Abstract
The highly conserved 14-3-3 protein family has risen to a position of importance in cell biology owing to its involvement in vital cellular processes, such as metabolism, protein trafficking, signal transduction, apoptosis and cell-cycle regulation. The 14-3-3 proteins are phospho-serine/phospho-threonine binding proteins that interact with a diverse array of binding partners. Because many 14-3-3 interactions are phosphorylation-dependent, 14-3-3 has been tightly integrated into the core phospho-regulatory pathways that are crucial for normal growth and development and that often become dysregulated in human disease states such as cancer. This review examines the recent advances that further elucidate the role of 14-3-3 proteins as integrators of diverse signaling cues that influence cell fate decisions and tumorigenesis.
Emergence of 14-3-3 proteins as regulators of diverse biological processes
The 14-3-3 proteins are abundant 28–33-kDa acidic poly-peptides found in all eukaryotic organisms (for a review, see Ref. [1]). Members of this protein family were first identified by Moore and Perez [2] in 1967 and acquired their unusual name based on the nomenclature used by these investigators during their systematic classification of proteins found in mammalian brain tissue. 14-3-3 proteins are highly conserved and seven family members are found in mammals – β, γ, ε, σ, ζ, τ and η. 14-3-3 proteins self-assemble into homo- and heterodimers, with some family members, such as σ and γ, preferring to homodimerize and other family members, such as ε, preferring to heterodimerize. 14-3-3 dimers can interact with a diverse array of cellular proteins including transcription factors, biosynthetic enzymes, cytoskeletal proteins, signaling molecules, apoptosis factors and tumor suppressors. 14-3-3 proteins are able to interact with many different proteins as a result of their specific phospho-serine/phospho-threonine binding activity. Two high-affinity 14-3-3 binding motifs have been described in 14-3-3 target proteins: RSXpSXP (mode 1) and RXXXpSXP (mode 2), where pS represents phospho-serine. However, it is important to note that not all phosphorylation-dependent sites conform to these motifs and not all interactions are phosphorylation-dependent.
Structural analyses of 14-3-3 dimers have revealed that each monomer contains an independent ligand-binding channel and, as a result, the dimer can interact with two motifs simultaneously, found either on a single target or on separate binding partners [3]. 14-3-3 dimers are highly rigid structures and binding can induce conformational changes in protein ligands that might alter the stability and/or catalytic activity of the ligand. In addition, by steric hindrance, 14-3-3 binding can hide intrinsic localization motifs, prevent molecular interactions and/or modulate the accessibility of a target protein to modifying enzymes such as kinases, phosphatases or proteases.
Phosphorylation is a key regulatory mechanism in cell biology and, because 14-3-3 interactions are primarily phosphorylation-dependent, the 14-3-3 proteins have emerged as important components of essential biological processes that are regulated by phosphorylation, such as signal transduction and cell-cycle regulation. This review focuses on recent advances further elucidating the function of 14-3-3 proteins as integrators of signaling cues that enable cells to make decisions that direct their fate. In particular, this review will examine how 14-3-3 contributes to survival and apoptotic signaling, cell growth, tumor suppression and cancer development.
Functions of 14-3-3 proteins in survival and apoptotic signaling
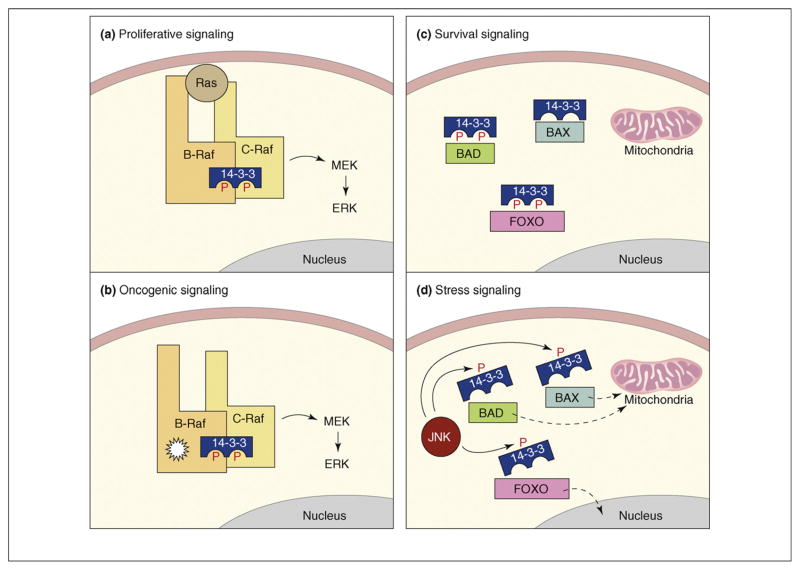
Regulating the balance between survival and apoptotic signaling is a key aspect of cell fate decisions and 14-3-3 proteins contribute to this process in multiple ways. For example, the binding of 14-3-3 has often been found to enhance the activity of proteins with proliferative or survival functions. A case in point is the Raf serine/threonine kinase family [4]. Members of the Raf family are key effectors of the Ras GTPase that initiate the activation of the extracellular signal-regulated kinase (ERK) cascade, which, in turn, promotes proliferative and survival signaling. Binding of 14-3-3 to a site in the C-terminal region of the Raf proteins (pS621 and pS729 on the C-Raf and B-Raf family members, respectively) is needed for full catalytic activity and is required for the heterodimerization of the C-Raf and B-Raf kinases [5,6]. Under normal signaling conditions, this heterodimerization of C- and B-Raf occurs in response to Ras activation and promotes maximal activation of C-Raf (Figure 1a). The B-Raf kinase is mutated in numerous human cancers and it is important to note that oncogenic B-Raf proteins have been found to heterodimerize constitutively with C-Raf [7,8]. This heterodimerization is also dependent on 14-3-3 binding and is required for the transforming function of B-Raf proteins with impaired kinase activity, thus, linking 14-3-3 with the oncogenic potential of B-Raf (Figure 1b).
