SUMMARY
To understand relationships between phosphorylation-based signaling pathways, we analyzed 150 deletion mutants of protein kinases and phosphatases in S. cerevisiae using DNA microarrays. Downstream changes in gene expression were treated as a phenotypic readout. Double mutants with synthetic genetic interactions were included to investigate genetic buffering relationships such as redundancy. Three types of genetic buffering relationships are identified: mixed epistasis, complete redundancy, and quantitative redundancy. In mixed epistasis, the most common buffering relationship, different gene sets respond in different epistatic ways. Mixed epistasis arises from pairs of regulators that have only partial overlap in function and that are coupled by additional regulatory links such as repression of one by the other. Such regulatory modules confer the ability to control different combinations of processes depending on condition or context. These properties likely contribute to the evolutionary maintenance of paralogs and indicate a way in which signaling pathways connect for multiprocess control.
INTRODUCTION
Protein kinases and protein phosphatases are key components of regulatory pathways, many of which have been studied in detail. This has revealed the pleiotropic role of signaling in cellular regulation, its involvement in disease and how pathway architecture underlies mechanistic aspects such as specificity. Understanding the complexity of cellular regulation also requires in depth knowledge about the ways in which different pathways work together.
Due to the extensive role of signaling, perturbation of different pathways leads to diverse phenotypes. Different pathways have therefore often been studied in isolation, frequently using different readouts for different pathways and thereby confounding systematic comparisons of pathways. This can be overcome by using a single assay that is detailed enough to reveal differences and at the same time comprehensive enough to reveal the workings of many different pathways simultaneously. Phenotypes are often accompanied by changes in gene expression and genome-wide mRNA expression profiling can reveal relationships between pathway components (Capaldi et al., 2008; Roberts et al., 2000). Here, we have applied expression profile phenotypes to systematically investigate relationships between many different signaling pathways that are simultaneously active under a single growth condition in the yeast Saccharomyces cerevisiae.
Analysis of pathway activity using mutants also requires buffering interactions between genes to be considered. Genetic buffering results in masking of the phenotypic consequences of mutations (Hartman et al., 2001). The best appreciated buffering relationship is redundancy, often defined as genes that can compensate for each other’s loss by their ability to share and takeover the exact same function. Redundancy is frequently associated with paralogs that are more likely to share an identical biochemical function (Prince and Pickett, 2002). Nonhomologous genes are less likely to share function but can still exhibit genetic buffering in the form of growth-rate compensation. The relative contribution of paralogs versus nonhomologs toward buffering is under debate (Gu et al., 2003; Ihmels et al., 2007; Papp et al., 2004; Wagner, 2000), but systematic analysis of synthetic genetic interactions (SGIs) is revealing extensive buffering between nonhomologs (Costanzo et al., 2010). How nonhomologous pairs compensate for loss of each other’s function is not well understood and the molecular mechanisms behind such genetic relationships are relatively uncharacterized. Also enigmatic is the question of why paralogs are stably maintained during evolution, often remaining redundant, despite evolutionary pressure against seemingly superfluous copies (Dean et al., 2008; Vavouri et al., 2008). Resolving these questions likely requires more detailed characterization of the mechanisms that underlie buffering interactions, including redundancy.
The yeast Saccharomyces cerevisiae has 141 genes encoding protein kinases and 38 genes encoding protein phosphatases. Here, kinase and phosphatase function is systematically compared by generating DNA microarray expression profiles for all 150 viable protein kinase and phosphatase knockout strains under a single growth condition. To take buffering interactions into account, SGI data is exploited by profiling double mutants that show greater than expected fitness reduction (Fiedler et al., 2009). This provides a detailed and systematic characterization of different genetic buffering relationships. The molecular mechanisms of each type are studied in detail, including analysis of a phosphatase that buffers kinase deletions. An important outcome is identification of a recurrent regulatory module for signaling pathways. This module consists of pairs of regulators that have partial overlap in function and that are also linked by additional regulatory relationships such as repression or inhibition of one partner by the other. The module offers insight into how signaling pathways may regulate different combinations of processes in a flexible yet coordinate manner and plausibly explains why apparently redundant components of regulatory pathways are maintained during evolution.
RESULTS
Expression Profiles of Kinase and Phosphatase Gene Deletions
To compare signaling pathways, DNA microarray gene expression profiles were generated for all 150 viable protein kinase/ phosphatase deletions in S. cerevisiae under a single growth condition (synthetic complete medium with 2% glucose). Each mutant was profiled four times, from two independent cultures on dual-channel microarrays using a batch of wild-type (WT) RNA as common reference. To further control for technical and biological variation, additional WT cultures were grown alongside sets of mutants on each day. These “same-day” WTs were processed in parallel to the mutants, all using automated, robotic procedures. Comparison of the many WT profiles yields insight into the expression variation of each gene. Statistical modeling results in an average profile for each mutant, consisting of p values and changes in mRNA expression for each gene, relative to the expression in the 200 WT cultures (Experimental Procedures). Throughout the manuscript “significant” indicates statistically significant. A p value of 0.05, in combination with a fold change (FC) of 1.7, is applied as a threshold for calling a change in mRNA expression significant. Aneuploidy, incorrect deletions, and spurious mutations were identified in 11% of the mutant strains (Experimental Procedures). These strains were remade and reprofiled.
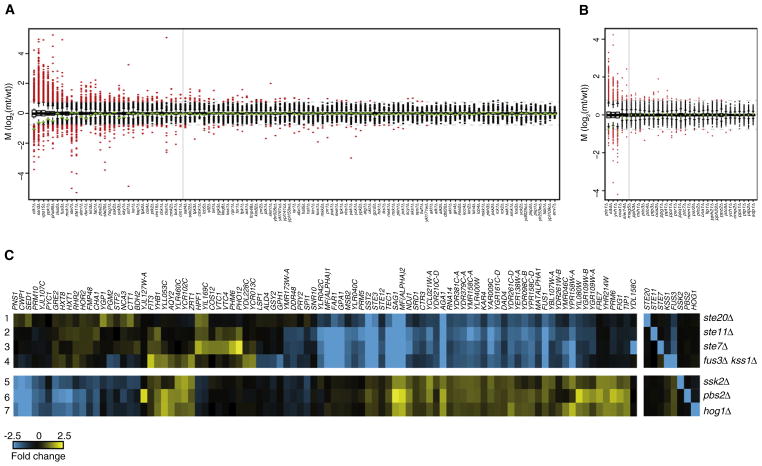
Individual mutants vary considerably with regard to the extent of gene expression changes (Figures 1A and 1B). None of the WT profiles exhibit more than eight genes changing significantly. Applying this threshold on the mutants indicates that 71% of the kinase deletions behave like WT under this growth condition (Figure 1A). For phosphatase deletions this number is even higher (85%, Figure 1B). Taking into account essential genes, this means that more than 60% of kinase/phosphatase genes can be individually removed under a single growth condition without defects in growth or in gene expression. Analysis of mutants with profiles that differ from WT indicates that lack of sensitivity is not the cause of apparent inactivity. For example, mutations in the kinase cascades that control mating and osmo-regulation result in significant changes in mRNA expression, related according to the pathways (Figure 1C). This reflects linear relationships between components of kinase cascades and indicates that the approach is sensitive enough to analyze pathways active even at uninduced basal levels (see Figure S1, available online, for all mutant profiles that differ from WT).
Figure 1. Expression Profiles of Kinase/Phosphatase Single Gene Deletions.
