Abstract
Background
The study of dendritic cell (DC) biology in the rhesus macaque is becoming increasingly important but is limited by incomplete characterization and the lack of rapid assay to quantify cells.
Methods
We characterized the surface phenotype of myeloid (mDC) and plasmacytoid DC (pDC) subsets in healthy rhesus macaque blood and developed a flow cytometry-based assay for absolute DC determinations.
Results
Rhesus CD11c+ mDC were CD16+ CD11b+ CD56lo CD8− CD1c− whereas CD123+ pDC lacked expression of these markers. Precise DC determinations were performed using a rapid two-step assay combining analysis of whole blood and peripheral blood leukocytes (PBL).
Conclusions
Antibodies to CD11b, CD56 and CD16 must be omitted from the lineage antibody cocktail to prevent inadvertent gating-out of DC when analyzing rhesus blood. The combined whole blood/PBL quantification assay will be invaluable for the rapid and repeated monitoring of blood DC counts in this species.
Keywords: nonhuman primate, flow cytometry, mononuclear cells
Introduction
Dendritic cells (DC) are bone marrow-derived leukocytes existing in blood and tissues that are critical in innate and adaptive immunity [1]. Identification of DC in human blood traditionally relies on expression of MHC class II and lack of expression of lineage markers, including CD3 (T cells), CD14 (monocytes), CD19 (B cells) and CD56 (NK cells) as well as the α-integrin CD11b, which is not expressed by any human DC subset [2]. Within this lineage MHC class+ fraction two major populations of DC have been described, the CD11c− CD123+ plasmacytoid DC (pDC) and the CD11c+ CD123− myeloid or ‘conventional’ DC (mDC) [3]. Human mDC can be further subdivided into three functionally distinct subsets based on differential expression of CD16, CD1b/c, and BDCA-3 [2].
Rhesus macaque monkeys are increasingly being used to study DC biology in models of infectious diseases and transplantation; however, blood DC subsets in this species remain incompletely defined [4–9]. Rhesus DC subsets frequently are defined based in part on lack of expression of CD11b, CD16 and/or CD56 [6, 7, 9], although whether they actually lack these markers has not been determined. The phenotype of rhesus monocytes and NK cells differs from their human counterparts particularly with respect to expression of CD56 [10], raising the possibility that differences exist between rhesus and human mDC as well. It is also not known whether rhesus blood mDC can be subdivided based on differential expression of surface markers.
Essential to studies of DC biology in vivo is the ability to accurately and repeatedly quantify DC subsets in blood. Studies in the rhesus macaque to date have relied on several different technologies to determine absolute DC counts, usually combining differential counts to make absolute determinations of mononuclear cells and flow cytometry to determine the proportion of mononuclear cells that are pDC or mDC [4, 8]. A rapid assay has been developed for enumeration of human DC and more recently cynomolgus macaque DC using single-platform technology whereby absolute cell counts are made from a single tube of whole blood using only the flow cytometer for analysis [11, 12]. This approach incorporates TruCOUNT tubes which contain a known quantity of fluorescent beads that serves as an internal calibrator and is the approved method for performing CD4+ T cell determinations in persons infected with human immunodeficiency virus [13]. A similar quantitative assay for rhesus macaque DC subsets would be invaluable but has yet to be described.
In this study we used multiparameter flow cytometry to characterize the cell surface phenotype of blood mDC and pDC subsets in healthy rhesus macaques and to investigate the potential for developing a reproducible and simple flow cytometry-based assay for performing absolute DC determinations. We found that rhesus macaque blood mDC but not pDC have a phenotype quite distinct from their counterpart in human blood. In addition, we found that DC could not be adequately resolved using whole rhesus macaque blood whereas both subsets were readily delineated when peripheral blood leukocytes (PBL) were analyzed. We therefore developed a modified single-platform enumeration assay using whole blood in TruCOUNT tubes to count mononuclear cells in parallel with analysis of PBL to determine the relative proportions of each DC subset.
Materials and methods
Animals and samples
Healthy adult rhesus macaques (Macaca mulatta) were used. Animals were housed and all procedures were conducted in compliance with the University of Pittsburgh Institutional Animal Care and Use Committee regulations. Peripheral blood mononuclear cells (PBMC) were isolated from anticoagulant citrate dextrose (ACD)-collected blood by density gradient centrifugation over Histopaque-1077 (Sigma). Alternatively, 50 or 100 μL EDTA-collected whole blood was treated with 1 or 2 mL ammonium-chloride-potassium (ACK) lysing buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM Na2EDTA in dH2O, pH: 7.2 – 7.4), respectively, for 10 min at room temperature, centrifuged and washed two times to generate peripheral blood leukocytes (PBL). For experiments with whole blood antibodies were added and blood was then treated with FACS Lysing Solution (BD Biosciences) to lyse red blood cells prior to analysis.
