Abstract
Staufen1 (STAU1)-mediated mRNA decay (SMD) degrades translationally active mRNAs that bind the double-stranded (ds)RNA binding protein STAU1 within their 3'-untranslated regions (3'UTRs)1,2. Earlier studies defined the STAU1 binding site (SBS) within ADP ribosylation factor 1 (ARF1) mRNA as a 19-base-pair stem with a 100-nucleotide apex2. However, we were unable to identify comparable structures within the 3'UTRs of other SMD targets. Here we report that SBSs can be formed by imperfect base-pairing between an Alu element within the 3'UTR of an SMD target and another Alu element within a cytoplasmic and polyadenylated long noncoding RNA (lncRNA). Individual lncRNAs can downregulate a subset of SMD targets, and distinct lncRNAs can downregulate the same SMD target. These are previously unappreciated functions for ncRNAs and Alu elements3–5. Not all mRNAs that contain a 3'UTR Alu element are targeted for SMD despite the presence of a complementary lncRNA that targets other mRNAs for SMD. Most known trans-acting RNA effectors consist of fewer than 200 nucleotides and include snoRNAs and microRNAs. Our finding that STAU1 binding to mRNAs can be transactivated by lncRNAs uncovers an unexpected strategy used by cells to recruit proteins to mRNAs and mediate their decay. We name these lncRNAs “half(½)-sbsRNAs”.
Our failure using Mfold6 to identify dsRNA structures similar to the SBS of ARF1 mRNA within the 3'UTRs of other SMD targets led us to notice that two well-characterized SMD targets – plasminogen activator inhibitor type 1 (SERPINE1) mRNA and hypothetical protein FLJ21870 mRNA1,2 – contain a single 3'UTR Alu element. We also found that ~13% of the ~1.6% of protein-encoding transcripts in human epithelial HeLa cells that are upregulated at least 1.8-fold upon STAU1 downregulation in three independently performed microarray analyses2 contain a single 3'UTR Alu element (Supplementary Table 1). This percentage is higher than the ~4% of HeLa-cell protein-encoding transcripts that contain one or more 3'UTR Alu elements7, indicating that 3'UTR Alu elements are enriched in SMD targets relative to the bulk of cellular mRNAs.
Alu elements are the most prominent repeats in the human genome: they constitute more than 10% of DNA sequences, are present at up to 1.4 million copies per cell, and share a 300-nucleotide consensus sequence of appreciable similarity among subfamilies8. To date, Alu elements have been documented to be cis-effectors of protein-encoding gene expression by influencing transcription initiation or elongation, alternative splicing, A-to-I editing or translation initiation3,5,9. Since ncRNAs that perfectly base-pair with mRNA can function in trans to generate endogenous siRNAs4, it seemed possible that imperfect base-pairing between the Alu element of a ncRNA and the Alu element of an mRNA 3'UTR could create an SBS so as to regulate mRNA decay. We focused on mRNAs that contain a single 3'UTR Alu-element to avoid the possibility of intramolecular base-pairing between inverted Alu elements, which could result in A-to-I editing and nuclear retention10.
Analysis of Antisense ncRNA Pipeline11,12 identified 378 lncRNAs that contain a single Alu element (Supplementary Table 2). Among them, the Alu element of lncRNA_AF087999 (NCBI) has the potential to base-pair with the Alu element within SERPINE1 and FLJ21870 3'UTRs (Fig. 1a; Supplementary Fig. 1a) with ΔG values of, respectively, −151.7 kcal/mol and −182.1 kcal/mol (Supplementary Table 2; where −151.7 kcal/mol defined the most stable duplex predicted to form between SERPINE1 mRNA and any of the 378 lncRNAs). lncRNA_AF087999, which for reasons that follow is designated ½-sbsRNA1, derives from chromosome 11. RT-semiquantitative (sq)PCR (Supplementary Fig. 2a) demonstrated that ½-sbsRNA1 is detected in cytoplasmic but not nuclear HeLa-cell fractions and is polyadenylated (Supplementary Fig. 2b,c). Downregulating the cellular abundance of the two major isoforms of STAU1 to <10% of normal (see, e.g., below) did not affect either the cellular distribution or the abundance of ½-sbsRNA1 (Supplementary Fig. 2b). ½-sbsRNA1 is present in every human tissue that was examined (Supplementary Fig. 2d). ½-sbsRNA1 is not a substrate for Dicer or AGO2 (Supplementary Fig. 2e) and thus is distinct from the lncRNAs that generate endogenous siRNAs.