Figure 1.
Functions of 14-3-3 in proliferative, oncogenic, survival and stress signaling. (a) Under proliferative signaling conditions, 14-3-3 binding is required for the Ras-dependent heterodimerization of B-Raf and C-Raf and contributes to the full kinase activation of C-Raf. (b) Under oncogenic B-Raf signaling conditions, 14-3-3 mediates the constitutive heterodimerization of B-Raf and C-Raf and is required for the transforming potential of kinase-impaired B-Raf mutants (indicated by starburst shape). (c) Under survival signaling conditions, 14-3-3 binding inactivates numerous pro-apoptotic proteins, such as BAD, BAX and the FOXO transcription factors, by sequestering them from their sites of action such as the mitochondria and the nucleus. (d) Under stress signaling conditions, activated JNK disrupts 14-3-3 binding to several pro-apoptotic proteins by directly phosphorylating the 14-3-3 proteins themselves, thus, enabling the apoptosis regulators to localize to their site of action.
Another way that 14-3-3 influences the balance between survival and apoptotic signaling is by antagonizing the activity of proteins that promote cell death and senescence (for a review, see Ref. [9]). 14-3-3 proteins can suppress apoptosis through interactions with core components of the mitochondrial apoptotic machinery, such as BCL-2 antagonist of cell death (BAD), BCL-2 interacting mediator of cell death (BIM) and BCL-2 associated x protein (BAX), and through interactions with proteins that transmit apop-totic signals, including the stress-responsive kinase ASK1 (MEKK5) and the forkhead box O1 (FOXO) transcription factors (Figure 1c). Kinases with pro-survival functions, such as AKT (protein kinase B, PKB), Rsk and PIM, are often responsible for generating the 14-3-3 docking sites on these molecules and the binding of 14-3-3 frequently promotes the relocalization of these pro-apoptotic proteins away from their site of action. For example, the BAD–14-3-3 interaction causes BAD to be retained in the cytoplasm, thus, preventing BAD from dimerizing with BCL2/XL at the mitochondria and mediating the release of BAX from BCL2/XL-mediated inhibition. For the FOXO proteins, the binding of 14-3-3 first facilitates the nuclear export of these transcription factors by exposing their intrinsic nuclear export sequences (NESs) and then retains them in the cytoplasm by masking their intrinsic nuclear localization sequences (NLSs).
When cells receive cues that tip the balance towards death, many of these 14-3-3 interactions are targeted for disruption. 14-3-3 binding can be disrupted by activation of phosphatases that abolish the docking site, by phosphorylation of the binding partner on sites that dislodge 14-3-3 and by direct phosphorylation of 14-3-3 proteins to abrogate their binding activity. For example, it has recently been shown that death of post-mitotic neurons can be induced when cyclin-dependent kinase 1 (Cdk1)-mediated phosphorylation of FOXO1 disrupts 14-3-3 binding, thus, enabling FOXO1 to enter the nucleus and activate the transcription of death-promoting proteins [10]. The stress-activated Jun-N-terminal kinase (JNK) has also been found to disrupt 14-3-3 interactions by phosphorylating either the 14-3-3 binding partner [11] or the 14-3-3 ζ, β, ε and σ proteins themselves [12–14]. Moreover, accumulating evidence indicates that 14-3-3 phosphorylation and ligand release is the primary mechanism by which activated JNK controls the pro-apoptotic function of BAD, BAX, FOXO and Abl [12–14] and represents a key step in JNK-mediated apoptosis (Figure 1d). Taken together, the above findings highlight the function of 14-3-3 as an integration point for proliferative, survival, apoptotic and stress signaling.