(A and B) Activity profiles of all deletion strains, ranked as box-whisker plots for kinases (A) and phosphatases (B), showing fold changes (vertical axis), with significantly changing genes (p < 0.05, FC > 1.7) as red dots and unresponsive genes as black dots. Green triangles indicate the doubling time of each mutant (-log2 relative to WT). Dashed gray lines indicate 1.7-fold change. The solid gray line is the threshold for distinguishing deletions with significant profiles (≥8 genes changing) versus deletions that behave similarly to WT (<8 genes changing). This threshold is based on the maximum number of changes observed in the 200 WT profiles, excluding the WT variable genes (Experimental Procedures).
(C) Lanes 1–7 are expression profiles of strains indicated to the right. All genes with significantly changed expression in any single mutant (p < 0.05, FC > 1.7) are depicted, with gene names on top. STE20, STE11, STE7 and FUS3 are the MAPK components of the mating pheromone response pathway. FUS3 is redundant with KSS1 and the profile of the double mutant is therefore shown in lane 4. Profiles of the single mutants are depicted in Figure 4C. SSK2, PBS2 and HOG1 are MAPK components of the HOG pathway. The opposite effects of the HOG pathway on some of the genes affected by the mating pathway agrees with inhibition of the mating pathway by the HOG pathway (Chen and Thorner, 2007).
See also Figure S1.
Profiling Negative Synthetic Genetic Interactions
For many mutants, similarity to WT is likely due to absence or inactivity of the protein under a single growth condition. The goal of comparing many pathways active under a single condition also requires genetic buffering interactions such as redundancy to be considered, since this may mask activity of components whereby deletion has no effect. To include redundancy relationships that influence fitness, we exploited SGI data for kinase/phosphatase genes (Fiedler et al., 2009). Selection was based on a greater than expected growth defect in a double mutant compared to the singles. An additional criterion was applied that consisted of one of the single mutants not showing an expression profile different from WT, resulting in 24 pairs. These double mutants were first remade in the genetic background used here and the SGIs were retested for the liquid culture growth used for expression profiling. Despite differences with colony growth (Fiedler et al., 2009), correspondence between the previous study is strong, with 20 of the 24 pairs also showing a greater than expected growth defect in liquid culture (Table S1). Two previously established redundant pairs (FUS3-KSS1, YPK1-YPK2) were added to the selection, and all viable double mutants were expression profiled.
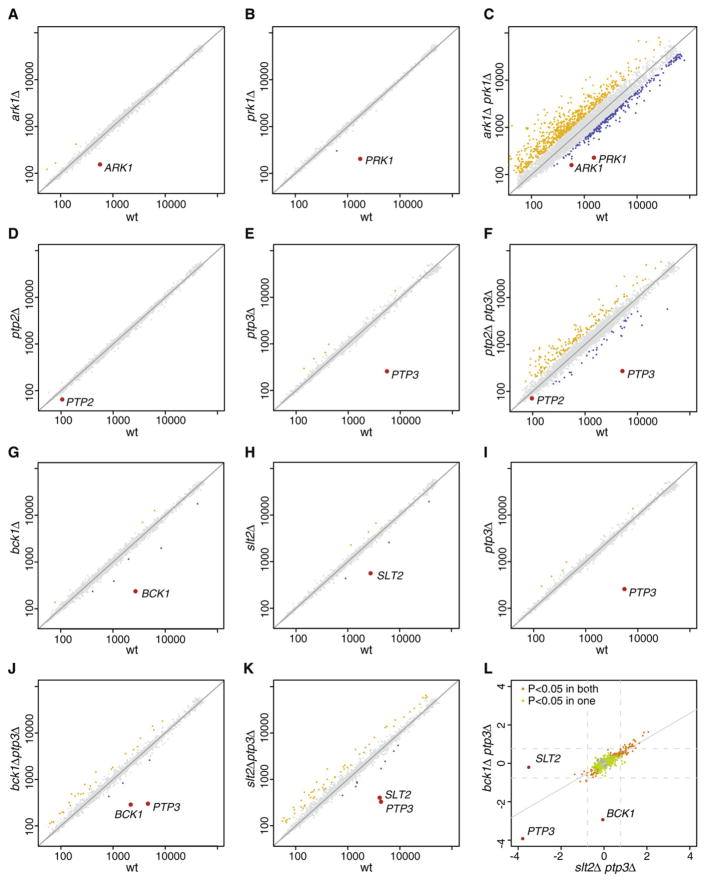
Genetically buffered gene pairs, such as redundant partners, were expected to show more gene expression changes as a double mutant compared to the two singles combined. Deletion of the kinase ARK1, shows an expression profile similar to WT (Figure 2A). Similarly, prk1Δ also has few genes changing significantly (Figure 2B). The ark1Δ prk1Δ double mutant has many genes with expression deviating significantly from WT (Figure 2C) and the profile therefore concurs with the previously reported redundancy (Cope et al., 1999). Likewise, the profile of the phosphatase double mutant ptp2Δ ptp3Δ also agrees with redundancy (Figures 2D–2F) (Jacoby et al., 1997; Wurgler-Murphy et al., 1997). Figure S2 depicts all scatter plots indicative of a buffering effect. Systematic analysis (Extended Experimental Procedures) shows that of the pairs successfully analyzed, 21 have expression profiles that support buffering (Table 1), with more genes changing expression in the double mutant versus the two single mutants combined. This includes all the pairs that showed a negative SGI in liquid culture (Table S1).
Figure 2. Expression Profiles of Genetically Buffered Pairs.
(A–K) Single and double deletion gene expression scatter plots of four genetically buffered pairs. In each scatter plot the normalized, dye-bias corrected and statistically modeled fluorescent intensity value is plotted for each gene. For each mutant this is the average of four measurements. For WT this is the average of 200 cultures grown throughout the project. Genes with significant increase or decrease in mRNA expression (p < 0.05, FC > 1.7) are represented by yellow and blue dots respectively. Gray dots are all other genes.
(L) Scatter plot of all genes that have a significant change in mRNA expression in either bck1Δ ptp3Δ (J), slt2Δ ptp3Δ (K) or in both double mutants. The log2 FC is plotted for each of these genes in both double deletions, showing that the same mRNAs are changing in both strains.
See also Figure S2.
Table 1.
Buffering Relationships between Kinases and Phosphatases
| Gene 1 | Gene 2 | Type | Duplication | Time (Years Ago) | Buffering Relationship |
|---|---|---|---|---|---|
| HAL5 | SAT4 | kk | old | 600 M – 2 G | complete redundancy |
| ARK1 | PRK1 | kk | whole genome | 125 M | complete redundancy |
| PTP2 | PTP3 | pp | recent | 125 M – 600 M | complete redundancy |
| YCK1 | YCK2 | kk | whole genome | 125 M | complete redundancya |
| PTC1 | PTC2 | pp | old | 600 M – 2 G | quantitative redundancy |
| PTC1 | PPH3 | pp | not homologous | quantitative redundancy | |
| PBS2 | PTK2 | kk | ancient | >2G | mixed epistasis |
| CLA4 | SLT2 | kk | ancient | >2G | mixed epistasis |
| CLA4 | HSL1 | kk | ancient | >2G | mixed epistasis |
| SNF1 | RIM11 | kk | ancient | >2G | mixed epistasis |
| BCK1 | PTP3 | kp | not homologous | mixed epistasis | |
| SLT2 | PTP3 | kp | not homologous | mixed epistasis | |
| FUS3b | KSS1 | kk | recent | 125 M – 600 M | mixed epistasis |
| ELM1 | MIH1 | kp | not homologous | mixed epistasisc | |
| CLA4 | BCK1 | kk | ancient | >2G | mixed epistasisc |
| DUN1 | PPH3 | kp | not homologous | mixed epistasisc | |
| CKA2 | CKA1 | kk | recent | 125 M – 600 M | not classifieda |
| YPK1b | YPK2 | kk | whole genome | 125 M | not classifieda |
| PTK1 | PTK2 | kk | whole genome | 125 M | not classifieda |
| HSL1 | MIH1 | kp | not homologous | not classifieda | |
| SKY1 | PTK2 | kk | ancient | >2G | not classifieda |
Double mutant is inviable, confirming a buffering effect.