Antibodies and flow cytometry
All antibodies were purchased from BD Biosciences unless otherwise noted. To identify DC subsets PBMC, PBL or whole blood depending on the experiment was labeled with antibodies to CD3 (clone SP34), CD14 (M5E2) and CD20 (2H7, eBioscience, all Pacific blue conjugates combined as a Lineage cocktail), HLA-DR (L243, FITC), CD11c (S-HCL-3, APC) and CD123 (7G3, PE) along with blue-fluorescent live/dead viability dye (Invitrogen). Alternatively, cells were stained as above except HLA-DR-PerCP and CD123-biotin with streptavidin QDot525 (Invitrogen) were used and co-stained with antibodies to CD11b (ICRF44, Alexa488), CD56 (MY31, PE), CD8α (B9.11, Coulter, FITC), CD16 (3G8, PE), CD33 (AC104.3E3, Miltenyi Biotech, PE) and/or CD1c (AD5-8E7, Miltenyi Biotech, FITC). In some experiments PBMC were stained as above except CD16-APC-Cy7 was used along with CD11b-Alexa488 and CD56-PE. Relevant isotype control antibodies were used as indicated. In some experiments monocytes (HLA-DR+ CD14+ cells in PBMC), NK cells (lineage− HLA-DR− cells in PBMC), granulocytes (CD45+ side scatter [SSC]hi cells in PBL) and B cells (CD20+ HLA-DR+ cells in PBMC) were used for comparison. To identify CD4+ T cells whole blood or PBL was stained with antibodies to CD45-PerCP, CD3-Pacific blue and CD4-FITC. Data were collected on a 4-laser BD LSRII flow cytometer and analyzed using FACSDiva software (BD Biosciences). Experiments with DC involved collection of at least 500 pDC and 1,000 mDC events. Histogram overlays were generated using FlowJo software (Tree Star).
Absolute cell determinations using TruCOUNT tubes
For absolute cell determinations 50 μL (T cells) or 100μL (DC) of EDTA-collected whole blood was precisely measured and placed into TruCOUNT tubes (BD Biosciences). Cells were then stained with monoclonal antibody mixtures as above for 15 minutes at room temperature followed by the addition of 450μL (T cells) or 900μL (DC) BD FACS Lysing Solution for 15 minutes at room temperature, placed at 4oC and analyzed within 4 hours.
Results
Surface phenotype of rhesus blood DC subsets
To investigate the phenotypic complexity of DC in healthy rhesus macaque blood we used multiparameter flow cytometry. mDC and pDC subsets were identified in PBMC as lineage− (using only markers for CD3, CD14, and CD20) HLA-DR+ cells that were CD11c+ CD123− or CD11c−CD123+, respectively (Fig. 1A), as previously described [4, 5]. Rhesus pDC were phenotypically homogeneous and lacked expression of CD11b, CD56 and CD16 (Fig. 1B). CD8 expression on pDC was difficult to visualize given the rarity of cells and the relative breadth of fluorescence but was judged to be negative as the histogram completely overlapped with that of the isotype control antibody (Fig. 1B). In contrast, blood mDC expressed both CD11b and CD56, although at levels considerably lower than on blood monocytes (Fig. 1C). Blood mDC lacked expression of CD8 but uniformly expressed high levels of CD16, antigens that were co-expressed on blood NK cells (Fig. 1C) [10]. Notably, rhesus mDC lacked surface expression of the myeloid marker CD33, which was expressed on circulating granulocytes (Fig. 1C) but not monocytes (data not shown). Rhesus mDC also lacked expression of CD1c, although this antigen was expressed at low levels by B cells (Fig. 1C). Multicolor staining of CD11c+ mDC within the lineage− HLA-DR+ gate gave no indication of the existence of distinct mDC subsets, as mDC were uniformly CD16+ CD11b+ CD56lo (Fig. 1D).
Figure 1. Surface phenotype of rhesus macaque blood DC subsets.