Functions of 14-3-3 proteins in cell growth signaling
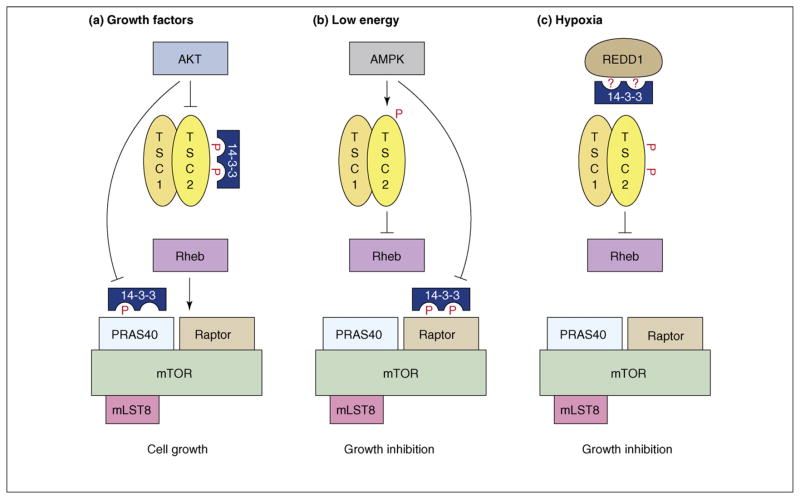
The mammalian target of rapamycin (mTOR) is a key effector in an important growth-promoting pathway that is commonly dysregulated in human cancers [15,16]. Under normal conditions, this pathway functions to interface growth factor signals with information regarding cellular nutrient, energy and oxygen levels. Signaling through this pathway converges on the mTOR complex 1 (TORC1) [15,17] and, strikingly, 14-3-3 binds four different regulators of TORC1 signaling. The TORC1 complex consists of the following components: (i) mTOR, a member of the phosphatidylinositol kinase-related protein kinase family; (ii) Raptor, a protein scaffold that recruits downstream substrates of mTOR, such as the S6 kinase and 4E-BP1; (iii) PRAS40, a repressor of mTOR activity; and (iv) mLST8, also known as G-protein β-subunit-like (GβL). Acting upstream of TORC1 is the Rheb GTPase, which directly stimulates TORC1 activity when Rheb is bound to GTP [18]. Also functioning upstream of TORC1 is the tuberous sclerosis complex TSC1–TSC2, which inhibits TORC1 signaling as a result of the GTPase-activating protein (GAP) activity that is intrinsic to TSC2 and that inactivates Rheb [19].
Two types of inputs can ‘turn on’ TORC1: the increased availability of amino acids in the cell and the activation of growth factor pathways such as the insulin-like growth factor 1 (IGF-1) and insulin pathways. In response to insulin treatment, both TSC2 and PRAS40 are phosphorylated on sites that relieve their inhibitory effects and these proteins have been shown to associate with 14-3-3 in a manner that correlates with their inactivation [20–26] (Figure 2a). With regard to TSC2, the function of 14-3-3 binding and the identities of the upstream kinases and TSC2 residues required for 14-3-3 binding have been the topic of much debate [21–23,27], perhaps indicating that multiple kinases and TSC2 sites might contribute to this interaction and that the kinases and sites involved might vary under different signaling contexts. Nevertheless, two recent studies examining the formation of complexes between endogenous 14-3-3 and TSC2 in insulin-treated cells report that the TSC2–14-3-3 interaction is induced by AKT signaling, with some combination of TSC2 residues pS939, pS981 and pT1462 serving as the 14-3-3 docking sites [24,28]. Moreover, in studies by Cai and colleagues, 14-3-3 was found to have an important role in the AKT-mediated inhibition of TSC2, in that 14-3-3 binding promoted the relocalization of TSC2 out of the endomembrane compartment that contains Rheb and into the cytoplasm [24]. With regard to PRAS40, 14-3-3 binding is mediated by pT246, a site phosphorylated by AKT [25,26]. From in vitro studies, it has been shown that the repressor activity of PRAS40 can be inactivated solely by phosphorylation of T246 [25], indicating that 14-3-3 binding per se is not required. However, in the context of an intracellular environment, 14-3-3 might contribute to PRAS40 inactivation either by preventing the dephosphorylation of PRAS40 residue pT246 or by destabilizing the PRAS40–TORC1 interaction, as has been indicated in studies by Vander Haar and colleagues [26].
Figure 2.
Functions of 14-3-3 in TORC1 signaling. (a) Under growth conditions, the AKT kinase is activated downstream of the insulin receptor (not depicted) and in turn activates TORC1 by phosphorylating TSC2 and PRAS40 on sites that mediate 14-3-3 binding and that relieve the inhibitory effects of these proteins. (b) AMPK is activated under conditions of low energy and inhibits TORC1 signaling by phosphorylating TSC2 and Raptor, thus, stimulating TSC2 activity and inhibiting Raptor function. AMPK-mediated phosphorylation of Raptor is required for the energy checkpoint induced by AMPK, and the sites that are phosphorylated mediate 14-3-3 binding. (c) REDD1 (DDIT4) expression is induced under hypoxic conditions and REDD1 inhibits TORC1 activity in a TSC2-dependent manner. REDD1 has been reported to reverse the inhibition of TSC2 by promoting the movement of 14-3-3 proteins from TSC2 to REDD1.
The above findings indicate a positive function for 14-3-3 in promoting TORC1 signaling. However, 14-3-3 proteins also contribute to the regulatory mechanisms that suppress TORC1 activity under conditions of cell stress. Before committing to cell growth, it is important that the cell possesses enough energy to drive the process. 5′-AMP-activated protein kinase (AMPK) is a kinase that senses cellular energy levels and becomes activated under energy stress. Activation of AMPK dominantly inhibits TORC1 signaling and one target of AMPK is TSC2, the phosphorylation of which at S1345 can stimulate its Rheb-GAP activity [29]. Another recently identified AMPK target is Raptor. Gwinn and colleagues found that AMPK phosphorylates Raptor at S722 and S792 and that phosphorylation of these sites mediates 14-3-3 binding and is required for TORC1 inactivation in vivo [30] (Figure 2b). Although the exact mechanism regarding how phosphorylation of Raptor inactivates TORC1 is unclear, experiments by Gwinn and coworkers demonstrate that Raptor phosphorylation, and perhaps 14-3-3 binding, is required for the metabolic checkpoint mediated by AMPK under conditions of energy stress [30].