Included based on previously reported redundancy.
Double mutant was aneuploid; aneuploid chromosomes were excluded from analysis.
Determination of paralogy relative to important radiations and events was performed by integration of information available in several orthology and homology databases. The timings in years are estimates derived from literature (Extended Experimental Procedures).
Redundancy involves overlap of function and is often associated with paralogs. Phylogenetic analysis reveals that less than one third of the buffering relationships observed here are derived from close paralogs, that is from duplication events that occurred less than approximately 600 million years ago (Table 1, Figure S3, Extended Experimental Procedures). More than half of the interactions are between pairs that arose from ancient duplications (an estimated 2 billion years ago) or between non-homologs, in five cases even between kinase-phosphatase pairs. Buffering between nonhomologs has been noted before (Gu et al., 2003; Ihmels et al., 2007; Papp et al., 2004; Wagner, 2000), but the underlying mechanisms are often not investigated. Therefore, we selected an example for further analysis, focusing on the intriguing buffering between kinases and phosphatases.
Buffering between a Kinase and Phosphatase Is due to Phosphatase-Mediated Inhibitory Crosstalk between Kinase Pathways
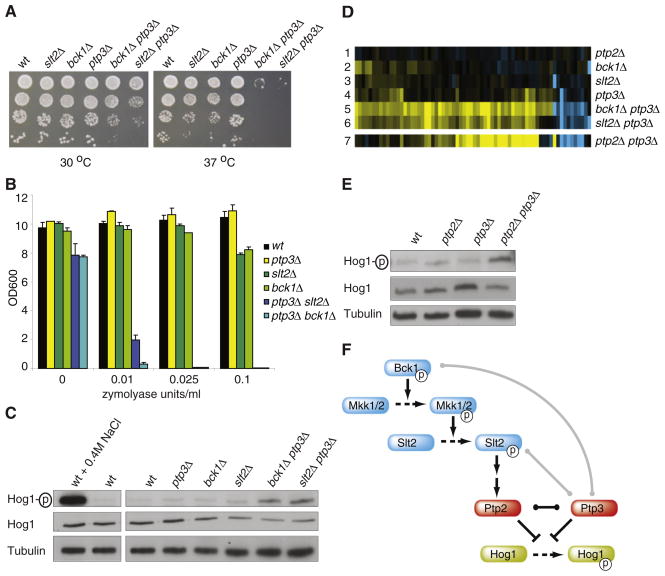
Bck1 and Slt2 are mitogen-activated protein kinase (MAPK) components of the cell-wall integrity (CWI) pathway (Chen and Thorner, 2007). Both kinases show buffering with the phosphatase PTP3, likely reflecting the fact that both kinases belong to the same MAPK cascade (Figures 2G–2K). In both kinase-phosphatase double mutants the same genes change (Figure 2L). Ptp3 dephosphorylates Hog1, resulting in inactivation of Hog1 (Jacoby et al., 1997). Most of the bck1Δ ptp3Δ and slt2Δ ptp3Δ double deletion profiles consist of upregulated genes (Figures 2J and 2K). This includes established Hog1 downstream target genes (Rodríguez-Peña et al., 2005), indicating that buffering may be related to defective inhibition of Hog1. To test this, the double deletion strains were first assayed for phenotypes associated with increased Hog1 activity such as elevated temperature (Figure 3A) (Winkler et al., 2002) and sensitivity to the cell wall disrupting agent zymolyase (Figure 3B) (Bermejo et al., 2008). That the buffering observed between the BCK1, SLT2 kinases and PTP3 phosphatase indeed involves Hog1 is confirmed by monitoring Hog1 phosphorylation, which is higher in both bck1Δ ptp3Δ and slt2Δ ptp3Δ double mutants compared to ptp3Δ or WT (Figure 3C).
Figure 3. Kinase-Phosphatase Buffering Is Caused by Phosphatase-Mediated Inhibitory Crosstalk between Kinase Pathways.
(A) The bck1Δ ptp3Δ and slt2Δ ptp3 kinase-phosphatase double mutants are sensitive to elevated temperature. Ten-fold dilutions of cultures were spotted onto plate and incubated at 30°C or 37°C.
(B) The bck1Δ ptp3Δ and slt2Δ ptp3 kinase-phosphatase double mutants show more sensitivity to zymolyase. Bars and standard deviations are based on the average of three.
(C) Active, phosphorylated Hog1 is increased in the bck1Δ ptp3Δ and slt2Δ ptp3 kinase-phosphatase double mutants. Immunoblots for phosphorylated Hog1 (top), all Hog1 (middle) and Tubulin (bottom). Lane 1 is a positive control of WT exposed to 0.4 M NaCl for five minutes prior to harvesting.
(D) All genes with significant changes in bck1Δ ptp3Δ or slt2Δ ptp3Δ (p < 0.05, FC > 1.7) are depicted. Lane 7 shows the same genes for the ptp2Δ ptp3Δ expression profile.
(E) As in (C).
(F) Model of interactions for the buffering observed between PTP3-SLT2 and PTP3-BCK1. Gray lines indicate buffering. Black line indicates redundancy. The two arrows between Slt2 and Ptp2 indicate that this activation may be direct or indirect.
Since it is unlikely that the kinases are directly responsible for dephosphorylation of Hog1, a second phosphatase was postulated to be involved. Candidates included Ptc1, Ptp2, and Ptc2, all also capable of dephosphorylating Hog1 (Jacoby et al., 1997; Warmka et al., 2001; Wurgler-Murphy et al., 1997; Young et al., 2002). PTP3-phosphatase double mutant expression profiles were analyzed. Only the ptp2Δ ptp3Δ double mutant expression profile shows a buffering effect whereby the majority of mRNAs that change in the CWI kinase-phosphatase double mutants are also similarly changing in the ptp2Δ ptp3Δ double phosphatase mutant (Figure 3D). In addition, Hog1 phosphorylation levels are increased in the ptp3Δ ptp2Δ double mutant (Figure 3E). Buffering between the CWI pathway kinases and the PTP3 phosphatase is therefore likely reflecting redundancy between PTP2 and PTP3 (Figure 3F) (Jacoby et al., 1997; Wurgler-Murphy et al., 1997). This agrees with the infrequently tested notion that SGIs arise from parallel pathways (Kelley and Ideker, 2005). In this case the parallel pathways converge on Hog1 through two redundant phosphatases, one of which, Ptp2, is likely activated by the CWI pathway.
