Abstract
Studies of bone genetics have addressed an array of related phenotypes, including various measures of biomechanical performance, bone size, bone, shape, and bone mineral density. These phenotypes are not independent, resulting in redundancy of the information they provide. Principal component (PC) analysis transforms multiple phenotype data to a new set of orthogonal “synthetic” phenotypes. We performed PC analysis on 17 femoral biomechanical, anatomic, and body size phenotypes in a reciprocal intercross of HcB-8 and HcB-23, accounting for 80% of the variance in 4 PCs. Three of the 4 PCs were mapped in the cross. The linkage analysis revealed a quantitative trait locus (QTL) with LOD = 4.7 for PC2 at 16 cM on chromosome 19 that was not detected using the directly measured phenotypes. The chromosome 19 QTL falls within a ~10 megabase interval, with Osf1 as a positional candidate gene. PC QTLs were also found on chromosomes 1, 2, 4, 6, and 10 that coincided with those identified for directly measured or calculated material property phenotypes. The novel chromosome 19 QTL illustrates the power advantage that attends use of PC phenotypes for linkage mapping. Constraint of the chromosome 19 candidate interval illustrates an important advantage of experimental crosses between recombinant congenic mouse strains.
Keywords: principal components, biomechanics, linkage, recombinant congenic mice, osteoporosis
INTRODUCTION
Bone biomechanical performance is complex, encompassing several different interrelated properties, each of which is subject to both genetic and environmental variability. Because fracture is an important health problem, there is great interest in gaining detailed understanding of the mechanisms by which bone biomechanical performance is determined. Studies of bone genetics have therefore addressed an array of related phenotypes, including various measures of biomechanical performance, bone size, bone shape, volumetric bone mineral density (BMD), and areal BMD. Covariation among these phenotypes is well-established [1, 2], leading some to assert that the biomechanical phenotypes of energy to failure, yield load, and maximum load are essentially equivalent [3].
While there is little doubt that biomechanical phenotypes are partially redundant, it is nevertheless useful to attempt to extract the unique information from each, as this can provide a more complete understanding of the mechanisms underlying fracture susceptibility. One approach to this challenge is to apply principal component (PC) analysis to the data [4]. PC analysis transforms the original phenotypes to an equal number of orthogonal PCs, each defined as a specific linear combination of the original phenotypes. Traditionally, further analysis is limited to the subset of PCs with Eigenvalues > 1, thus reducing the dimensionality of the data from k directly measured phenotypes to m (< k) PCs. Furthermore, in the best case scenario, the PCs are more robust as phenotypes for genetic analysis than any of the contributing phenotypes, thus allowing identification of genes that could not be found using directly measured phenotypes. These advantages have a price, however, as the PCs are “virtual” phenotypes whose biological interpretation may not be obvious.
We recently performed a reciprocal intercross of the recombinant congenic strains HcB-8 and HcB-23 [5], in which we studied a group of phenotypes encompassing body size, femoral diaphysis size and shape, and femoral biomechanical performance [6]. Here, we report PC analysis of the original data and linkage mapping of the PCs. This analysis was undertaken to gain insight into bone biology that we could not obtain from studying the directly measured phenotypes themselves. We achieve substantial dimensional reduction of the data, accounting for 80% of the phenotypic variance within the first 4 PCs. Linkage mapping identifies a QTL on chromosome 19 that was undetected in the original study. Finding a QTL on chromosome 19 that was not detected with any of the directly measured phenotypes demonstrates that we were successful in this endeavor.