Oxygen deprivation, or hypoxia, is another cell stress that inhibits mTORC1 activity and, although hypoxia can cause energy stress and impact TORC1 through AMPK activation, the predominant means by which hypoxia inhibits TORC1 is through the induced expression of the DNA-damage-inducible transcript 4 (DDIT4; also known as REDD1) protein [31–35]. REDD1 has been shown to function in a dominant manner downstream of AKT signaling and to inhibit TORC1 in a TSC2-dependent manner, indicating that REDD1 might regulate TSC2. A report by DeYoung and colleagues describes a mechanism for how expression of REDD1 might modulate TSC2 activity and this mechanism involves 14-3-3 [28]. The authors found that 14-3-3 proteins interact with REDD1 at a motif surrounding S137 of REDD1 and that REDD1 can reverse the AKT-mediated inhibition of TSC2 by promoting the movement of 14-3-3 proteins from TSC2 to REDD1 (Figure 2c). Thus, according to this model, REDD1 activates the TSC1–TSC2 complex, which inhibits Rheb, by functioning as a ‘binding sink’ for 14-3-3.
Functions of 14-3-3 proteins in tumor suppression
Regulation of cytokinesis
In addition to their well-known pro-proliferative and anti-apoptotic effects, 14-3-3 proteins have also been found to suppress cell growth and cell-cycle progression, especially after DNA damage, indicating functions in tumor suppression. Of the 14-3-3 proteins, tumor-suppressor activity has most clearly been defined for 14-3-3σ. 14-3-3σ is unique among the 14-3-3 proteins in that it is expressed primarily in epithelial cells and forms homodimers almost exclusively [36]. Expression of 14-3-3σ is coordinately upregulated by the cellular tumor antigen p53 and the breast cancer type 1 susceptibility protein (BRCA1) and contributes to the DNA-damage-induced cell-cycle checkpoint mediated by these tumor suppressors [37,38]. When induced, 14-3-3σ binds to and sequesters cyclin-B–cell division cycle 2 (cdc2) complexes in the cytoplasm, thus, enabling DNA damage to be repaired before the cell cycle progresses [39,40]. Further indicating tumor-suppressor activity, the expression of 14-3-3σ is frequently lost in tumors of epithelial origin, including most breast cancers [41]. This loss can be caused either by hypermethylation of the promoter for the gene encoding 14-3-3σ or by targeted ubiquitin-mediated degradation of 14-3-3σ. Downregulation of 14-3-3σ occurs not only in the tumor itself but also in surrounding pre-dysplastic tissue, indicating that loss of 14-3-3σ function might be an early step in tumor development [42].
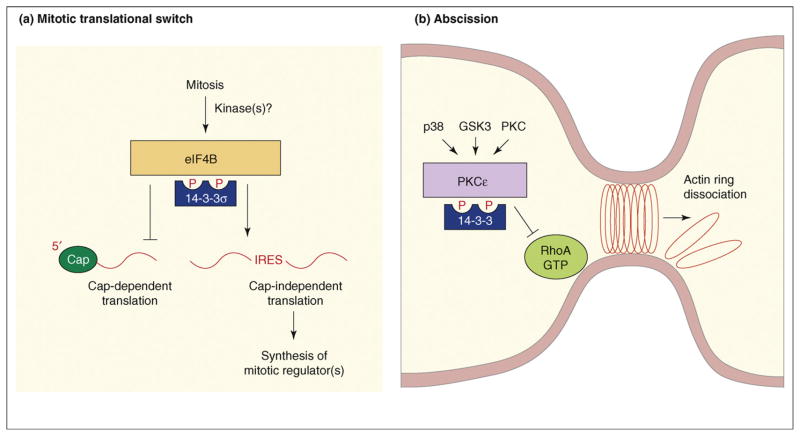
Further insight as to why loss of 14-3-3σ might facilitate tumor formation comes from the discovery by Wilker and colleagues that 14-3-3σ is a crucial regulator of translation during mitosis and that 14-3-3σ function is required for proper mitotic exit and cytokinesis [43] (Figure 3a). In eukaryotic cells, most mRNA translation occurs via a cap-dependent mechanism in which ribosome recruitment begins with the binding of eukaryotic initiation factors, such as eIF4B, to a modified guanosine residue (known as a ‘cap’) at the 5′ end of the mRNA. However, some mRNAs contain internal ribosome entry sites (IRESs) and are translated in a cap-independent manner. During mitosis, cap-dependent translation is suppressed and cap-independent translation is stimulated, allowing for the translation of key cell-cycle regulators such as cell division cycle 2-like 1 (CDC2L1; also known as Cdk11-p58 and PITSLRE). Experiments by Wilker and coworkers found that 14-3-3σ is needed for the mitotic switch from cap-dependent to cap-independent translation and that 14-3-3σ seems to mediate this switch by binding to eIF4B and perhaps other factors involved in cap-dependent translation [43]. When cells are depleted of 14-3-3σ, cap-dependent translation is not suppressed and cytokinesis is impaired, resulting in the generation of binucleated cells, a phenotype observed in early stages of tumor formation. Yet to be determined is the kinase(s) that regulates the mitosis-specific binding of 14-3-3σ to eIF4B.