Expression Profiling Reveals Three Different Genetic Buffering Relationships
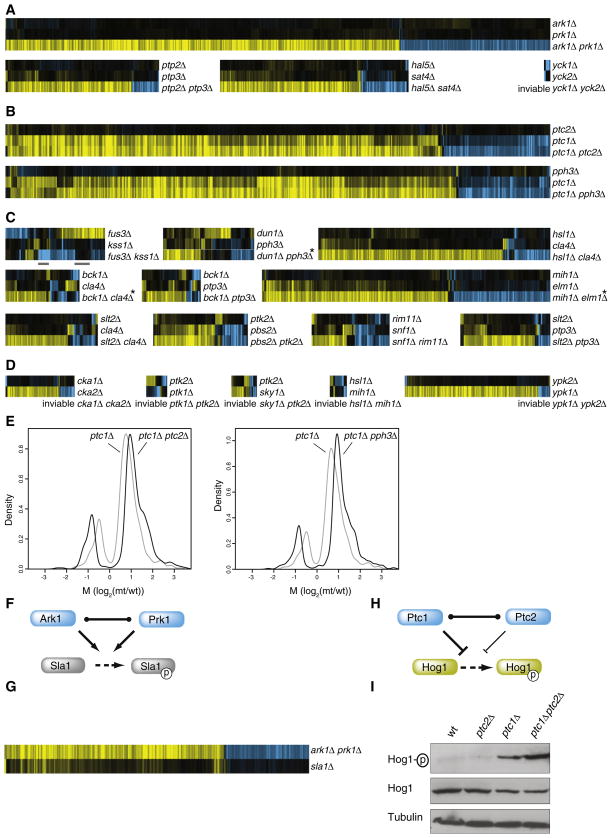
Division into paralogous and nonhomologous pairs is one type of classification that can be applied to genetic buffering. The data also prompted a new characterization of genetic buffering relationships, based on the single- and double mutant expression profiles. Intriguingly, these can be classified into three types: complete redundancy, quantitative redundancy and mixed epistasis (Figure 4, systematic classification is described in detail in Extended Experimental Procedures). Complete redundancy is exemplified by the ark1Δ, prk1Δ scatter plots (Figures 2A–2C). There are no changes in single deletions (less than eight genes changing significantly compared to WT), but an effect is observed in the double mutant. Four redundant pairs show complete redundancy (Figure 4A). Besides ARK1-PRK1, this includes the kinase pairs HAL5-SAT4, YCK1-YCK2 and the phosphatase pair PTP2-PTP3.
Figure 4. Expression Profiling Reveals Three Different Genetic Buffering Interactions.
For each set of three profiles all genes with changes in mRNA expression in any single profile are shown (p < 0.05, FC > 1.7).
(A) Complete redundancy whereby the single mutants have less than eight genes changing significantly and the double have more than eight.
(B) Quantitative redundancy, whereby one single mutant shows no significant profile (<8 genes p < 0.05, FC > 1.7), the other single mutant has a significant profile and in the double the same genes change to a higher degree.
(C) Mixed epistasis. Here at least 8 more genes change significantly in the double versus the two singles, with at least 8 genes behaving in other ways than in complete or quantitative phenotypes. The two bars below the FUS3-KSS1 profiles indicate the two gene sets selected for modeling (Figure 5).
(D) Unclassified buffering interactions due to inviability of the double mutant (Table 1).
(E) Quantification of the profiles shown in B, plotted for all genes with significant (p < 0.05, FC > 1.7) changes in mRNA expression in any one single or double mutant strain. M is the log2 ratio of normalized fluorescent mRNA expression in the mutant divided by WT. Asterisks indicate strains showing aneuploidy in the double mutant whereby all genes on aneuploid chromosomes were excluded from analyses.
(F) Complete redundancy can result from two proteins able to directly substitute for all of each other’s activity.
(G) Expression profiles of the ark1Δ prk1Δ double mutant and the target sla1Δ. All genes are depicted with significant changes (p < 0.05, FC > 1.7) in mRNA expression in any profile.
(H) Quantitative redundancy resulting from the ability of two proteins to directly substitute for each others activity qualitatively, but not quantitatively.
(I) Immunoblot as described in Figure 3C.
A second type of redundancy is evident from the quantitative effects observed in the phosphatase pairs PTC2-PTC1 and PPH3-PTC1 (Figure 4B). Here, one single mutant shows no effect (less than eight gene changes), but the other single mutant does. The term quantitative is applicable because the effect observed in the single mutant is amplified in the double mutant (see also Figure 4E) but without involving additional gene sets.
Complete and quantitative redundancy are intuitive in their classification and as is demonstrated below, both can be understood through simple molecular mechanisms. This is not true because of the third buffering relationship, which we call mixed epistasis for the different types of epistatic effects observed on different gene sets (Figure 4C). Whereas some gene sets respond as in complete or quantitative redundancy, other gene sets behave in completely different ways. These typically show expression changes in single mutants that disappear or even show an opposite effect in the double mutant. The classification scheme (Extended Experimental Procedures) depends on thresholds for identification of differently behaving gene sets. Changing thresholds would result in a different classification for some of the pairs. The thresholds were kept identical to those used for identification of which mutants behave as WT (Figure 1). In this way sixteen of the twenty-one gene pairs exhibiting genetic buffering are classified: four as complete, two as quantitative and ten as mixed epistatic. In six cases, the double mutant is inviable (Table 1), hindering classification of CKA1-CKA2, PTK2-PTK1, PTK2-SKY1, HSL1-MIH1, and YPK1-YPK2 (Figure 4D). One case of inviability (YCK1-YCK2) can be unambiguously classified as complete redundancy (Figure 4A).
The ten pairs showing mixed epistasis are the kinase pairs KSS1-FUS3, HSL1-CLA4, SNF1-RIM11, BCK1-CLA4, SLT2-CLA4 and the kinase-phosphatase pairs PBS2-PTK2, ELM1-MIH1, DUN1-PPH3, BCK1-PTP3, SLT2-PTP3. Mixed epistasis is therefore exhibited by paralogous as well as nonhomologous pairs. Besides the mixed epistasis itself, it is striking that this buffering interaction is the most common. Redundancy is not necessarily complete. Partial overlap in function is expected to result in single mutants exhibiting effects on their own, with these same effects reflected in the double mutant, alongside additional genes changing due to loss of the shared function. It is remarkable that no very clear example of this expected partial redundancy pattern is observed. As is made clear below, this is related to the finding of mixed epistasis.
Mechanisms Underlying Complete and Quantitative Redundancy
We next considered molecular mechanisms. Complete and quantitative redundancy can be explained by similar models whereby redundant partners function on the same targets (Figures 4F and 4H). As an example, Ark1 and Prk1 are previously established redundant kinases that regulate endocytosis and the actin cytoskeleton (Smythe and Ayscough, 2003). ARK1-PRK1 demonstrate complete redundancy (Figures 2A–2C). The endocytic adaptor protein Sla1 is an established direct target of both kinases (Zeng et al., 2001). The sla1Δ expression profile reflects this, with the changes in mRNA expression forming a perfect subset of the ark1Δ prk1Δ expression profile (Figure 4G). This illustrates that kinase targets can in some cases be identified by comparative expression profiling and indicates here that Ark1 and Prk1 likely have more than one target.
It is similarly intuitive that pairs showing quantitative redundancy have identical targets, since the same genes are affected in single and double mutants, but to different degrees (Figures 4B and 4E). Quantitative redundancy may reflect a quantitatively different effect on the target. To test this, we investigated the phosphatase pair PTC1-PTC2 (Figure 4B). Hog1 is a shared target of Ptc1 and Ptc2 (Young et al., 2002). In agreement with the hypothesis, the degree to which Ptc1 and Ptc2 dephosphorylate Hog1 differs (Figure 4I). Levels of phosphorylated Hog1 in the different mutants match the quantitative effects observed in the expression profiles (Figure 4B). This supports the proposal that quantitative redundancy is caused by identical target specificity combined with a quantitatively different effect on the target. This could be due to differences in enzyme efficiency or through differences in expression levels of redundant partners. Due to the selection criteria, the effects observed here always involve one single mutant showing an expression profile similar to WT. This implies that the enzyme that does show a single-deletion phenotype is overabundantly active under this growth condition.
