after electroporation, Id1 protein expression was significantly reduced in Mo7e cells treated with increasing amounts of Id1 siRNA, but not in those cells treated with control siRNA (Figure 6a). Total viable cell growth of Mo7e cells was also decreased by more than 90% compared to the cells transfected with nonspecific siRNA 72 h after transfection (Mo7e + CTRL: 54.67 ± 5.4, Mo7e + siRNA: 3.37 ± 0.5 × 105 cells, *P < 0.01; Figure 6b). Thus, Id1 expression is required to maintain the pathologic hyperproliferation of Mo7e cells, and Id1 may represent a valid molecular target to inhibit the growth of Id1 overexpressing AML.
Figure 6.
Effect of Id1 on myeloid leukemic cell line proliferation and expression of cell cycle regulators in Id1 overexpressing bone marrow cells (BMC). (a) Mo7e cells were electroporated with either Id1 siRNA oligo or control oligo with designated concentration in 100 µl buffer. Immediately following electroporation, cells were seeded at a concentration of 1 × 106 cells per ml in culture containing RPMI 1640, 10% fetal calf serum (FCS), 20 ng/ml granulocyte monocyte colony-stimulating factor (GM-CSF) and 100 ng/ml stem cell factor (SCF). Id1 protein knockdown was confirmed by western blotting 48 h after electroporation. (b) The viable cell number was determined each day over a 3-day period by trypan blue exclusion (*P < 0.01). (c) Protein levels of cell cycle regulators in 5FU-Id1 BMC were measured by western blotting. 5FU-treated Id1 overexpressing BMC were maintained with CYTOKINES (SCF + Flt3-L + IL-6 + TPO) over 9 months (M), and cells were harvested at the times indicated. Western blot of the total cellular extracts from the cells was probed with p15, p16, p19 and p21 antibodies. The blot was stripped and reprobed with anti-actin antibody to verify equal protein loading.
Cdk inhibitor expression is reduced during Id1-mediated immortalization of hematopoietic progenitor cells
We found that overexpression of Id1 leads to the immortalization of SCF-dependent CMP-like cells (5FU-Id1 clones). To investigate the mechanism(s) by which Id1 immortalizes hematopoietic progenitors, we evaluated the expression levels of the cyclin-dependent kinase (Cdk) inhibitors, which have been shown to be decreased by Id1 in other cell systems (Prabhu et al., 1997; Pagliuca et al., 2000; Alani et al., 2001). To do this, cell lysates from bulk cultures of 5FU-Id1 or 5FU-MSCV were obtained, and compared p15, p16, p19 and p21 expressions over time. However, as 5FU-MSCV BMC did not proliferate after 1 month, the expression level of p15, p16, p19 and p21 in 5FU-Id1 BMC was compared to 1 month 5FU-MSCV BMC. The expressions of p15, p16, p19 and p21 were reduced or lost during Id1-mediated immortalization of SCF-dependent BMC. Specifically, p15, p21 and p19 expression were not detected after 1, 3 and 7 months, respectively, and p16 was reduced to low levels after 9 months (Figure 6c). Thus, reduced Cdk inhibitor expression could contribute to the immortalization of BMC progenitors. However, additional studies will be required to determine if these effect are direct, and if other pathways are involved.
Discussion
Id genes normally function to inhibit the differentiation and enhance cell proliferation (Jen et al., 1992; Kreider et al., 1992; Alani et al., 1999). Thus, deregulation of Id gene expression could readily contribute to cancer initiation or promotion by disrupting these normal cellular processes. In this report, we show that constitutive Id1 gene expression immortalizes murine hematopoietic progenitors in vitro, and promotes an MPD in vivo suggesting that deregulated Id gene expression may contribute to human AML, because MPD often precedes AML. In support of this hypothesis, we found that Id1 protein levels were elevated in human myeloid cell lines and AML patient samples in comparison to normal BM and PB. In addition, we found that AML patients expressing Id1 were highly represented in a cohort of 285 AML patient samples, further indicating that deregulated Id1 genes may contribute to the progression or the initiation of AML.