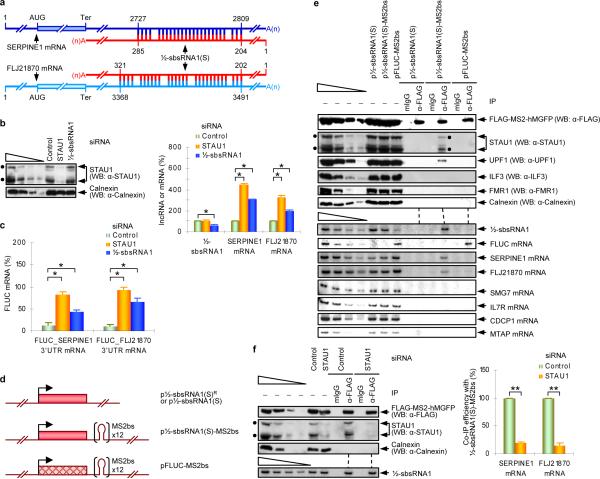
Figure 1. lncRNA_AF087999 (½-sbsRNA1) binds to and reduces the abundance of specific SMD targets.
a, Predicted base-pairing between the Alu element within the SERPINE1 or FLJ21870 3'UTR and the Alu element within ½-sbsRNA1, where 1 was defined as the first transcribed nucleotide of each mRNA or ½-sbsRNA1(S). b, Left: Western blotting (WB), using the designated antibody (α), of lysates of HeLa cells treated with the specified siRNA, where Calnexin serves as a loading control. Right: Representation of RT-sqPCR analyses of ½-sbsRNA1, SERPINE1 or FLJ21870 mRNA from the same lysates, where the normalized level of each transcript in the presence of Control siRNA was defined as 100. c, Representation of RT-sqPCR analyses of FLUC-SERPINE1 3'UTR or FLUC-FLJ21870 3'UTR SMD reporter mRNA in cells that had been transiently transfected with the specified siRNA, where the normalized level of each transcript in the presence of Control siRNA was defined as 100. d, Diagrams of expression vectors encoding ½-sbsRNA1(S)R, which is siRNA-resistant, ½-sbsRNA1(S) or FLUC with or without 12 copies of the MS2 coat protein binding site (MS2bs). e, Western blot (upper) or RT-sqPCR (lower) before (−) or after immunoprecipitation (IP) using anti-FLAG or, as a control for nonspecific IP, mouse(m) IgG of lysates of formaldehyde-crosslinked HeLa cells that had been transiently transfected with pFLAG-MS2-hMGFP and either the denoted ½-sbsRNA1(S) expression vector or pFLUC_MS2bs. f, As in e, except cells were treated with Control or STAU1 siRNA. Left: Western blotting. Right: RT-sqPCR, where the co-IP efficiency indicates the level of each mRNA-derived product after IP relative to before IP. Each ratio in the presence of Control siRNA was defined as 100%. See Supplementary Fig. 5 for phosphorimages and evaluations of RT-sqPCR data shown here as histograms. Error bars indicate s.e.m. Single asterisk, n = 6, P < 0.01; double asterisks, n = 3, P < 0.05.
Two forms of ½-sbsRNA1 have been reported (NCBI). They differ at their 5' end but share a common Alu element and a common 3' end that contains a putative polyadenylation signal (AUUAAA) situated 13 nucleotides upstream of a poly(A) tract. RNase protection assays confirmed the presence of one short (S) and one long (L) form of ½-sbsRNA1 that have a different 5' end and a relative abundance in HeLa cells of 3:1 (Supplementary Fig. 3a). Primer extension (Supplementary Fig. 3b) and RT-sqPCR (Supplementary Fig. 3c) mapped the 5' end of ½-sbsRNA1(S) to a C residue. Therefore, ½-sbsRNA1(S) consists of 688 nucleotides excluding the poly(A) tract (Supplementary Fig. 3d). While some transcripts that are annotated as ncRNAs may be translated4, data indicate that ½-sbsRNA1(S) is not translated (Supplementary Fig. 4).