Figure 3.
Functions of 14-3-3 in cytokinesis. (a) 14-3-3σ function is required for a mitotic translational switch. During mitosis, binding of 14-3-3σ to eIF4B and possibly to other factors involved in cap-dependent translation is required for the mitotic switch from cap-dependent to cap-independent translation. This translational switch is needed for the synthesis of key mitotic regulators required for proper mitotic exit and cytokinesis. mRNA shown as red line. Abbreviations: IRES, internal ribosome entry site. (b) 14-3-3 function in abscission. During mitosis, phosphorylation mediated by p38, GSK3 and PKCε generates 14-3-3 binding sites on PKCε. Binding of 14-3-3 activates PKCε and formation of the active PKCε–14-3-3 complex is needed to limit RhoA activity at the midbody, which in turn enables the actomyosin ring to dissociate during the final stages of abcission.
Strikingly, 14-3-3 proteins have been found to function at another key regulatory point during cytokinesis – abscission (Figure 3b). In a recent study, Saurin and colleagues [44] report that binding of 14-3-3 to protein kinase C epsilon (PKCε) activates PKCε in a lipid-independent manner and that formation of this complex is required for the final stage of cell separation during cytokinesis. PKCε interacts with multiple 14-3-3 family members and binding is mediated by two phosphorylation sites – pS368, a PKCε auto-phosphorylation site and pS346, a site phosphorylated by glycogen synthase kinase 3 (GSK3) following a priming phosphorylation event mediated by p38 mitogen-activated protein kinase (MAPK). Both pS346 phosphorylation and 14-3-3 binding peak during mitosis and formation of the active PKCε–14-3-3 complex is apparently needed during late telophase to limit RhoA activity at the midbody, which in turn enables the actomyosin ring to dissociate during the final stages of abscission. Saurin and colleagues show that disruption of the PKCε–14-3-3 interaction, loss of PKCε expression, inactivation of PKCε catalytic activity or overexpression of a dominant-negative 14-3-3 protein all promote defects in cytokinesis that result in the formation of binucleated cells [44]. Moreover, as indicated earlier, the formation of binucleated cells due to failed cytokinesis has been reported to be an early event in tumorigenesis and underlies the subsequent development of genomic instability [45–47].
A role in Hippo pathway signaling
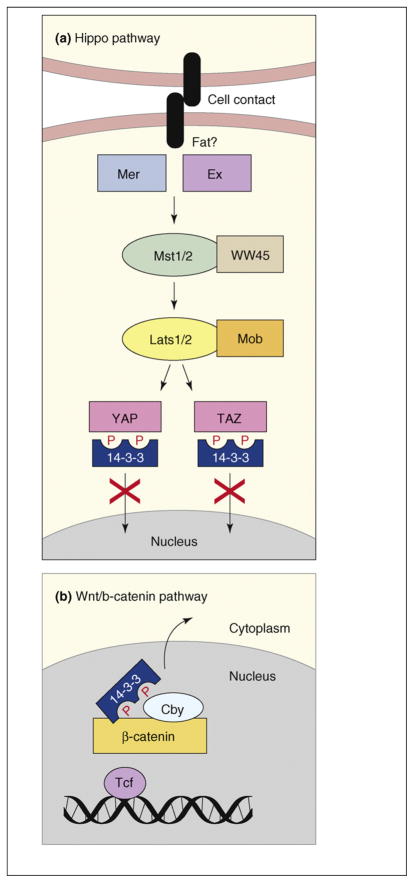
In the past year, several studies have revealed new tumor-suppressor activities for the 14-3-3 proteins. Of particular interest is the function of 14-3-3 in the Hippo pathway, a novel tumor-suppressor pathway first discovered in Drosophila melanogaster (for reviews, see Refs [48–50]). In flies, the Hippo pathway controls tissue and organ size by regulating cell proliferation and apoptosis. The Drosophila Hippo (Hpo) protein is a member of the Ste20 serine/threonine kinase family that complexes with a regulatory scaffold protein Salvador (Sav) to phosphorylate and activate Warts (Wrts), a member of the nuclear DBf2-related (NDR) serine/threonine kinase family. Wrts associates with its activating subunit, Mats, and phosphorylates the transcriptional coactivator Yorkie (Yki), thus, inhibiting Yki function. Acting upstream of Hpo are two actin-binding cytoskeletal proteins Merlin (Mer) and Expanded (Ex) and the cell surface protocadherin Fat. In mammals, Mst1/2 and Lats1/2 function as the Hpo and Wrts homologues, respectively, and signaling through the pathways inactivates two transcriptional coactivators, YAP (Yki homolog) and TAZ. The mammalian Hippo pathway also contributes to organ size control [51,52] and dysregulation of numerous Hippo pathway components, including Mer, Sav, Lats and YAP, has been observed in human tumors [48,53].