In previous studies, T-cell hyperplasia and T-cell lymphomas have been observed in transgenic mouse models that express high levels of Id1 in developing T cells (Kim et al., 1999). Because this phenotype is also observed in E2A−/− mice, deregulated Id gene expression is considered to contribute to the development of T-cell lymphoma by inhibiting the transcriptional activity of E proteins, or E protein silencing (Bain et al., 1997). In comparison, we found that mice transplanted with Id1 overexpressing BMC do not develop lymphomas, but rather an MPD. We have shown that long-term repopulating myeloid cells express significantly higher levels of Id1 than T cells in mice transplanted with Id1 overexpressing HSC (Leeanansaksiri et al., 2005). Therefore, it is likely that T cells with the highest levels of Id1 would undergo apoptosis, whereas T cells that express lower levels of Id1 would survive and repopulate in the host. We speculate that the difference between the lymphoma development in Id1 transgenic mouse model and MPD in our transplantation model may be due to the levels of Id1 expression. E protein silencing has also been implicated in myeloid hyperplasia and malignancies resulting from deregulated expression of SCL/TAL-1, a bHLH transcription factor that binds E2A, and AML1-ETO fusion proteins produced by t(8;21) (O’Neil et al., 2004; Zhang et al., 2004). Specifically, deregulated expression of SCL/TAL-1 in HSC/HPC leads to increased myeloid proliferation in vitro and in vivo, suggesting that SCL/TAL-1 and Id1 may function similarly to induce myeloid hyperplasia (Kunisato et al., 2004). AML1-ETO also induces a myeloid leukemia via E protein silencing by a mechanism distinct from Id proteins, which involves in aberrant cofactor exchange (Zhang et al., 2004). Thus, E protein silencing may represent a common mechanism that contributes to myeloid hyperplasia and disease.
Acute leukemia is characterized by enhanced proliferation and survival with impaired differentiation, and develops as a result of a multistep process involving several gene mutations. To date, no single mutation is sufficient to cause acute leukemia in humans (Grisolano et al., 1997; He et al., 1997; Castilla et al., 1999; Higuchi et al., 2002). We found that mice transplanted with 5FU-Id1 BMC show an increase in the percentage of immature donor (GFP+) myeloid cells in BM, and develop an MPD after a long latency, suggesting that Id1 overexpression sustains active cell cycling, expands the primitive progenitor compartment, but does not prevent differentiation in vivo in which hematopoietic cells are continuously stimulated to differentiate by extrinsic signals. Consistent with this observation, we found that cloned Id1 overexpressing CMP-like immortalized cells could be induced differentiate into neutrophils and macrophages when cultured in GM-CSF, G-CSF or M-CSF, demonstrating these progenitors were not completely blocked in differentiation. Thus, the ability of Id1 to prevent hematopoietic progenitor differentiation can be overridden by specific myeloid growth factors including G-CSF, GM-CSF and M-CSF, suggesting that the BM microenvironment may contribute to the leukemogenic process in vivo.
No chromosomal translocations, deletions or mutations involving Id1 gene, have been identified to date. However, in a separate study to identify the molecular signature of stem cells in patients with MDS, which also progress to AML in some cases, Id1 gene expression was upregulated in purified human CD34+CD38+Thy-1+ stem cells from MDS patients with 5q deletion compared to normal stem cells, suggesting that Id genes may contribute to the enhanced proliferation of 5q-MDS stem/progenitor cells (Nilsson et al., 2007). Id genes are induced by growth factors including VEGF, IGF1 and cytokines that activate the RAS-mitogen-activated protein kinase and phosphatidylinositol 3-kinase signal transduction pathways, and bone morphogenetic protein (BMP)2/4 and transforming growth factor-β (TGF-β) that activate SMAD signal transduction pathways, suggesting that oncogenic activation of these signal transduction pathways could result in deregulated Id gene expression and contribute to cancer (Hollnagel et al., 1999; Lyden et al., 2001; Belletti et al., 2002; Kang et al., 2003). We found that AML patients’ BMC-expressing Id1 are correlated with activating FLT3 mutations, elevated NRAS, and Evi-1 gene expression, which could lead to increased Id1 expression in these patients. Although the precise molecular mechanism(s) by which deregulated Id gene expression contributes to MPD and tumor progression are not completely known, deregulated Id gene expression can affect key cell proliferation pathways, which lead to enhanced cell proliferation (Prabhu et al., 1997; Pagliuca et al., 2000; Alani et al., 2001). We found that the levels of p15, p16, p19 and p21 were decreased during the immortalization of CMP-like BM progenitors. These changes occurred over a long period in culture, suggesting these effects might be indirect.