(A) PBMC from healthy rhesus macaques were labeled with antibodies to lineage markers (CD3, CD14 and CD20, all Pacific blue), HLA-DR-FITC, CD11c-APC and CD123-PE along with blue-fluorescent live/dead viability dye and mDC and pDC identified as shown. (B, C) PBMC were stained as above except HLA-DR-PerCP and CD123-biotin/streptavidin QDot525 was used and co-stained with antibodies to CD11b-Alexa488, CD56-PE, CD8α-FITC, CD16-PE, CD33-PE and/or CD1c-FITC and pDC and mDC analyzed as shown. Monocytes, NK cells, granulocytes and B cells were used for comparison as indicated. Dotted lines and solid histograms represent staining with control and specific antibodies, respectively. Numbers represent percentage of cells in bracketed regions. (D) PBMC were stained as in (B) except CD16-APC-Cy7 was used along with CD11b-Alexa488 and CD56-PE or relevant isotype control antibodies and DC gated as indicated. Data represent between 2 and 5 experiments from different monkeys. FSC = forward scatter; SSC = side scatter.
Poor discrimination of lineage− MHC class II+ cells and CD11c+ mDC in whole blood
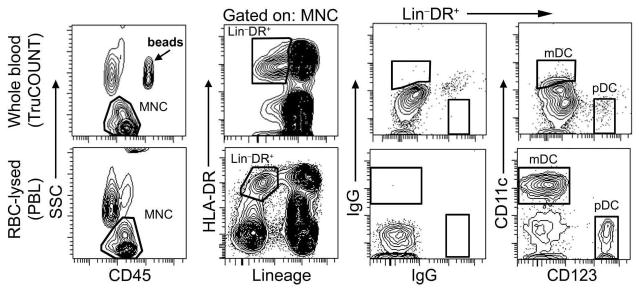
We next determined whether rhesus macaque blood DC subsets could be discriminated in lysed whole blood in the absence of washing, a prerequisite to establishing an accurate single-platform step assay for absolute DC determinations [12]. Mononuclear cells characterized by low SSC and expression of CD45 were readily identified along with fluorescent beads when whole blood was stained in TruCOUNT tubes prior to red blood cell lysis, as expected (Fig. 2). However, the lineage− HLA-DR+ fraction of cells was poorly delineated in lysed whole blood, and CD11c staining within this fraction was almost absent, making accurate enumeration of mDC in particular unlikely (Fig. 2). To determine whether the presence of red blood cells and/or plasma in whole blood may be interfering with antibody staining we next stained PBL, in which whole blood was first lysed of red blood cells and washed prior to addition of antibodies. In contrast to staining of whole blood, lineage− HLA-DR+ cells and both DC subsets were readily discriminated when PBL were analyzed (Fig. 2). The lineage− HLA-DR+ fraction of PBL represented approximately 4% (3.9 ± 1.5% [mean ± SD for 8 animals]) of mononuclear cells with mDC and pDC being 60% (62 ± 18%) and 3% (2.5 ± 1.5%) respectively of cells in this fraction, similar to the proportions in PBMC (data not shown) [4, 5, 8].
Figure 2. DC subsets can be discriminated when PBL but not whole blood are stained.
Whole blood was stained in a TruCOUNT tube with antibodies to CD45-PerCP, CD3/CD20/CD14-Pacific blue (lineage), HLA-DR-FITC and either CD11c-APC and CD123-PE or isotype control antibodies followed by lysis of red blood cells and analysis by flow cytometry (top). Alternatively, whole blood was treated with ACK lysing buffer to generate PBL before addition of antibodies (bottom). Fluorescence from beads is identified by the arrow, and individual regions define cells as indicated. Data are representative of experiments in 8 different animals.
Validation of a two-step quantification assay using whole blood and PBL
To get around the inability to adequately discriminate DC subsets in whole blood we next determined if we could accurately count rhesus blood cells using a modified single-platform assay that combined absolute mononuclear cell determinations in whole blood using TruCOUNT tubes with multiparameter analysis of PBL. To test this approach we turned to CD4+ T cell enumeration, for which the TruCOUNT assay is the gold standard [13]. For the traditional single-platform assay, whole blood from 9 different rhesus macaques was labeled with antibodies to CD45, CD3 and CD4 in TruCOUNT tubes followed by red blood cell lysis and analysis by flow cytometry. The number of CD4+ T cells/μL blood was then calculated using the formula: number of CD45+ SSClo CD3+ CD4+ T cells collected (R1)/number of beads collected × number of beads per TruCOUNT tube/blood volume (Fig. 3). For our combined whole blood/PBL assay, we made absolute mononuclear cell determinations using whole blood in TruCOUNT tubes by gating only on CD45+ SSClo cells, and in parallel we labeled PBL with antibodies to CD45, CD3 and CD4 to determine the proportion of CD45+ SSClo cells that were CD3+ CD4+ (Fig. 3). This proportion was multiplied by the calculated number of CD45+ SSClo cells to reach the absolute number of CD4+ T cells. The conventional, single-platform TruCOUNT assay and the combined whole blood/PBL assay resulted in equivalent CD4+ T cell determinations for 9 different animals (Fig. 3), validating the combined whole blood/PBL approach for cell determinations.