Recently, three studies have identified a molecular mechanism by which Hippo signaling inactivates Yki, YAP and TAZ [52,54,55]. Specifically, for all three transcriptional coactivators, phosphorylation by the Wrts and Lats kinases promotes 14-3-3 binding and their relocalization to the cytoplasm (Figure 4a). 14-3-3ε, and probably other 14-3-3 family members, bind to the transcriptional coactivators [52,54,55] and 14-3-3 binding is indicated to be an important regulatory mechanism in that several activating mutations in Yki result in decreased 14-3-3 binding [54]. With regard to YAP and TAZ, mutants unable to bind to 14-3-3 localize constitutively in the nucleus and exhibit constitutive activity that can result in the loss of cell contact inhibition, epithelial-to-mesenchymal transition and/or anchorage-independent growth [52,54,55], all of which are properties that contribute to tumorigenesis.
Figure 4.
Functions of 14-3-3 in tumor-suppressor pathways. (a) The mammalian Hippo pathway transmits signals received at the cell surface, such as cell–cell contact, through the sequential activation of the Mst1/2–WW45 and Lats1/2–Mob kinase complexes. The Lats kinases phosphorylate YAP and TAZ on sites that mediate 14-3-3 binding, thus, inactivating the function of these transcriptional coactivators by means of their cytoplasmic retention. (b) β-catenin functions as a transcriptional coactivator in the Wnt signaling pathway. Cby antagonizes β-catenin signaling by preventing the interaction between β-catenin and the Tcf transcription factor enhancer and by mediating the formation of a stable β-catenin–Cby–14-3-3 tripartite complex that results in the nuclear export of β-catenin.
A role in β-catenin signaling
Another tumor-suppressor pathway in which 14-3-3 has recently been shown to function is the Wnt–β-catenin pathway. Wnt–β-catenin signaling serves crucial functions during embryonic development, is required for tissue homeostasis and is aberrantly activated in numerous human tumors, including >70% of colorectal cancers (for reviews, see Refs [56–58]). β-catenin contributes to cell-adhesion processes and functions as a transcriptional coactivator in the Wnt pathway. In the absence of Wnt signals, β-catenin is maintained at low levels in the cytoplasm owing to ubiquitin-mediated degradation that is facilitated by the tumor suppressors adenomatous poly-posis coli (APC) and axin. β-catenin function can also be antagonized by a conserved 14-kDa protein named Chibby (Cby) [59]. Previous studies show that Cby competes with Tcf enhancer factors for binding to β-catenin in the nucleus, thus, inhibiting Wnt target-gene transcription [59]. In a recent report, Li and colleagues describe another mechanism by which Cby antagonizes β-catenin signaling [60]. They identify multiple 14-3-3 family members (ε, ζ and η) as Cby binding partners and find that binding of 14-3-3 to pS20 of Cby results in the cytoplasmic relocalization of Cby. However, as opposed to sequestering Cby from β-catenin, Cby, β-catenin and 14-3-3 form a stable tripartite complex that facilitates the nuclear export of β-catenin (Figure 4b). The implication from these findings is that 14-3-3 functions as a tumor suppressor with Cby to limit β-catenin signaling.
Investigating the function of 14-3-3ζ in tumorigenesis
The impact of 14-3-3 function on tumor development is best understood for 14-3-3σ, where 14-3-3 has been shown to have specific tumor-suppressing activities and its loss seems to be an early event in tumorigenesis. Whether and how other 14-3-3 family members contribute to human cancer is less clear. 14-3-3 proteins are undoubtedly important regulatory components in many signaling pathways; however, if a particular 14-3-3 family member was found to function primarily in survival signaling, upregulation of this 14-3-3 protein might provide a survival advantage to tumor cells. Moreover, because resistance to apoptosis is a characteristic feature of cancer cells, an approach towards therapy might be to abolish the activity of a ‘survival-promoting’ 14-3-3 family member to force cells into the default death pathway. To begin to address these issues, two recent reports have investigated the contribution of 14-3-3ζ to oncogenesis. 14-3-3ζ is a ubiquitously expressed 14-3-3 family member and increased expression of 14-3-3ζ has been observed in several human tumors, including human hepatocellular carcinoma, stomach cancer, seminoma, squamous carcinoma, pancreatic adenocarcinoma, breast cancer and several types of lung carcinoma [61–63]. In addition, the gene encoding 14-3-3ζ maps to a chromosome region (8q23) that is frequently amplified in metastatic cancer [64,65]. In a study by Li and colleagues, depletion of 14-3-3ζ in A549 lung adenocarcinoma cells was found to inhibit the anchorage-independent growth of these cells and to cause increased apoptosis when cells were detached from the plate matrix – a phenomenon known as anoikis [66]. The enhanced anoikis correlated with upregulation of the pro-apoptotic proteins BAD and BIM and with increased BAX activity. Work by Niemantsverdriet and colleagues also observed an effect of 14-3-3ζ depletion on the levels of apoptosis [61]. In this case, depletion of 14-3-3ζ in human keratinocytes dramatically increased the rate of apoptosis induced by UV stress. In addition, the expression of cell-adhesion proteins, including epithelial E-cadherin, T-cadherin and γ-catenin, was elevated in 14-3-3ζ-depleted cells, indicating that 14-3-3ζ might be inhibitory towards cell adhesion. The overriding conclusion from these studies is that upregulation of 14-3-3ζ can help tumor cells resist apoptosis and might provide them with an increased potential for tumor invasion and metastasis.