In summary, using a murine BMC transplantation model and cell line models, we provide evidence that deregulated Id1 expression results in an MPD. Furthermore, we found that Id1 protein expression is upregulated in AML and MDS patient samples, and knock down of Id1 expression inhibits AML cell line growth. Collectively, these results suggest that Id1 may represent a potential therapeutic target for early stage intervention in the treatment of hematopoietic malignancy.
Materials and methods
Mice
C57BL/6-CD45. 1, C57BL/6-CD45.2 mice (8- to 12-week old) were obtained from the animal production area at Charles Rivers Laboratories, NCI-Frederick (Frederick, MD, USA). Animal care was provided in accordance with the procedures outline in the ‘Guide for Care and Use of Laboratory Animals’ (National Institute of Health, Bethesda, MD, USA, 1996).
Retroviral transduction, transplantation and FACS analysis
Recombinant MSCV biscistronic retroviral vectors expressing murine Id1 and GFP were generated, transduced, sorted as previously described (Suh et al., 2006). GFP+ cells were cultured in vitro or transplanted to 8- to 12-week-old recipient mice (C57BL/6-CD45.1). Repopulation of donor BMC was determined by analysing GFP+ cells in the PB or BM using FACScan (BD Biosciences, San Diego, CA, USA). BD CELLQuest software was used for fluorescence-activated cell sorting (FACS) analysis. All lineage specific antibodies and Tri-color streptavidin were from BD Biosciences. Gr-1, c-Kit, B220 and TER119 were PE-conjugated antibodies, whereas Sca-1, Mac-1, CD3 and CD71 were biotin-conjugated antibodies.
Soft agar colony assays and 3H-thymidine incorporation assay
Myeloid colony assay was performed as described (Suh et al., 2006). To measure proliferation, transduced cells were plated at 5 × 103 cells per well per 100 µL in triplicate in a 96-well plate (Corning, Corning, NY, USA). Cells were pulsed with 1 µCi of 3H-thymidine per well (6.7 Ci/mmol, PerkinElmer Life Sciences Inc. Boston, MA, USA), and harvested onto a glass-fiber filtermats (Filtermat A; Wallac Oy, Turku, Finland). Incorporated 3H-thymidine was counted by 1205 BetaPlate liquid scintillation counter (LKB Wallac, PerkinElmer Life Sciences Inc.).
Human hematopoietic cells and cell lines
DB, HF-urba (B cell); A301, Jurkat, kit225, HUT78, CEM-SS (T cell); K562 (erythroleukemia); HL60 (promyelocytic cell); MBO2, Mo7e, TF-1, UT7 (GM-CSF-dependent myeloid cell); THP1, U937 (myelomonocytic cell); KG1a (myeloblast cell) were chosen to examine the expression of Id1 protein. All cell lines were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) with 10% fetal calf serum (Atlanta Biologicals, Atlanta, GA, USA) and P/S (Invitrogen; except UT-7 cells in cIMDM) with/without additional cytokines required for growth at 37 °C, with 5% CO2. Cryopreserved BM or PB cells from patients were used to check the expression of Id1 protein (18 AML, 2 MDS). All patients provided written informed consent according to The Johns Hopkins Medical Institutional Review Boards and guidelines.