Figure 3. Validation of a combined whole blood/PBL counting assay using CD4+ T cells.
Whole blood was either stained with antibodies to CD45-PerCP, CD3-Pacific blue and CD4-FITC in a TruCOUNT tube and then treated with BD FACS Lysing Solution (top) or treated with ACK lysing buffer before staining with the same antibodies (bottom) and analyzed by flow cytometry. The number of CD4+ T cells/μL blood for 9 healthy rhesus macaques was calculated using either the single-platform TruCOUNT assay (designated TruCOUNT) or the combined whole blood/PBL assay (designated PBL) as described in the text. Symbols represent values from individual animals. A paired Wilcoxon rank sum test was used to compare CD4+ T cell counts using the two methods. MNC = mononuclear cells; NS = not significant.
Quantification of rhesus blood DC subsets using the combined whole blood/PBL assay
We next adapted the whole blood/PBL assay to enumerate DC in healthy rhesus macaque blood. For this assay, whole blood was stained in TruCOUNT tubes with antibody to CD45 and the number of CD45+ SSClo mononuclear cells/μL calculated using the formula described above. In parallel, PBL were stained with antibodies to CD45, lineage cocktail, HLA-DR, CD11c and CD123 and the proportion of mononuclear cells that were either lineage− HLA-DR+ CD11c+ CD123− mDC or lineage− HLA-DR+ CD11c− CD123+ pDC determined (Fig. 4A). The percentage of mDC and pDC in the mononuclear cell fraction was then multiplied by the number of mononuclear cells/μL blood to calculate the number of mDC/μL and pDC/μL, respectively, as for the CD4+ T cell counts previously. Using this whole blood/PBL assay we performed absolute mDC and pDC determinations for 9 healthy rhesus macaques taking 4 to 6 measurements over a 2 to 3 month period. The within animal variability in mean pDC number over time was relatively constant for pDC but was quite marked for mDC determinations, with some animals having very little change in mDC count over time (for example animals 1 and 7) and other animals having marked variability (for example animals 3 and 4) (Fig. 4B). Across all animals the mean number of each DC subset also varied with the most variability again noted in the mDC subset (Fig. 4B, C). The intra-and inter-animal differences in mDC and pDC numbers could not be explained solely by differences in mononuclear cell counts, as there was no consistent pattern within or between animals for all 3 cell types (Fig. 4B). The median number of mDC and pDC in blood for all 9 rhesus macaques was 50 cells/μL and 3 cells/μL, respectively (Fig. 4C). This result was similar to that determined using a white blood cell differential count to enumerate mononuclear cells together with flow cytometric analysis to determine the proportion of mDC and pDC in PBMC (data not shown), and is consistent with data reported previously by us and others in rhesus macaques using this conventional approach [4, 5, 8].
Figure 4. Enumeration of rhesus blood DC subsets using the combined whole blood/PBL assay.
(A) Whole blood was stained with antibodies to CD45-PerCP in a TruCOUNT tube or treated with ACK lysing buffer and washed to generate PBL prior to staining with antibodies to CD45-PerCP, CD3/CD20/CD14-Pacific blue (lineage), HLA-DR-FITC, CD11c-APC and CD123-PE. (B) The number of mDC, pDC and mononuclear cells in blood of 9 rhesus macaques was calculated by taking 4–6 measurements over a 2–3 month period using the combined whole blood/PBL assay as described in the text. Shown are the mean ± SD for individual animals. (C) Box plots showing mDC and pDC counts for all animals. The horizontal line in the box represents the median, the top and bottom borders of the box represent 25th and 75th percentiles, and the bars represent maximum and minimum values. MNC = mononuclear cells.