Mixed Epistasis of FUS3-KSS1 Is a Result of Partial Redundancy Coupled to Unidirectional Repression
Mixed epistasis is the most frequently observed buffering interaction (Figure 4C, Table 1). To investigate mechanism, we first focused on the FUS3-KSS1 kinase pair (reviewed in Chen and Thorner, 2007). The Fus3 MAPK is responsible for activation of mating genes in response to pheromone. Kss1 is the MAPK of the filamentous growth pathway that activates a nutrient starvation response whereby yeast cells change polarity and shape, resulting in filamentous colony outgrowth that enables foraging for nutrients. The fus3Δ, kss1Δ and fus3Δ kss1Δ profiles consist of several responder gene sets that behave in different ways in the three strains (Figure 4C). To understand mixed epistasis, we focused on two such gene sets. The first set behaves as in complete redundancy, with downregulation only in the double mutant (Figure 5A). The second set shows upregulation in fus3Δ only. Together, these two gene sets form a minimal mixed epistasis pattern, shared by the majority of pairs classified as such (Figure 4C).
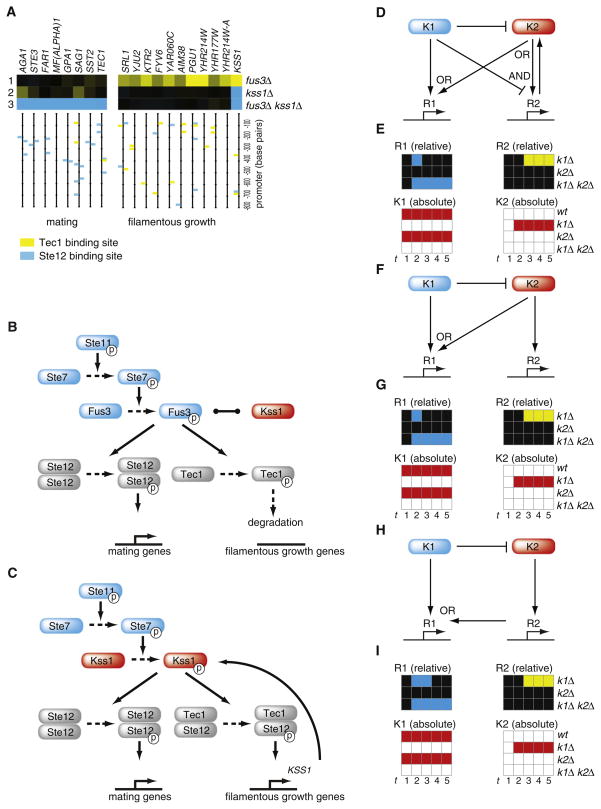
Figure 5. Mechanisms of Mixed Epistasis: Partial Overlap in Function Coupled to Unidirectional Repression.
(A) A minimal mixed epistasis pattern consisting of two gene sets selected from the FUS3-KSS1 profiles (Figure 4C). The names “mating” and “filamentous growth” are based on the enrichment for Ste12 and Tec1 transcription factor binding sites respectively, upstream of each gene, as indicated in the vertical bars.
(B) Experimentally-derived/literature-based model for regulation of the mating and filamentous growth gene sets under basal, unactivated conditions in WT cells. The model omits details such as activation of Ste12 and Tec1 transcription factor complexes through phosphorylation of the Dig1, Dig2 repressors (Chen and Thorner, 2007). The black line between Kss1 and Fus3 indicates redundancy.
(C) Model for fus3Δ.
(D, F, and H) Boolean solution models for a minimal mixed epistasis pattern.
(E, G, and I) The accompanying state transitions for one of the eight simulated initial states (Experimental Procedures). R1 and R2 indicates the activities of the two responder gene sets, depicted for the mutants relative to WT, similarly to the expression profiles, with blue indicating decrease, black no change and yellow increase in expression. K1 and K2 indicate the absolute activities of the regulator nodes with red for True and white for False. The numbers at the bottom indicate the first five time steps of simulation.
A model that explains the different epistatic behavior of the two responder gene sets (Figures 5B and 5C) is based on data presented here (Figure 5A) as well as on many previous studies of these pathways (Chen and Thorner, 2007). FUS3 and KSS1 are redundant paralogs but the redundancy is only partial (Elion et al., 1991). The two pathways work through two downstream transcription factors, Ste12 and Tec1 (Chen and Thorner, 2007; Chou et al., 2006; Madhani and Fink, 1997). The promoters of the two gene sets are differentially enriched for Ste12 and Tec1 binding sites (Figure 5A). The first gene set consists of mating genes, enriched for pheromone response elements that bind homodimerized Ste12. The second gene set is enriched for the filamentation response element that binds the Ste12-Tec1 heterodimer. In agreement with previous studies (Chen and Thorner, 2007), Kss1 is inactive under noninducing conditions and kss1Δ has virtually no effect (Figure 5A). The mating pathway (Fus3) is active at low basal levels under noninducing conditions. Fus3 is an activating kinase for Ste12 and an inactivating kinase for Tec1, whereby Tec1 phosphorylation leads to its degradation (Chen and Thorner, 2007; Chou et al., 2004). KSS1 is a target of Tec1 in this model. Upon deletion of FUS3, Tec1 is no longer degraded. KSS1 becomes upregulated and because of their redundancy, Kss1 can (partially) take over the role of Fus3 (Figure 5C). Kss1 takes over the role of activating Ste12 (Madhani et al., 1997). No change is therefore observed in the mating genes, which remain active at basal levels (Figure 5A). Kss1 does not take over the inactivating role of Fus3 toward Tec1 (Chou et al., 2004), leading to activation of the filamentous gene cluster in fus3Δ (Figure 5A). This effect is lost in the double mutant and the filamentous gene set reverts back to WT levels (Figure 5A). The mating gene set is down in the double mutant (Figure 5A) because neither Kss1 nor Fus3 are present to activate Ste12.
The two pivotal elements that explain the mixed epistatic effects are therefore partial redundancy and the negative regulation of KSS1 by Fus3. A negative effect of Fus3 on KSS1 has been described for activating conditions (Chou et al., 2006). The promoter of KSS1 contains binding sites for Tec1 (Figure 5A) and, as predicted, KSS1 indeed becomes upregulated in fus3Δ (Figure 5A). The involvement of the two downstream transcription factors (Chen and Thorner, 2007) is supported by the differential enrichment of binding sites (Figure 5A) and was tested by analyzing tec1Δ and ste12Δ (Figure S4).
Boolean Modeling Reveals Two General Properties of Mixed Epistasis: Partial Overlap in Function and Regulatory Coupling
Mixed epistasis similar to FUS3-KSS1 occurs in 10 out of the 16 pairs that can be classified (Figure 4C). To determine whether similar mechanisms underlie all such cases, we asked which regulatory network topologies lead to such phenotypes. By definition, all the cases of mixed epistasis contain at least two differently responding gene sets. We therefore considered models consisting of four nodes: two gene sets and two regulators. To arrive at all possible solution models rather than a single optimized solution, modeling was performed with Boolean operators (Albert et al., 2008; Ma et al., 2009). Since two nodes can be linked by different combinations of positive and negative regulatory edges going in different directions, any two nodes can be connected in nine different ways. This leads to 794,176 models (Experimental Procedures), of which 106 result in the minimal mixed epistasis pattern (Figure 5A, Table S5). These steady-state solution models were pruned by removing superfluous edges (Figure S4C), revealing 28 root models that all exhibit the experimentally observed mixed epistasis (Table S2).