Id1 knockdown with siRNA
siRNA duplexes directed against the human Id1 RNA sequence (NM_002165) were synthesized by Dharmacon Inc. (Lafayette, CO, USA) using the Dharmacon SMART technology software. Four SMART-selected siRNA duplexes were synthesized and provided in a single pool (SiGENOME SMARTpool catalogue no.: M-005051-00). Nonspecific sense strands were used as negative control (SiCONTROL nontargeting siRNA Pool; D-001206-13-05). siRNAs were mixed with ‘Human CD34+ cell Nucleofactor’ solution and Mo7e human leukemic cells were transfected using the Nucleofactor I transfection device according to the protocol (program: U-008) provided by Amaxa Inc. (Gaithersburg, MD, USA).
Western blot analysis
Denatured cell lysates were loaded on gradient SDS–polyacrylamide gel electrophoresis (Invitrogen) and transferred to Immobilon-P membranes (Millipore, Billerica, MA, USA). The membranes were blocked at room temperature in 5% milk/Tris-buffered saline Tween solution, and then probed with anti-mouse Id1 (sc-488), anti-mouse p15 (sc-611), p16 (sc-1207), p21 (sc-6246) or anti-mouse α-actin antibodies (sc-8432; all from Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) or anti-mouse p19 (ab 80–100; Abcam, Cambridge, MA, USA).
RT–PCR
Total RNA was purified using RNeasy (Qiagen, Valencia, CA, USA) and reverse transcribed to cDNA with Moloney murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, CA, USA). The PCR was conducted using Ampli-Taq DNA polymerase (PerkinElmer Life Sciences Inc.). PCR amplification cycles consisted of 30 s at 94 °C, 30 s at 58 °C and 1 min at 72 °C, 40 cycles. The primer sets used were as given in Supplementary Table S4.
Supplementary Material
Acknowledgements
We gratefully acknowledge the technical support of Steve Stull, Kathleen Noer, Roberta Matthai and Samantha Bauchiero. We also thank Dr Nancy Colburn, Dr Sandra Ruscetti and Dr Kristbjorn Gudmundsson for their critical review of this article. This project has been funded, in part, with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number NO1-CO-12400. This research was supported, in part, by the Intramural Research Program of NIH, National Cancer Institute, Center for Cancer Research. Clinical specimens were provided by the Sidney Kimmel Cancer Center at Johns Hopkins Tumor and Cell Procurement Bank, supported by the Regional Oncology Research Center Grant no. 5 P30 CA06973.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Akashi K, He X, Chen J, Iwasaki H, Niu C, Steenhard B, et al. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood. 2003;101:383–389. doi: 10.1182/blood-2002-06-1780. [DOI] [PubMed] [Google Scholar]
- Akashi K, Reya T, Dalma-Weiszhausz D, Weissman IL. Lymphoid precursors. Curr Opin Immunol. 2000a;12:144–150. doi: 10.1016/s0952-7915(99)00064-3. [DOI] [PubMed] [Google Scholar]
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000b;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Alani RM, Hasskarl J, Grace M, Hernandez MC, Israel MA, Munger K. Immortalization of primary human keratinocytes by the helix-loop-helix protein, Id-1. Proc Natl Acad Sci USA. 1999;96:9637–9641. doi: 10.1073/pnas.96.17.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani RM, Young AZ, Shifflett CB. Id1 regulation of cellular senescence through transcriptional repression of p16/Ink4a. Proc Natl Acad Sci USA. 2001;98:7812–7816. doi: 10.1073/pnas.141235398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Engel I, Robanus Maandag EC, Te Riele HP, Voland JR, Sharp LL, et al. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belletti B, Drakas R, Morrione A, Tu X, Prisco M, Yuan T, et al. Regulation of Id1 protein expression in mouse embryo fibroblasts by the type 1 insulin-like growth factor receptor. Exp Cell Res. 2002;277:107–118. doi: 10.1006/excr.2002.5542. [DOI] [PubMed] [Google Scholar]