Remarkably, not only STAU1 siRNA but also ½-sbsRNA1 siRNA increased the levels of SERPINE1 and FLJ21870 mRNAs to 2-to-4.5-fold above normal (Fig. 1b; Supplementary Fig. 5; Supplementary Fig. 6a; Supplementary Table 3). Furthermore, experiments that employed cycloheximide indicated that the ½-sbsRNA1-mediated reduction in SERPINE1 and FLJ21870 mRNA abundance depends on translation (Supplementary Fig. 6b), as does SMD13. The reduction in SERPINE1 and FLJ21870 mRNA abundance is attributable to their respective 3'UTR sequences since ½-sbsRNA1 siRNA also increased the levels of FLUC-SERPINE1 3'UTR and FLUC-FLJ21870 3'UTR reporter mRNAs relative to FLUC-No SBS mRNA (Fig. 1c; Supplementary Fig. 5; Supplementary Table 3). The ½-sbsRNA1 siRNA-mediated increase in the abundance of SERPINE1 or FLJ21870 mRNA was reversed by co-expressing ½-sbsRNA1(S)R, which is resistant to siRNA (Supplementary Fig. 6c), arguing against siRNA-mediated off-target effects. Significantly, ½-sbsRNA1 siRNA did not affect the expression of other FLUC reporter mRNAs that contain the 3'UTR of SMD targets not predicted to base-pair with ½-sbsRNA1 (Supplementary Fig. 7).
If ½-sbsRNA1 were to create an SBS by base-pairing with the 3'UTR of SERPINE1 or FLJ21870 mRNA, then it should be possible to co-immunoprecipitate complexes of the lncRNA and each mRNA. To test this possibility, lysates of HeLa cells that transiently expressed (i) ½-sbsRNA1(S)-MS2bs, which contains 12 copies of the MS2 coat protein binding site (MS2bs)14 upstream of the lncRNA polyadenylation signal or, as a negative control, ½-sbsRNA1(S) or FLUC-MS2bs mRNA (Fig. 1d) and (ii) FLAG-MS2-hMGFP, which consists of FLAG-tagged MS2 coat protein fused to hMGFP, were immunoprecipitated using anti-FLAG. As expected, prior to IP ½-sbsRNA1(S) as well as ½-sbsRNA1(S)-MS2bs decreased the abundance of SERPINE1 and FLJ21870 mRNAs but not SMD targets that encode interleukin 7 receptor (IL7R), CUG domain-containing protein 1 (CDCP1) or methylthioadenosine phosphorylase (MTAP) (Fig. 1e; see below). In support of our hypothesis that ½-sbsRNA1 creates an SBS with partially complementary mRNA sequences, using lysates of cells expressing ½-sbsRNA1(S)-MS2bs, the anti-FLAG IP of FLAG-MS2-hMGFP bound to ½-sbsRNA1(S)-MS2bs co-immunoprecipitated endogenous STAU1, SERPINE1 mRNA and FLJ21870 mRNA as well as the UPF1 SMD factor (Fig. 1e). In contrast, irrelevant proteins, such as Calnexin, the dsRNA binding protein ILF315, the single-stranded RNA binding protein FMR116, and mRNAs that are not predicted to base-pair with ½-sbsRNA1, such as those encoding SMG7, IL7R, CDCP1 or MTAP, were not co-immunoprecipitated (Fig. 1e). STAU1 siRNA reduced the co-IP of the SERPINE1 mRNA as well as FLJ21870 mRNA with ½-sbsRNA1(S)-MS2bs to, respectively, ~19% or ~15% of normal (Fig. 1f; Supplementary Fig. 5), indicating that STAU1 stabilizes the duplex formed between SERPINE1 or FLJ21870 mRNA and ½-sbsRNA1.
As additional evidence that ½-sbsRNA1 creates an SBS by base-pairing with the SERPINE1 or FLJ21870 3'UTR, only STAU1-HA3 but not ILF3 or FMR1 co-immunoprecipitated with ½-sbsRNA1 (Supplementary Fig. 8).