Two important general properties emerge from these models. The first is inhibition or repression of one regulator by the other (Table S2 and Table S3). Different ways of achieving these unidirectional negative effects are exemplified by the model solution that most closely resembles the literature-derived model for FUS3-KSS1 (Figures 5D and 5E). Besides encompassing all the regulatory edges contained in the experimentally derived scheme, including repression of kinase 2 expression by kinase 1, in this Boolean model, kinase 1 also inhibits kinase 2. Previous experiments have suggested the existence of an inhibitory effect of Fus3 toward Kss1, albeit indirectly through Fus3-mediated activation of a Kss1-inhibitory phosphatase (Chen and Thorner, 2007). Although this Boolean solution closely resembles the experimentally derived model (Figure 5C), it should be noted that this is not a root model and can be pruned by removal of two edges without affecting outcome (Figures 5F and 5G). That the experimentally derived model contains seemingly superfluous edges indicates that these features are required for aspects of FUS3-KSS1 not modeled here, such as regulatory dynamics and the different behavior of other gene sets (Figure 4C).
A second general property of all the Boolean solutions is partial overlap in function. As with the negative effects, the models indicate that partial overlap in function can also be achieved in different ways. The least complex models, the two solutions that consist of only four edges, illustrate direct (Figure 5F) and indirect ways (Figures 5H and 5I) in which partial overlap in function can be achieved. In the first root model (Figure 5F) both kinases have activating edges toward the first responder gene set. This indicates redundancy and fits best with the expected action of redundant paralogs. The partial nature of the redundancy is represented by different edges to the other responder gene set. In the second simple Boolean root model (Figure 5H), partial overlap in function is achieved in a different, indirect way, with kinase 2 indirectly acting on one responder gene set through the other. This indirect manner of achieving overlap in function explains how functionally distinct nonhomologous pairs such as kinase-phosphatase pairs, can nevertheless still have buffering effects. That the Boolean solutions encompass both direct and indirect ways of achieving overlapping function fits well with the observation that mixed epistasis is exhibited by paralogous as well as nonhomologous pairs (Table 1).
Modeling shows that mixed epistasis arises through partial overlap in function combined with regulatory links from one partner to the other. The majority of genetic buffering interactions are mixed epistatic (Table 1). This indicates that the majority of genetically buffered kinase/phosphatase pairs have partial overlap in function and regulatory links. As is explained below, this has implications for understanding multiprocess control and for explaining the evolutionary maintenance of redundant paralogs.
Regulatorily Linked Pairs with Partial Overlap in Function Form Modules for Controlling Different Combinations of Processes
A consequence of the network topologies that explain the minimal mixed epistasis pattern is that two distinct responses can be regulated in either coupled or uncoupled manners. Depending on which regulator is active, a single process, or a second process in combination with the first, can be coordinately regulated. This feature is illustrated by FUS3-KSS1. Although the mating pheromone response (Fus3) and the filamentous growth starvation response (Kss1) are often treated as distinct, it has been reported that Kss1 is briefly activated during pheromone treatment (Ma et al., 1995). Furthermore, under low mating pheromone concentrations, yeast cells display a Kss1-dependent filamentation response that allows outgrowth toward cells of the opposite mating type (Erdman and Snyder, 2001). This is similar to Kss1-dependent filamentous growth during nutrient starvation and suggests that under certain conditions, such as low pheromone concentration, aspects of filamentous growth are indeed regulatorily coupled to the mating response.
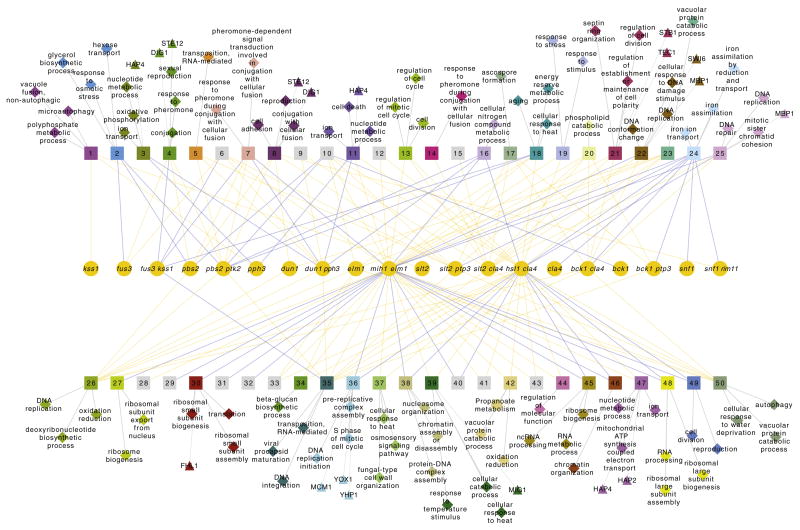
It is not well understood why redundant pairs such as paralogs are evolutionarily maintained (Vavouri et al., 2008). The ability to flexibly couple and uncouple regulation of distinct processes is intuitively advantageous as a multiprocess control mechanism for responding to a large variety of different (combinations of) conditions. If this ability is a driving force behind the evolutionary maintenance of redundant pairs, then one prediction is that the gene sets that behave in different ways in mixed epistatic interactions should correspond to distinct processes. This prediction is confirmed by Gene Ontology (GO) analysis of the groups of genes contained within the mixed epistasis profiles (Figure 6). Presentation of this enrichment analysis as a network also illustrates the potential advantage of wiring together several such regulatory-coupled redundancy modules for multiprocess control. Many different responses, represented by the square nodes of coregulated genes, are influenced by several different regulatorily coupled regulators (Figure 6). In this way a large number of distinct combinations of processes can be regulated through different activity mixes of a relatively small number of pathways.
Figure 6. Multiprocess Control through Signaling Components with Mixed Epistasis.
Yellow circular nodes represent the single and double mutant profiles for the pairs with mixed epistasis (Table 1). Single mutants with no significant changes are not shown. Square nodes (numbered 1–50) indicate gene sets that show differential expression patterns across this set of mutants, obtained by QT clustering all genes with a significant change (p < 0.05, FC > 1.7) in any one profile. Yellow edges between mutants and gene sets indicate that a gene set is upregulated in the mutant, blue indicates downregulation. Diamonds indicate significant (p < 0.05) enrichment of a particular GO category in the gene set. Only the top three categories are shown. Three-quarters of the gene sets are significantly enriched for at least one GO category. Triangles depict enrichment for transcription factor binding sites in the gene set, indicating which transcription factor may be mediating the response. See also Figure S5.
DISCUSSION
Mixed Epistasis and Synthetic Genetic Interactions
In model organisms, genetic buffering interactions are most readily uncovered by measuring fitness under a standard growth condition. Systematic determination of SGIs across all genes has only recently been initiated (Costanzo et al., 2010) and the molecular mechanisms underlying such interactions are relatively uncharacterized (Kelley and Ideker, 2005). Expression profiling provides detailed insight into the consequences of mutations. This is exemplified here by the classification of a single type of SGI into three classes. Mixed epistasis is the most unanticipated and it is also striking that it is the most common. The term epistasis is applied here in the broad, Fisherian definition of any genetic interaction (Roth et al., 2009). To the best of our knowledge, the simultaneous occurrence of different types of epistatic interactions between two genes has not been generally described before. This is likely because the phenotypical readout used here is more detailed than a fitness defect.
Paralogous versus Nonhomologous Buffering
Redundancy is often associated with pairs of highly related genes (Prince and Pickett, 2002). One outcome of recently initiated genome-wide mapping of genetic interactions is the contribution of nonhomologous genes toward buffering (Costanzo et al., 2010). The relative contributions of nonhomologs versus duplicate pairs is under debate (Gu et al., 2003; Papp et al., 2004; Wagner, 2000), with a recent estimate as high as 75% for nonhomologs (Ihmels et al., 2007). The gene pairs investigated here were selected from a comprehensive kinase/phosphatase genetic interaction study (Fiedler et al., 2009). Half are either unambiguously nonhomologous or have arisen from ancient duplication events (over two billion years ago, Table 1). This agrees with a strong contribution of nonhomologous pairs toward genetic buffering (Ihmels et al., 2007) and indicates that redundancy is not merely the transient by-product of gene duplications, since overlaps in cellular function have evolved from nonhomologous genes too.