- Buitenhuis M, Van Deutekom HW, Verhagen LP, Castor A, Jacobsen SE, Lammers JW, et al. Differential regulation of granulopoiesis by the basic helix-loop-helix transcriptional inhibitors Id1 and Id2. Blood. 2005;105:4272–4281. doi: 10.1182/blood-2004-12-4883. [DOI] [PubMed] [Google Scholar]
- Bullinger L, Valk PJ. Gene expression profiling in acute myeloid leukemia. J Clin Oncol. 2005;23:6296–6305. doi: 10.1200/JCO.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Castilla LH, Garrett L, Adya N, Orlic D, Dutra A, Anderson S, et al. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat Genet. 1999;23:144–146. doi: 10.1038/13776. [DOI] [PubMed] [Google Scholar]
- Cooper CL, Brady G, Bilia F, Iscove NN, Quesenberry PJ. Expression of the Id family helix-loop-helix regulators during growth and development in the hematopoietic system. Blood. 1997;89:3155–3165. [PubMed] [Google Scholar]
- Fong S, Debs RJ, Desprez PY. Id genes and proteins as promising targets in cancer therapy. Trends Mol Med. 2004;10:387–392. doi: 10.1016/j.molmed.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Grisolano JL, Wesselschmidt RL, Pelicci PG, Ley TJ. Altered myeloid development and acute leukemia in transgenic mice expressing PML-RAR alpha under control of cathepsin G regulatory sequences. Blood. 1997;89:376–387. [PubMed] [Google Scholar]
- He LZ, Tribioli C, Rivi R, Peruzzi D, Pelicci PG, Soares V, et al. Acute leukemia with promyelocytic features in PML/RARalpha transgenic mice. Proc Natl Acad Sci USA. 1997;94:5302–5307. doi: 10.1073/pnas.94.10.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, O’Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1:63–74. doi: 10.1016/s1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem. 1999;274:19838–19845. doi: 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- Ishiguro A, Spirin K, Shiohara M, Tobler A, Norton JD, Rigolet M, et al. Expression of Id2 and Id3 mRNA in human lymphocytes. Leuk Res. 1995;19:989–996. doi: 10.1016/0145-2126(95)00084-4. [DOI] [PubMed] [Google Scholar]
- Jen Y, Weintraub H, Benezra R. Overexpression of Id protein inhibits the muscle differentiation program: in vivo association of Id with E2A proteins. Genes Dev. 1992;6:1466–1479. doi: 10.1101/gad.6.8.1466. [DOI] [PubMed] [Google Scholar]
- Kang Y, Chen CR, Massague J. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell. 2003;11:915–926. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- Kim D, Peng XC, Sun XH. Massive apoptosis of thymocytes in T-cell-deficient Id1 transgenic mice. Mol Cell Biol. 1999;19:8240–8253. doi: 10.1128/mcb.19.12.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan SC, Ward JM, Anver MR, Berman JJ, Brayton C, Cardiff RD, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in the mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Koschmieder S, Rosenbauer F, Steidl U, Owens BM, Tenen DG. Role of transcription factors C/EBPalpha and PU.1 in normal hematopoiesis and leukemia. Int J Hematol. 2005;81:368–377. doi: 10.1532/ijh97.05051. [DOI] [PubMed] [Google Scholar]
- Kreider BL, Benezra R, Rovera G, Kadesch T. Inhibition of myeloid differentiation by the helix-loop-helix protein Id. Science. 1992;255:1700–1702. doi: 10.1126/science.1372755. [DOI] [PubMed] [Google Scholar]
- Kunisato A, Chiba S, Saito T, Kumano K, Nakagami-Yamaguchi E, Yamaguchi T, et al. Stem cell leukemia protein directs hematopoietic stem cell fate. Blood. 2004;103:3336–3341. doi: 10.1182/blood-2003-06-1935. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Uo T, Iavarone A. Id proteins at the cross-road of development and cancer. Oncogene. 2001;20:8326–8333. doi: 10.1038/sj.onc.1205093. [DOI] [PubMed] [Google Scholar]