Concluding remarks and future perspectives
Research in the past few years has revealed exciting advances that further our understanding of the complex roles that 14-3-3 proteins have in cell biology and signal transduction. Diverse signaling cues are transmitted through regulated cycles of phosphorylation and dephosphorylation and, because most 14-3-3 interactions are phosphorylation-dependent, 14-3-3 proteins have emerged as key targets capable of integrating multiple signaling inputs. 14-3-3 proteins function in pathways that modulate cell survival and proliferation and in those that control apoptosis and tumor suppression. These pathways are crucial for normal growth and development and their dysregulation often results in the onset of tumorigenesis. Recent studies have clearly demonstrated the involvement of 14-3-3 proteins in cellular processes that directly impact tumor development, such as cytokinesis, cell-contact inhibition, anchorage-independent growth and cell adhesion. The challenge that continues for 14-3-3 research is the further elucidation of specific functions for individual 14-3-3 family members. As progress is made in this area, it is possible that alterations in the expression of certain 14-3-3 proteins might provide new avenues for the diagnosis and treatment of human cancer. In this regard, the development of small molecule inhibitors that globally target 14-3-3 interactions or that specifically inhibit the interactions of certain 14-3-3 proteins could prove useful in the design of new anti-cancer therapies.
References
- 1.Aitken A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Moore BW, Perez VJ. Specific Acid Proteins in the Nervous System. Prentice-Hall; 1967. [Google Scholar]
- 3.Gardino AK, et al. Structural determinants of 14-3-3 binding specificities and regulation of subcellular localization of 14-3-3-ligand complexes: a comparison of the X-ray crystal structures of all human 14-3-3 isoforms. Semin Cancer Biol. 2006;16:173–182. doi: 10.1016/j.semcancer.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Tzivion G, et al. 14-3-3 proteins as potential oncogenes. Semin Cancer Biol. 2006;16:203–213. doi: 10.1016/j.semcancer.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Rushworth LK, et al. Regulation and role of Raf-1/B-Raf heterodimerization. Mol Cell Biol. 2006;26:2262–2272. doi: 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garnett MJ, et al. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol Cell. 2005;20:963–969. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 7.Wan PT, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 8.Wellbrock C, et al. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 9.Porter GW, et al. Dynamic 14-3-3/client protein interactions integrate survival and apoptotic pathways. Semin Cancer Biol. 2006;16:193–202. doi: 10.1016/j.semcancer.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Yuan Z, et al. Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science. 2008;319:1665–1668. doi: 10.1126/science.1152337. [DOI] [PubMed] [Google Scholar]
- 11.Donovan N, et al. JNK phosphorylation and activation of BAD couples the stress-activated signaling pathway to the cell death machinery. J Biol Chem. 2002;277:40944–40949. doi: 10.1074/jbc.M206113200. [DOI] [PubMed] [Google Scholar]
- 12.Sunayama J, et al. JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J Cell Biol. 2005;170:295–304. doi: 10.1083/jcb.200409117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuruta F, et al. JNK promotes Bax translocation to mitochondria through phosphorylation of 14-3-3 proteins. EMBO J. 2004;23:1889–1899. doi: 10.1038/sj.emboj.7600194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida K, et al. JNK phosphorylation of 14-3-3 proteins regulates nuclear targeting of c-Abl in the apoptotic response to DNA damage. Nat Cell Biol. 2005;7:278–285. doi: 10.1038/ncb1228. [DOI] [PubMed] [Google Scholar]
- 15.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 16.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Bhaskar PT, Hay N. The two TORCs and Akt. Dev Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Manning BD, Cantley LC. Rheb fills a GAP between TSC and TOR. Trends Biochem Sci. 2003;28:573–576. doi: 10.1016/j.tibs.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoki K, et al. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, et al. Regulation of TSC2 by 14-3-3 binding. J Biol Chem. 2002;277:44593–44596. doi: 10.1074/jbc.C200510200. [DOI] [PubMed] [Google Scholar]
- 22.Nellist M, et al. Identification and characterization of the interaction between tuberin and 14-3-3ζ. J Biol Chem. 2002;277:39417–39424. doi: 10.1074/jbc.M204802200. [DOI] [PubMed] [Google Scholar]
- 23.Shumway SD, et al. 14-3-3β binds to and negatively regulates the tuberous sclerosis complex 2 (TSC2) tumor suppressor gene product, tuberin. J Biol Chem. 2003;278:2089–2092. doi: 10.1074/jbc.C200499200. [DOI] [PubMed] [Google Scholar]
- 24.Cai SL, et al. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol. 2006;173:279–289. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sancak Y, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Vander Haar E, et al. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, et al. The p38 and MK2 kinase cascade phosphorylates tuberin, the tuberous sclerosis 2 gene product, and enhances its interaction with 14-3-3. J Biol Chem. 2003;278:13663–13671. doi: 10.1074/jbc.M300862200. [DOI] [PubMed] [Google Scholar]