To determine if ½-sbsRNA1 is required for the co-IP of STAU1 with SERPINE1 or FLJ21870 mRNA, HeLa cells that transiently expressed STAU1-HA3 and Control siRNA or ½-sbsRNA1 siRNA in the presence or absence of ½-sbsRNA1(S)R were immunoprecipitated using anti-HA. Compared to Control siRNA, ½-sbsRNA1 siRNA, which reduced the level of ½-sbsRNA1 to ~50% of normal, reduced by ~2-fold the co-IP of STAU1-HA3 with SERPINE1 or FLJ21870 mRNA (Fig. 2a; Supplementary Fig. 5). In contrast, restoring the level of ½-sbsRNA1 to ~100% of normal by expressing ½-sbsRNA1 siRNA together with ½-sbsRNA1(S)R restored the co-IP of STAU1-HA3 with SERPINE1 or FLJ21870 mRNA to near normal (Fig. 2a; Supplementary Fig. 5). As expected, the level of IL7R mRNA, which binds STAU12 but does not contain sequences complementary to ½-sbsRNA1, was unaffected by any condition either before or after IP (Fig. 2a; Supplementary Fig. 5).
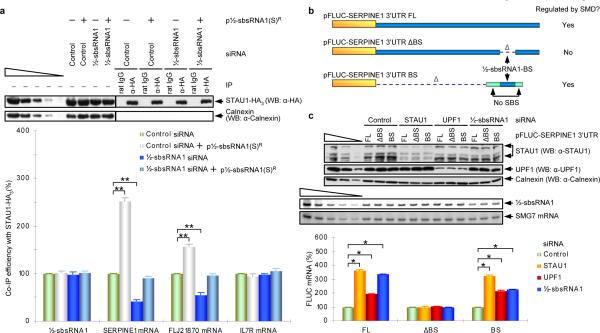
Figure 2. ½-sbsRNA1 co-immunoprecipitates with STAU1 and is required for STAU1 binding to specific SMD targets.
a, Western blotting (upper) or RT-sqPCR (lower) of lysates of formaldehyde-crosslinked HeLa cells that had been transiently transfected with the specified siRNA and either empty vector (−) or p½-sbsRNA1(S)R (+) before or after IP with anti-HA or rat IgG. After IP, each sample was spiked with in vitro-synthesized E. coli LACZ mRNA. The co-IP efficiency provides the level of each mRNA RT-sqPCR product after IP relative to before IP, where each ratio in the presence of Control siRNA was defined as 100%. b, Diagrams of pFLUC-SERPINE1 3'UTR FL, which contains the full-length (FL) SERPINE1 3'UTR, and 3'UTR deletion variants. Yellow boxes, FLUC sequences; blue bars, SERPINE1 3'UTR sequences; Δ, deletion; pale green boxes, 3'UTR of FLUC No SBS, which does not bind STAU1. The 5'-most pale green box ensures that ribosomes translating to the FLUC termination codon do not displace STAU1 that had been recruited to the ½-sbsRNA1-binding site (BS, which is 86 nucleotides as shown in Supplementary Fig. 1a). c, Western blotting (upper) and RT-sqPCR (middle and lower) of lysates of HeLa-cells that had been transiently transfected with the noted pFLUC-SERPINE1 3'UTR test construct and the phCMV-MUP reference plasmid. Lower: The normalized level of each FLUC mRNA in the presence of Control siRNA was defined as 100%. See Supplementary Fig. 5 for phosphorimages and evaluation of RT-sqPCR data shown here as histograms. Error bars indicate s.e.m. Single asterisk, n = 6, P < 0.01; double asterisks, n = 3, P < 0.05.
We conclude that the SMD of SERPINE1 or FLJ21870 mRNA involves base-pairing between their 3'UTR Alu element and the Alu element within ½-sbsRNA1. Base-pairing creates an SBS that is stabilized by STAU1. Furthermore, the level of STAU1 and, thus, the efficiency of SMD does not alter the level of ½-sbsRNA1. Our finding that downregulating SERPINE1 or FLJ21870 mRNA to 50% and 25% of normal, respectively, failed to detectably decrease the co-IP of STAU1-HA3 with ½-sbsRNA1 (Supplementary Fig. 9) indicates that ½-sbsRNA1 may bind to more than SERPINE1 and FLJ21870 mRNAs to recruit STAU1 if not trigger SMD.