- Leeanansaksiri W, Wang H, Gooya JM, Renn K, Abshari M, Tsai S, et al. IL-3 induces inhibitor of DNA-binding protein-1 in hemopoietic progenitor cells and promotes myeloid cell development. J Immunol. 2005;174:7014–7021. doi: 10.4049/jimmunol.174.11.7014. [DOI] [PubMed] [Google Scholar]
- Lyden D, Hattori K, Dias S, Costa C, Blaikie P, Butros L, et al. Impaired recruitment of bone-marrow-derived endothelial and hematopoietic precursor cells blocks tumor angiogenesis and growth. Nat Med. 2001;7:1194–1201. doi: 10.1038/nm1101-1194. [DOI] [PubMed] [Google Scholar]
- Nilsson L, Eden P, Olsson E, Mansson R, Astrand-Grundstrom I, Strombeck B, et al. The molecular signature of MDS stem cells supports a stem cell origin of 5q- myelodysplastic syndromes. Blood. 2007;110:3005–3014. doi: 10.1182/blood-2007-03-079368. [DOI] [PubMed] [Google Scholar]
- Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. 2000;113(Pt 22):3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, et al. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409:1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- O’Neil J, Shank J, Cusson N, Murre C, Kelliher M. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer Cell. 2004;5:587–596. doi: 10.1016/j.ccr.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Pagliuca A, Gallo P, De Luca P, Lania L. Class A helix-loop-helix proteins are positive regulators of several cyclin-dependent kinase inhibitors’ promoter activity and negatively affect cell growth. Cancer Res. 2000;60:1376–1382. [PubMed] [Google Scholar]
- Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- Prabhu S, Ignatova A, Park ST, Sun XH. Regulation of the expression of cyclin-dependent kinase inhibitor p21 by E2A and Id proteins. Mol Cell Biol. 1997;17:5888–5896. doi: 10.1128/mcb.17.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesenberry PJ, Iscove NN, Cooper C, Brady G, Newburger PE, Stein GS, et al. Expression of basic helix-loop-helix transcription factors in explant hematopoietic progenitors. J Cell Biochem. 1996;61:478–488. doi: 10.1002/(SICI)1097-4644(19960601)61:3%3C478::AID-JCB15%3E3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Rosenbauer F, Koschmieder S, Steidl U, Tenen DG. Effect of transcription-factor concentrations on leukemic stem cells. Blood. 2005;106:1519–1524. doi: 10.1182/blood-2005-02-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosmarin AG, Yang Z, Resendes KK. Transcriptional regulation in myelopoiesis: hematopoietic fate choice, myeloid differentiation, and leukemogenesis. Exp Hematol. 2005;33:131–143. doi: 10.1016/j.exphem.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell. 2003;3:525–530. doi: 10.1016/s1535-6108(03)00141-7. [DOI] [PubMed] [Google Scholar]
- Suh HC, Gooya J, Renn K, Friedman AD, Johnson PF, Keller JR. C/EBPalpha determines hematopoietic cell fate in multipotential progenitor cells by inhibiting erythroid differentiation and inducing myeloid differentiation. Blood. 2006;107:4308–4316. doi: 10.1182/blood-2005-06-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- Wilson RB, Kiledjian M, Shen CP, Benezra R, Zwollo P, Dymecki SM, et al. Repression of immunoglobulin enhancers by the helix-loop-helix protein Id: implications for B-lymphoid-cell development. Mol Cell Biol. 1991;11:6185–6191. doi: 10.1128/mcb.11.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, Wang X, Ling MT. Id-1 expression and cell survival. Apoptosis. 2004;9:279–289. doi: 10.1023/b:appt.0000025804.25396.79. [DOI] [PubMed] [Google Scholar]
- Yokota Y, Mori S, et al. Role of Id family proteins in growth control. J Cell Physiol. 2002;190:21–28. doi: 10.1002/jcp.10042. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science. 2004;305:1286–1289. doi: 10.1126/science.1097937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.