The Selective Advantage of Kinase/Phosphatase Redundancy Is Superior Regulatory Systems
Other arguments in favor of an important functional role for redundancy include the stable evolutionary maintenance of paralogs and the persistent nature of redundancy (Dean et al., 2008; Vavouri et al., 2008). Different types of selective advantages have been proposed for the maintenance of redundant paralogs, including robustness against mutation and robustness against stochastic fluctuations in gene expression (Kafri et al., 2006; Nowak et al., 1997; Prince and Pickett, 2002). Backup models lack explanation of why only some genes have backups and why redundancy is present in diploid organisms too. The partial nature of most redundancy, observed here and elsewhere (Ihmels et al., 2007), as well as the condition-dependence of paralogous redundancy (Musso et al., 2008), also argue against backup function. Instead, the results favor superior control mechanisms as a selective advantage. The lack of phenotypes expected for simple partial functional redundancy relationships (Figure 4) is particularly interesting since this indicates that pairs with partial overlap in function are always connected through additional links. One property of such modules is that dependent on which member of a pair is active, distinct processes can be regulated in coupled or uncoupled manners.
The formation of regulatory modules with superior control potential may also have other implications for understanding the evolution of gene duplications. Models explaining the maintenance of paralogs include neo- and subfunctionalisation of duplicate copies (DeLuna et al., 2008; Innan and Kondrashov, 2010). Recent systematic studies indicate that neofunctionalisation does not play a large role (Dean et al., 2008). The regulatory modules described here fit best with subfunctionalisation, but the finding that partially redundant pairs are also coupled by regulatory links to each other may require additional subclassification of these models (Innan and Kondrashov, 2010).
Quantitatively redundant pairs may also confer superior regulatory properties or may simply indicate requirement for a higher enzymatic capacity than can otherwise be reached with only a single copy. Complete redundancy phenotypes are a minority (Table 1). The selective advantage of such pairs remains enigmatic. Growth condition dependency of redundancy (Musso et al., 2008) suggests that if profiled under other conditions, such pairs may exhibit one of the other phenotypes.
Recurrent Modules and Pathway Connectivity
Recurrent motifs with important properties have previously been described for transcription regulatory networks (Alon, 2007). The extent of signaling pathway connectivity has recently been highlighted by systematic analysis of protein interactions (Breitkreutz et al., 2010). Common regulatory motifs within signaling networks are not well established and little is known in general about multiprocess control. Our analyses indicate that regulatorily coupled pairs with partial overlap in function form a common module for contributing to the control of different combinations of processes (Figure 6).
One of the regulatory links is repression of one regulator by the other, as exemplified by FUS3-KSS1. The dataset contains other examples where inactivation of one redundant gene leads to increase in expression of its partner (Figure S5). This regulatory link contributes to differential expression of paralogs (Kafri et al., 2005) and to paralog-responsiveness (DeLuna et al., 2010). The minimal mixed epistasis pattern modeled here consists of only two gene sets (Figure 5). Besides such gene sets, most mixed epistasis profiles also have additional gene sets behaving in different epistatic ways (Figure 4C). This implies that wiring of such pairs also occurs in more ways than unidirectional repression and likely involves other mechanisms, including differential dose-response effects for other gene sets. The data forms a basis for unraveling such modules further and will be useful for engineering different types of combinatorial control in synthetic signaling pathways (Kiel et al., 2010). Although the number of pairs described here is likely an underestimate, it should be noted that these were selected based on SGIs and form only a distinct subset of all possible kinase/phosphatase pairs. Connectivity between signaling pathways therefore occurs in more ways. It can be anticipated that besides regulatorily coupled pairs with partial overlapping function, more recurrent modules will be uncovered by combinatorial analyses (Kelley and Ideker, 2005), especially of datasets that are starting to reveal the full scale of pathway connectivity (Breitkreutz et al., 2010; Costanzo et al., 2010).
EXPERIMENTAL PROCEDURES
All procedures are described in detail in the Extended Experimental Procedures.
Expression Profiling and Deletion Strains
Each mutant strain, BY4742 (Table S4), was profiled four times from two independently inoculated cultures. Sets of mutants were grown alongside WT cultures, all processed in parallel. Dual-channel 70-mer oligonucleotide arrays were employed with a common reference WT RNA. All steps after RNA isolation were automated using robotic liquid handlers. These procedures were first optimized for accuracy (correct fold change) and precision (reproducible result), using spiked-in RNA calibration standards (van Bakel and Holstege, 2004). After quality control, normalization and dye-bias correction (Margaritis et al., 2009), statistical analysis was performed for each mutant versus the collection of 200 WT cultures. The reported fold change is the average of the four replicate mutant profiles versus the average of all WTs. 76 genes showed stochastic changes in WT profiles and were excluded from the analyses. Incorrect strains from the collection as indicated by aneuploidy (5%), incorrect deletion (3%) or additional spurious mutation affecting the profile (3%), were remade and reprofiled (Table S4). None of the WT profiles had more than eight genes changing compared to the average WT (p < 0.05, FC > 1.7). A threshold of fewer than eight genes changing was therefore applied to determine whether a mutant had a significant profile.
Double Mutants
SGI data (Fiedler et al., 2009) were converted to Z-scores and double mutants were selected based on exhibiting a negative SGI, a Z-score significance of p < 0.05 after multiple testing correction (46 pairs) and one of the mutants having an expression profile similar to WT (24 pairs). Double mutants were all remade in an identical genetic background as the single mutants. Six were inviable, consistent with buffering. One double mutant (dun1Δ chk1Δ) had different degrees of aneuploidy in different isolates and buffering could not be confidently determined from the profile (Table S1).
Boolean Modeling
Given four nodes and no self-edges, topologies were constrained to be completely connected and have at least two edges from the regulator nodes (K1, K2) to the responder nodes (R1, R2). The number of incoming edges on any node was limited to two. Influence of two incoming edges could be Boolean AND or OR. Synchronous Boolean simulations were run for all possible initial states of K2, R1, and R2. The initial state of K1 was True. Solution models were those that converged to a steady state under all initial state settings and had the final states of wild-type: R1 = True, R2 = False; k1Δ: R1 = True, R2 = True; k2Δ: R1 = True, R2 = False; k1Δ k2Δ: R1 = False, R2 = False.
Supplementary Material
Acknowledgments
This work was supported by the Netherlands Bioinformatics Centre (NBIC) and the Netherlands Organization of Scientific Research (NWO), grants 016.108.607, 817.02.015, 050.71.057, 911.06.009, 021.002.035 (T.L.L.), 863.07.007 (P.K.), 700.57.407 (J.J.B.).
Footnotes
ACCESSION NUMBERS
All microarray gene expression data have been deposited in the public data repositories ArrayExpress (accession numbers E-TABM-907 [mutants] and E-TABM-773 [200 WT replicates]) and GEO (GSE25644 [mutants]). The data are also available as flat-file or in TreeView format from http://www.holstegelab.nl/publications/sv/signaling_redundancy/.
Supplemental Information includes Extended Experimental Procedures, five figures, and five tables and can be found with this article online at doi:10.1016/j.cell.2010.11.021.