The presence of UPF1 in the anti-FLAG IP of FLAG-MS2-hMGFP (Fig. 1e) is consistent with the idea that STAU1 that is bound to a ½-sbsRNA1-created SBS associates with UPF1, analogously to how STAU1 that is bound to the ARF1 SBS associates with UPF12,13. Furthermore, downregulating UPF1, like downregulating STAU1, increases the abundance of SERPINE1 mRNA, FLJ21870 mRNA and FLUCSERPINE1 3'UTR mRNA by increasing mRNA half-life1,2. To test for UPF1 function in conjunction with ½-sbsRNA1, we analyzed the effects of various siRNAs on the production of FLUC-SERPINE1 3'UTR mRNA in which the 3'UTR was intact, precisely lacked the region that was partially complementary to ½-sbsRNA1, or contained solely this region (Fig. 2b). Relative to Control siRNA, STAU1 siRNA, UPF1 siRNA or ½-sbsRNA1 siRNA did not affect the level of FLUC-SERPINE1 3'UTR mRNA that lacked the ½-sbsRNA1 binding site (BS; Fig. 2c; Supplementary Fig. 5), but each siRNA increased the levels of FLUC-SERPINE1 3'UTR mRNA and FLUC mRNA that contained only the ½-sbsRNA1-BS (Fig. 2c; Supplementary Fig. 5). We conclude that, as indicated by its name, ½-sbsRNA1 base-pairs with the 3'UTR of SERPINE1 mRNA and, by analogy, FLJ21870 mRNA so as to recruit STAU1 and its binding partner UPF1 in a way that triggers a reduction in mRNA abundance. Consistent with previous studies of SMD2,13, the STAU1- and ½-sbsRNA1-mediated reduction in mRNA abundance is due to a decrease in mRNA half-life (Supplementary Fig. 10). With regard to function, scrape injury repair assays revealed that ½-sbsRNA1 contributes toward reducing cell migration by targeting SERPINE1 and RAB11FIP1 mRNAs for SMD (Supplementary Fig. 11).
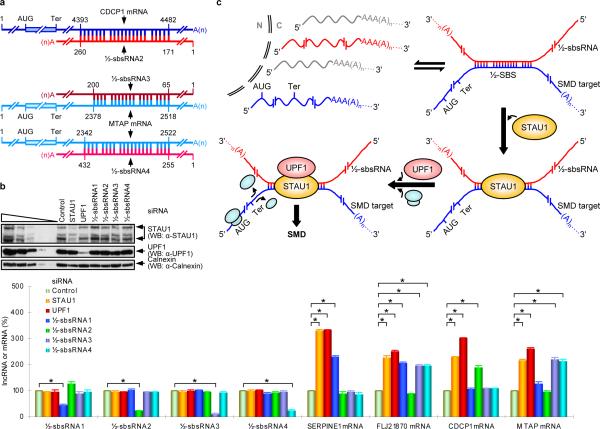
Characterizing seven other lncRNAs that contain a single Alu element and consist of <1000 nucleotides (Supplementary Table 2) confirmed that they, too, are largely cytoplasmic and polyadenylated (Supplementary Fig. 2b,c; data not shown) and have the potential to base-pair with the single Alu element within at least one mRNA 3'UTR (Fig. 3a; Supplementary Fig. 1b,c,d; Supplementary Table 2; data not shown). Individually downregulating three of these lncRNAs – lncRNA_BC058830 (½-sbsRNA2), lncRNA_AF075069 (½-sbsRNA3) or lncRNA_BC009800 (½-sbsRNA4) – upregulated those tested mRNAs that (i) contain a partially complementary Alu element and (ii) are upregulated upon STAU1 or UPF1 downregulation; each lncRNA failed to upregulate mRNAs that lack a partially complementary Alu element (Fig. 3b; Supplementary Fig. 5; data not shown). While ½-sbsRNA2 targeted the 3'UTR Alu element of CDCP1 mRNA (Fig. 3b; Supplementary Fig. 5; Supplementary Table 2, where ΔG = −153.7 kcal/mol), ½-sbsRNA3 and ½-sbsRNA4 targeted the 3'UTR Alu element of MTAP mRNA (Fig. 3b; Supplementary Fig. 5; Supplementary Table 2, where ΔG = −203.1 and −264.2 kcal/mol, respectively). Furthermore, none of the three lncRNAs downregulated SERPINE1 mRNA (Fig. 3b; Supplementary Fig. 5; Supplementary Table 2, where ΔG = 0, −66.4 and −108.2 kcal/mol, respectively) but two downregulated FLJ21870 mRNA ~2-fold (Fig. 3b; Supplementary Fig. 5; Supplementary Table 2, where ΔG = −261.9 and −444.2 kcal/mol).