Discussion
Our results have significant implications for the study of DC in the rhesus macaque and highlight some key similarities and differences between human and rhesus DC subsets. Rhesus blood CD123+ pDC lacked expression of CD16, CD8, CD11b and CD56, as in the human [2]. In contrast, rhesus CD11c+ mDC were uniformly CD16+ CD8− CD11b+ CD56lo CD33− CD1c−, a phenotype quite distinct from human mDC [2]. These findings clearly show that inclusion of antibodies to CD11b, CD16 and/or CD56 in the lineage cocktail will result in inadvertent exclusion of mDC from the lineage− MHC class II+ gate when analyzing rhesus blood, as indicated by others previously [14]. Moreover, given that rhesus mDC expressed CD16 at intensities equivalent to NK cells when stained with the 3G8 clone, in vivo delivery of this antibody designed to deplete NK cells would likely also deplete mDC [15].
The lack of CD1c staining on rhesus macaque mDC is notable as it defines an apparent phenotypic difference between rhesus and cynomolgus macaques, where CD1c has been used as an alternative marker to CD11c to identify blood mDC [11]. CD1c+ mDC in cynomolgus macaque blood express high levels of HLA-DR [11], as do CD1b/c+ CD16− human mDC [2]. In previous studies we have identified a small population of Lineage− HLA-DRhi cells that are CD11clo CD123−, consistent with mDC [4], however this minor subset lacks expression of both CD16 and CD1c (data not shown). We have previously reported that CD1c is expressed by rhesus macaque PBMC [5] and our current data confirm this expression to be on B cells, as in the human [16]. It should be noted that our antibody panel was limited by the fact that antibodies to BDCA-2 and BDCA-3, which have been used to define discrete mDC subsets in human blood [2, 16], are not cross-reactive in the rhesus macaque [5]. It is therefore possible that additional subsets of mDC do exist in rhesus macaque blood but cannot be identified based on available reagents.
The inability to identify mDC based on CD11c staining when whole blood was analyzed has been previously noted in other nonhuman primates [11] but not humans [12], for reasons that are not clear. We circumvented this problem by developing a whole blood/PBL assay to perform absolute DC determinations using the flow cytometer, which allowed us to use conventional markers including CD11c to determine proportions of mDC and pDC within the mononuclear cell fraction. The advantage of using PBL over PBMC to define DC subsets lies in the fact that PBL can be rapidly prepared from a minimal quantity of blood without the risk of cell loss associated with density gradient purification. In addition, we frequently find superior discrimination of DC subsets when PBL are analyzed as compared to PBMC (data not shown).
The whole blood/PBL quantification assay involves two simple steps that can be performed in parallel: (1) Antibody staining and analysis of a precise volume of whole blood in TruCOUNT tubes to identify SSClo CD45+ mononuclear cells that are then quantified based on a known number of internal calibrator beads, and (2) Multiparameter analysis of PBL to determine the percentage of mononuclear cells that are pDC and mDC. The pDC and mDC percentages are then multiplied by the absolute number of mononuclear cells to determine blood pDC and mDC counts, respectively. We believe this assay will be of great benefit to in vivo studies of DC biology in the rhesus macaque and potentially other nonhuman primate species. Using the whole blood/PBL assay we found that the number of mDC and to a lesser extent pDC fluctuated over time depending on the individual animal, which is in apparent contrast to findings in the cynomolgus macaque [11]. The same fluctuations were not as apparent in the total mononuclear cell compartment, suggesting that the variability was not due to loss of cells during processing. Given this natural fluctuation we recommend that at least 4 independent determinations for each DC subset be made to establish a baseline level prior to studying DC kinetics in response to infection or other experimental manipulation in this model.
Acknowledgments
We would like to thank N. Banichar, A. Trichel and C. Janssen for excellent veterinary support. These studies were supported by training grant AI065380 to K.N.B. and research grant AI071777 to S.M.B.B. from the US National Institutes of Health.