- 28.DeYoung MP, et al. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inoki K, et al. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 30.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brugarolas J, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corradetti MN, et al. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J Biol Chem. 2005;280:9769–9772. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- 33.Ellisen LW. Growth control under stress: mTOR regulation through the REDD1-TSC pathway. Cell Cycle. 2005;4:1500–1502. doi: 10.4161/cc.4.11.2139. [DOI] [PubMed] [Google Scholar]
- 34.Schwarzer R, et al. REDD1 integrates hypoxia-mediated survival signaling downstream of phosphatidylinositol 3-kinase. Oncogene. 2005;24:1138–1149. doi: 10.1038/sj.onc.1208236. [DOI] [PubMed] [Google Scholar]
- 35.Sofer A, et al. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 2005;25:5834–5845. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilker EW, et al. A structural basis for 14-3-3σ functional specificity. J Biol Chem. 2005;280:18891–18898. doi: 10.1074/jbc.M500982200. [DOI] [PubMed] [Google Scholar]
- 37.Hermeking H, et al. 14-3-3σ is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 38.Aprelikova O, et al. BRCA1 is a selective co-activator of 14-3-3σ gene transcription in mouse embryonic stem cells. J Biol Chem. 2001;276:25647–25650. doi: 10.1074/jbc.C100265200. [DOI] [PubMed] [Google Scholar]
- 39.Chan TA, et al. 14-3-3σ is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 40.Laronga C, et al. Association of the cyclin-dependent kinases and 14-3-3σ negatively regulates cell cycle progression. J Biol Chem. 2000;275:23106–23112. doi: 10.1074/jbc.M905616199. [DOI] [PubMed] [Google Scholar]
- 41.Lodygin D, Hermeking H. Epigenetic silencing of 14-3-3σ in cancer. Semin Cancer Biol. 2006;16:214–224. doi: 10.1016/j.semcancer.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 42.Umbricht CB, et al. Hypermethylation of 14-3-3σ (stratifin) is an early event in breast cancer. Oncogene. 2001;20:3348–3353. doi: 10.1038/sj.onc.1204438. [DOI] [PubMed] [Google Scholar]
- 43.Wilker EW, et al. 14-3-3σ controls mitotic translation to facilitate cytokinesis. Nature. 2007;446:329–332. doi: 10.1038/nature05584. [DOI] [PubMed] [Google Scholar]
- 44.Saurin AT, et al. The regulated assembly of a PKCε complex controls the completion of cytokinesis. Nat Cell Biol. 2008;10:891–901. doi: 10.1038/ncb1749. [DOI] [PubMed] [Google Scholar]
- 45.Fujiwara T, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 46.King RW. When 2+2=5: The origins and fates of aneuploid and tetraploid cells. Biochim Biophys Acta. 2008;1786:4–14. doi: 10.1016/j.bbcan.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature. 2005;437:1038–1042. doi: 10.1038/nature03958. [DOI] [PubMed] [Google Scholar]
- 48.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway -an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 49.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 50.Saucedo LJ, Edgar BA. Filling out the Hippo pathway. Nat Rev Mol Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- 51.Camargo FD, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 52.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 54.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lei QY, et al. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the Hippo pathway. Mol Cell Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 57.Gavert N, Ben-Ze’ev A. β-catenin signaling in biological control and cancer. J Cell Biochem. 2007;102:820–828. doi: 10.1002/jcb.21505. [DOI] [PubMed] [Google Scholar]
- 58.Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 59.Takemaru K, et al. Chibby, a nuclear β-catenin-associated antagonist of the Wnt/Wingless pathway. Nature. 2003;422:905–909. doi: 10.1038/nature01570. [DOI] [PubMed] [Google Scholar]
- 60.Li FQ, et al. Chibby cooperates with 14-3-3 to regulate β-catenin subcellular distribution and signaling activity. J Cell Biol. 2008;181:1141–1154. doi: 10.1083/jcb.200709091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Niemantsverdriet M, et al. Cellular functions of 14-3-3ζ in apoptosis and cell adhesion emphasize its oncogenic character. Oncogene. 2008;27:1315–1319. doi: 10.1038/sj.onc.1210742. [DOI] [PubMed] [Google Scholar]
- 62.Arora S, et al. Identification of differentially expressed genes in oral squamous cell carcinoma. Mol Carcinog. 2005;42:97–108. doi: 10.1002/mc.20048. [DOI] [PubMed] [Google Scholar]
- 63.Jang JS, et al. The differential proteome profile of stomach cancer: identification of the biomarker candidates. Oncol Res. 2004;14:491–499. doi: 10.3727/0965040042380441. [DOI] [PubMed] [Google Scholar]
- 64.Ghadimi BM, et al. Gain of chromosome 8q23-24 is a predictive marker for lymph node positivity in colorectal cancer. Clin Cancer Res. 2003;9:1808–1814. [PubMed] [Google Scholar]
- 65.Tada K, et al. Gains of 8q23-qter and 20q and loss of 11q22-qter in esophageal squamous cell carcinoma associated with lymph node metastasis. Cancer. 2000;88:268–273. doi: 10.1002/(sici)1097-0142(20000115)88:2<268::aid-cncr4>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 66.Li Z, et al. Down-regulation of 14-3-3ζ suppresses anchorage-independent growth of lung cancer cells through anoikis activation. Proc Natl Acad Sci U S A. 2008;105:162–167. doi: 10.1073/pnas.0710905105. [DOI] [PMC free article] [PubMed] [Google Scholar]