Figure 3. Evidence that ½-sbsRNA2, ½-sbsRNA3 and ½-sbsRNA4 base-pair with particular mRNA 3'UTRs and decrease mRNA abundance, as do STAU1 and UPF1.
a, Predicted base-pairing between the 3'UTR Alu element of CDCP1 mRNA (Acc#: NM_022842) and ½-sbsRNA2, or MTAP mRNA (Acc#: NM_002451) and ½-sbsRNA3 as well as ½-sbsRNA4, where 1 was defined as the first nucleotide listed in the NCBI data base for each mRNA or lncRNA. b, Essentially as in Fig. 1b. See Supplementary Fig. 5 for phosphorimages and evaluation of RT-sqPCR data shown here as histograms. Error bars indicate s.e.m. Asterisk, n = 6, P < 0.01. c, Model for how an Alu element-containing ½-sbsRNA that is polyadenylated and largely cytoplasmic (red) base-pairs with a partially complementary Alu element, i.e., a half-STAU1 binding site (½-SBS), within the 3'UTR of a particular mRNA (blue) to trigger SMD. Base-pairing forms a functional SBS. The STAU1-bound SBS triggers SMD in a UPF1-dependent mechanism when translation terminates sufficiently upstream of the SBS so that translating ribosomes do not remove bound STAU1. The ½-sbsRNA is not destroyed in the process. N, nucleus; C, cytoplasm; AUG, translation initiation codon; Ter, termination codon (which is generally, but not necessarily, a normal termination codon).
These findings illustrate the potentially complex network of regulatory events that are controlled by lncRNA–mRNA duplexes that bind STAU1 and is reminiscent of the web of regulatory mechanisms that are mediated by miRNAs17. Notably, both CDCP1 mRNA and MTAP mRNA were upregulated at least 2-fold upon STAU1 downregulation in experiments reported here (Fig. 3b), and indeed CDCP1 mRNA is among those mRNAs that were upregulated minimally 1.8-fold upon STAU1 downregulation2(Supplementary Table 1). However, since MTAP mRNA was upregulated only ~1.5-fold2, it is not included in Supplementary Table 1. Thus, this Table must be considered as providing only a partial list of mRNAs that are modulated by one or more ½-sbsRNAs. Conceivably, the degree of modulation could vary in different cell types (Supplementary Fig. 2d) or developmental stages depending on the abundance of the ½-sbsRNA(s) and on proteins that inhibit or enhance base-pairing.
It is important to note that ΔG values are not in themselves absolute predictors of SBS function. For example, while ½-sbsRNA2 is predicted to base-pair with the 3'UTR Alu element of BAG5 mRNA with a ΔG of – 416 kcal/mol, BAG5 mRNA is not targeted for SMD in HeLa cells (Supplementary Fig. 12). The 3'UTR Alu element of BAG5 mRNA may be physically inaccessible to base-pairing with ½-sbsRNA2. Nevertheless, base-pairing per se may not be sufficient for SBS function since converting the 100-nt apex of the intramolecular ARF1 SBS to a 4-nt loop that is not predicted to disrupt the adjacent 19-bp stem of the ARF1 SBS reduces STAU1 binding in vivo by 50%2.
Here, we report an unforeseen role for some of the lncRNAs that contain Alu elements: the creation of SBSs by intermolecular base-pairing with an Alu element within the 3'UTR of one or more mRNAs. We conclude that SBSs can form either through intramolecular base-pairing, as exemplified by the ARF1 SBS, or intermolecular base-pairing between a ½-SBS within an mRNA 3'UTR and a complementary ½-sbsRNA in the form of a largely cytoplasmic lncRNA (Fig. 3c).