References
- 1.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 2.MacDonald KP, Munster DJ, Clark GJ, Dzionek A, Schmitz J, Hart DN. Characterization of human blood dendritic cell subsets. Blood. 2002;100:4512–20. doi: 10.1182/blood-2001-11-0097. [DOI] [PubMed] [Google Scholar]
- 3.Shortman K, Liu YJ. Mouse and human dendritic cell subtypes. Nat Rev Immunol. 2002;2:151–61. doi: 10.1038/nri746. [DOI] [PubMed] [Google Scholar]
- 4.Brown KN, Trichel A, Barratt-Boyes SM. Parallel loss of myeloid and plasmacytoid dendritic cells from blood and lymphoid tissue in simian AIDS. J Immunol. 2007;178:6958–67. doi: 10.4049/jimmunol.178.11.6958. [DOI] [PubMed] [Google Scholar]
- 5.Coates PT, Barratt-Boyes SM, Zhang L, Donnenberg VS, O’Connell PJ, Logar AJ, Duncan FJ, Murphey-Corb M, Donnenberg AD, Morelli AE, Maliszewski CR, Thomson AW. Dendritic cell subsets in blood and lymphoid tissue of rhesus monkeys and their mobilization with Flt3 ligand. Blood. 2003;102:2513–21. doi: 10.1182/blood-2002-09-2929. [DOI] [PubMed] [Google Scholar]
- 6.Koopman G, Niphuis H, Haaksma AG, Farese AM, Casey DB, Kahn LE, Mann D, MacVittie TJ, Woulfe SL, Heeney JL. Increase in plasmacytoid and myeloid dendritic cells by progenipoietin-1, a chimeric Flt-3 and G-CSF receptor agonist, in SIV-Infected rhesus macaques. Hum Immunol. 2004;65:303–16. doi: 10.1016/j.humimm.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Pichyangkul S, Saengkrai P, Yongvanitchit K, Limsomwong C, Gettayacamin M, Walsh DS, Stewart VA, Ballou WR, Heppner DG. Isolation and characterization of rhesus blood dendritic cells using flow cytometry. J Immunol Methods. 2001;252:15–23. doi: 10.1016/s0022-1759(01)00327-1. [DOI] [PubMed] [Google Scholar]
- 8.Reeves RK, Fultz PN. Disparate effects of acute and chronic infection with SIVmac239 or SHIV-89.6P on macaque plasmacytoid dendritic cells. Virology. 2007;365:356–68. doi: 10.1016/j.virol.2007.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teleshova N, Jones J, Kenney J, Purcell J, Bohm R, Gettie A, Pope M. Short-term Flt3L treatment effectively mobilizes functional macaque dendritic cells. J Leukoc Biol. 2004;75:1102–10. doi: 10.1189/jlb.1103588. [DOI] [PubMed] [Google Scholar]
- 10.Carter DL, Shieh TM, Blosser RL, Chadwick KR, Margolick JB, Hildreth JE, Clements JE, Zink MC. CD56 identifies monocytes and not natural killer cells in rhesus macaques. Cytometry. 1999;37:41–50. [PubMed] [Google Scholar]
- 11.Malleret B, Karlsson I, Maneglier B, Brochard P, Delache B, Andrieu T, Muller-Trutwin M, Beaumont T, McCune JM, Banchereau J, Le Grand R, Vaslin B. Effect of SIVmac infection on plasmacytoid and CD1c+ myeloid dendritic cells in cynomolgus macaques. Immunology. 2008;124:223–33. doi: 10.1111/j.1365-2567.2007.02758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vuckovic S, Gardiner D, Field K, Chapman GV, Khalil D, Gill D, Marlton P, Taylor K, Wright S, Pinzon-Charry A, Pyke CM, Rodwell R, Hockey RL, Gleeson M, Tepes S, True D, Cotterill A, Hart DN. Monitoring dendritic cells in clinical practice using a new whole blood single-platform TruCOUNT assay. J Immunol Methods. 2004;284:73–87. doi: 10.1016/j.jim.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Mandy FF, Nicholson JK, McDougal JS. Guidelines for performing single-platform absolute CD4+ T-cell determinations with CD45 gating for persons infected with human immunodeficiency virus. Centers for Disease Control and Prevention. MMWR Recomm Rep. 2003;52:1–13. [PubMed] [Google Scholar]
- 14.Lore K. Isolation and immunophenotyping of human and rhesus macaque dendritic cells. Methods Cell Biol. 2004;75:623–42. doi: 10.1016/s0091-679x(04)75026-8. [DOI] [PubMed] [Google Scholar]
- 15.Choi EI, Wang R, Peterson L, Letvin NL, Reimann KA. Use of an anti-CD16 antibody for in vivo depletion of natural killer cells in rhesus macaques. Immunology. 2008;124:215–22. doi: 10.1111/j.1365-2567.2007.02757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S, Buck DW, Schmitz J. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol. 2000;165:6037–46. doi: 10.4049/jimmunol.165.11.6037. [DOI] [PubMed] [Google Scholar]