References
- Albert I, Thakar J, Li S, Zhang R, Albert R. Boolean network simulations for life scientists. Source Code Biol Med. 2008;3:16. doi: 10.1186/1751-0473-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Bermejo C, Rodríguez E, García R, Rodríguez-Peña JM, Rodríguez de la Concepción ML, Rivas C, Arias P, Nombela C, Posas F, Arroyo J. The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Mol Biol Cell. 2008;19:1113–1124. doi: 10.1091/mbc.E07-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi AP, Kaplan T, Liu Y, Habib N, Regev A, Friedman N, O’Shea EK. Structure and function of a transcriptional network activated by the MAPK Hog1. Nat Genet. 2008;40:1300–1306. doi: 10.1038/ng.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RE, Thorner J. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1773:1311–1340. doi: 10.1016/j.bbamcr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Huang L, Liu H. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell. 2004;119:981–990. doi: 10.1016/j.cell.2004.11.053. [DOI] [PubMed] [Google Scholar]
- Chou S, Lane S, Liu H. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:4794–4805. doi: 10.1128/MCB.02053-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cope MJ, Yang S, Shang C, Drubin DG. Novel protein kinases Ark1p and Prk1p associate with and regulate the cortical actin cytoskeleton in budding yeast. J Cell Biol. 1999;144:1203–1218. doi: 10.1083/jcb.144.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean EJ, Davis JC, Davis RW, Petrov DA. Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet. 2008;4:e1000113. doi: 10.1371/journal.pgen.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuna A, Springer M, Kirschner MW, Kishony R. Need-based up-regulation of protein levels in response to deletion of their duplicate genes. PLoS Biol. 2010;8:e1000347. doi: 10.1371/journal.pbio.1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuna A, Vetsigian K, Shoresh N, Hegreness M, Colón-González M, Chao S, Kishony R. Exposing the fitness contribution of duplicated genes. Nat Genet. 2008;40:676–681. doi: 10.1038/ng.123. [DOI] [PubMed] [Google Scholar]
- Elion EA, Brill JA, Fink GR. FUS3 represses CLN1 and CLN2 and in concert with KSS1 promotes signal transduction. Proc Natl Acad Sci USA. 1991;88:9392–9396. doi: 10.1073/pnas.88.21.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S, Snyder M. A filamentous growth response mediated by the yeast mating pathway. Genetics. 2001;159:919–928. doi: 10.1093/genetics/159.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, Silva AC, Shales M, Collins SR, van Wageningen S, et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- Hartman JL, 4th, Garvik B, Hartwell L. Principles for the buffering of genetic variation. Science. 2001;291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- Ihmels J, Collins SR, Schuldiner M, Krogan NJ, Weissman JS. Backup without redundancy: genetic interactions reveal the cost of duplicate gene loss. Mol Syst Biol. 2007;3:86. doi: 10.1038/msb4100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- Jacoby T, Flanagan H, Faykin A, Seto AG, Mattison C, Ota I. Two protein-tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J Biol Chem. 1997;272:17749–17755. doi: 10.1074/jbc.272.28.17749. [DOI] [PubMed] [Google Scholar]
- Kafri R, Bar-Even A, Pilpel Y. Transcription control reprogramming in genetic backup circuits. Nat Genet. 2005;37:295–299. doi: 10.1038/ng1523. [DOI] [PubMed] [Google Scholar]
- Kafri R, Levy M, Pilpel Y. The regulatory utilization of genetic redundancy through responsive backup circuits. Proc Natl Acad Sci USA. 2006;103:11653–11658. doi: 10.1073/pnas.0604883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nat Biotechnol. 2005;23:561–566. doi: 10.1038/nbt1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel C, Yus E, Serrano L. Engineering signal transduction pathways. Cell. 2010;140:33–47. doi: 10.1016/j.cell.2009.12.028. [DOI] [PubMed] [Google Scholar]
- Ma D, Cook JG, Thorner J. Phosphorylation and localization of Kss1, a MAP kinase of the Saccharomyces cerevisiae pheromone response pathway. Mol Biol Cell. 1995;6:889–909. doi: 10.1091/mbc.6.7.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Trusina A, El-Samad H, Lim WA, Tang C. Defining network topologies that can achieve biochemical adaptation. Cell. 2009;138:760–773. doi: 10.1016/j.cell.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Styles CA, Fink GR. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- Margaritis T, Lijnzaad P, van Leenen D, Bouwmeester D, Kemmeren P, van Hooff SR, Holstege FC. Adaptable gene-specific dye bias correction for two-channel DNA microarrays. Mol Syst Biol. 2009;5:266. doi: 10.1038/msb.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso G, Costanzo M, Huangfu M, Smith AM, Paw J, San Luis BJ, Boone C, Giaever G, Nislow C, Emili A, Zhang Z. The extensive and condition-dependent nature of epistasis among whole-genome duplicates in yeast. Genome Res. 2008;18:1092–1099. doi: 10.1101/gr.076174.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, Boerlijst MC, Cooke J, Smith JM. Evolution of genetic redundancy. Nature. 1997;388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- Papp B, Pál C, Hurst LD. Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature. 2004;429:661–664. doi: 10.1038/nature02636. [DOI] [PubMed] [Google Scholar]
- Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Nelson B, Marton MJ, Stoughton R, Meyer MR, Bennett HA, He YD, Dai H, Walker WL, Hughes TR, et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Peña JM, Pérez-Díaz RM, Alvarez S, Bermejo C, García R, Santiago C, Nombela C, Arroyo J. The ‘yeast cell wall chip’-a tool to analyse the regulation of cell wall biogenesis in Saccharomyces cerevisiae. Microbiology. 2005;151:2241–2249. doi: 10.1099/mic.0.27989-0. [DOI] [PubMed] [Google Scholar]
- Roth FP, Lipshitz HD, Andrews BJ. Q&A: epistasis. J Biol. 2009;8:35. doi: 10.1186/jbiol144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe E, Ayscough KR. The Ark1/Prk1 family of protein kinases. Regulators of endocytosis and the actin skeleton. EMBO Rep. 2003;4:246–251. doi: 10.1038/sj.embor.embor776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Holstege FC. In control: systematic assessment of microarray performance. EMBO Rep. 2004;5:964–969. doi: 10.1038/sj.embor.7400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri T, Semple JI, Lehner B. Widespread conservation of genetic redundancy during a billion years of eukaryotic evolution. Trends Genet. 2008;24:485–488. doi: 10.1016/j.tig.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Wagner A. Robustness against mutations in genetic networks of yeast. Nat Genet. 2000;24:355–361. doi: 10.1038/74174. [DOI] [PubMed] [Google Scholar]
- Warmka J, Hanneman J, Lee J, Amin D, Ota I. Ptc1, a type 2C Ser/Thr phosphatase, inactivates the HOG pathway by dephosphorylating the mitogen-activated protein kinase Hog1. Mol Cell Biol. 2001;21:51–60. doi: 10.1128/MCB.21.1.51-60.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler A, Arkind C, Mattison CP, Burkholder A, Knoche K, Ota I. Heat stress activates the yeast high-osmolarity glycerol mitogen-activated protein kinase pathway, and protein tyrosine phosphatases are essential under heat stress. Eukaryot Cell. 2002;1:163–173. doi: 10.1128/EC.1.2.163-173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurgler-Murphy SM, Maeda T, Witten EA, Saito H. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol. 1997;17:1289–1297. doi: 10.1128/mcb.17.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C, Mapes J, Hanneman J, Al-Zarban S, Ota I. Role of Ptc2 type 2C Ser/Thr phosphatase in yeast high-osmolarity glycerol pathway inactivation. Eukaryot Cell. 2002;1:1032–1040. doi: 10.1128/EC.1.6.1032-1040.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng G, Yu X, Cai M. Regulation of yeast actin cytoskeleton-regulatory complex Pan1p/Sla1p/End3p by serine/threonine kinase Prk1p. Mol Biol Cell. 2001;12:3759–3772. doi: 10.1091/mbc.12.12.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.