There are estimated to be tens of thousands of human lncRNAs that have little or no ability to direct protein synthesis and that are distinct from rRNAs, tRNAs, snRNAs, snoRNAs, small interfering RNAs or microRNAs18. Thus, the paradigm that partially complementary ncRNA–mRNA duplexes can form SBSs may extend to the creation of binding sites for other dsRNA binding proteins. Since only 23% of lncRNAs were found to contain one or more Alu elements, lncRNA–mRNA duplexes that do not involve Alu elements could expand the number of ncRNAs that regulate gene expression via SMD or a different dsRNA binding protein-dependent pathway.
METHODS SUMMARY
The Perl program “Alu_Mask” was written and used together with REPEATMASKER (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) to define Alu elements. The Perl program “RNA_RNA_anneal” was generated to predict intermolecular duplexes between Alu elements within lncRNAs and proven or putative SMD targets. Duplexes were then validated using RNA structure 4.6 (http://rna.urmc.rochester.edu/rnastructure.html), which provides folding free energy changes. Human HeLa or HaCaT cells were transiently transfected with the specified plasmids or siRNAs as described1. For mRNA half-life measurements, HeLa Tet-Off cells (Clontech) were utilized. If formaldehyde-crosslinked, cells were sonicated six times for 30 sec to facilitate lysis, and crosslinks were subsequently reversed by heating at 65°C for 45 min after IP. IP was performed as described1. Protein was purified and Western blotting was performed as noted1. RNA was purified from total, nuclear or cytoplasmic cell fractions or immunoprecipitated from total-cell lysates as reported1. Poly(A)+ RNA was extracted from total-cell RNA using the Oligotex mRNA Mini Kit (Qiagen). RT-sqPCR and RT-qPCR were as described1, except when RT was primed using oligo(dT)18 rather than random hexamers. The RNase protection assay employed the RPA III RNase Protection Assay Kit (Ambion) and uniformly labeled RNA probes that were generated by transcribing linearized pcDNA3.1(+)/Zeo_Chr11_66193000-66191383 in vitro using α-[P32]-UTP (Perkin Elmer) and the MAXIscript Kit (Ambion). Cells were visualized using a Nikon Eclipse TE2000-U inverted fluorescence microscope and, for phase microscopy, a 480-nm excitation spectra. Images were captured utilizing TILLVISION software (TILL Photonics). Scrape injury repair assays were essentially as published21,22. All data derive from at least three independently performed experiments that did not vary by more than the amount shown, and p values for all RT-sqPCR results were <0.05.
Supplementary Material
Acknowledgements
We thank Dave Mathews and Alan Grossfield for use of computer clusters; Dave Matthews for access to Structure 5.0; Keith Nerhrke for fluorescence microscope time; Susana de Lucas and Juan Ortíz for anti-STAU1; Myriam Gorospe for pcDNA3-MS2bsX12; Stephen and Paul Higgins for HaCaT cells and advice on the scrape injury repair assay; Jiashi Wang for initial BAG5 mRNA assays; and Olaf Isken and Michael Gleghorn for comments on the manuscript. This work was supported by NIH GM074593 (L.E.M.) and an Elon Huntington Hooker Graduate Student Fellowship (C.G.).
Footnotes
Full Methods and any associated references are available in Supplementary Information
References
- 1.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim YK, et al. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walters RD, Kugel JF, Goodrich JA. InvAluable junk: the cellular impact and function of Alu and B2 RNAs. IUBMB Life. 2009;61:831–837. doi: 10.1002/iub.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yulug IG, Yulug A, Fisher EM. The frequency and position of Alu repeats in cDNAs, as determined by database searching. Genomics. 1995;27:544–548. doi: 10.1006/geno.1995.1090. [DOI] [PubMed] [Google Scholar]
- 8.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 9.Hasler J, Strub K. Alu elements as regulators of gene expression. Nucleic Acids Res. 2006;34:5491–5497. doi: 10.1093/nar/gkl706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pang KC, et al. RNAdb 2.0--an expanded database of mammalian non-coding RNAs. Nucleic Acids Res. 2007;35:D178–182. doi: 10.1093/nar/gkl926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engstrom PG, et al. Complex Loci in human and mouse genomes. PLoS Genet. 2006;2:e47. doi: 10.1371/journal.pgen.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 14.Kim HH, et al. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuwano Y, et al. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res. 38:225–238. doi: 10.1093/nar/gkp861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashley CT, Jr., Wilkinson KD, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.