MATERIALS AND METHODS
Mice
The parental mice in this study were the recombinant congenic strains HcB-8 and HcB-23, produced by inbreeding N3 C3H/DiSnA (C3H) x C57BL/10ScSnA (B10) mice to fixation [5]. This breeding program resulted in inbred strains harboring alleles of B10 origin at approximately 1/8 of the genome on a C3H background. Therefore, only approximately ¼ of the genome segregates in an intercross of HcB-8 and HcB-23, with the remaining portions of the genome fixed for the same allele in both parental strains. We performed a reciprocal F2 intercross and maintained the animals to an age of 17 + 1 weeks, as this is the age at which mice achieve peak bone mass [7]. Mice were housed 2–5 mice per 500cm2 cages, with 12h light-dark cycling, given autoclaved tap water and fed laboratory rodent chow 5001 (PMI Nutrition International, Richmond, IN) ad lib. Animals were euthanized by CO2 inhalation, following AVMA recommendation. Immediately following sacrifice, animals were weighed and measured (rostro-anal length), viscera were harvested for DNA isolation (Puregene), and femora and humeri were dissected free of soft tissue for additional phenotyping. Bones were wrapped in phosphate buffered saline-saturated gauze and stored frozen at −70°. The animal protocol was approved by the University of Wisconsin and the William S. Middleton Memorial Veterans’ Hospital IACUCs.
Phenotyping
We measured areal BMD of isolated femora by DXA as previously described [8]. Briefly, each bone was scanned twice with repositioning. Measured BMD was adjusted for position on the scanning grid and the average of the duplicate measurements was used for linkage analysis.
We tested femoral diaphysis biomechanical performance by quasi-static 3-point bending under displacement control at a rate of 0.3 mm/sec, with a support span of 7.5 mm as previously described [6]. This produces a mid-diaphyseal fracture directly below the crosshead. By using the femoral condyles and the 3rd trochanter as anatomical landmarks to position bones consistently, the testing protocol produces highly reproducible fractures. We obtained the periosteal perimeter, cortical cross-sectional area, outer and inner major and minor axis lengths, shape factor (ratio of outer major axis length to outer minor axis length), and cross-sectional moment of inertia from digital photographs [6].
PC analysis
Body mass, rostro-anal length, femoral BMD, elastic deflection, post-yield deflection, yield load, maximum load, stiffness, femoral cross-sectional area, femoral periosteal perimeter, femoral inner major and minor axis lengths, femoral outer major and minor axis lengths, femoral diaphyseal shape factor, and femoral diaphyseal cross-sectional moment of inertia were included as input variables. Prior to analysis, normalizing transformations were applied to the directly measured phenotypes and the resulting data were standardized as Z-scores. The PC analysis was performed with the R function prcomp [9–11].
Genotyping and linkage analysis
HcB-8 and HcB-23 share the same alleles over approximately ¾ of the genome. We genotyped the F2 progeny at 41 microsatellite markers where HcB-8 and HcB-23 harbor different alleles [12], using standard methods. Markers and complete reaction conditions are listed in table 1. Map positions of markers and genes are based the Mouse Genome Database (build 37) (http://www.informatics.jax.org/genes.shtml) [13]. We adjusted the locations of D1Mit105 and D3Mit199 to reflect the observed recombination frequencies in our cross [6]. We further refined the genetic map with the est.map function within R/qtl.
Table 1.
Correlations Among Phenotypes1
| Mass | Length | InMin | InMaj | OutMin | OutMaj | Perim | CSA | SF | CSMI | ED | PYD | Stiff | YLoad | MLoad | Energy | fBMD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mass | - | 0.59 10−57 |
0.11 NS |
0.33 10−15 |
0.43 10−28 |
0.40 10−24 |
0.47 10−33 |
0.53 10−44 |
0.14 .001 |
0.48 10−34 |
0.12 0.004 |
−0.01 NS |
0.49 10−37 |
0.35 10−18 |
0.54 10−46 |
0.28 10−11 |
0.50 10−38 |
| Length | 0.59 10−57 |
- | 0.05 NS |
0.15 0.001 |
0.30 10−13 |
0.27 10−10 |
0.30 10−13 |
0.36 10−19 |
0.08 NS |
0.33 10−16 |
−0.16 10−4 |
0.06 NS |
0.36 10−18 |
0.24 10−8 |
0.35 10−17 |
0.24 10−8 |
0.38 10−20 |
| InMin | 0.46 10−32 |
0.20 10−5 |
- | 0.59 10−57 |
0.57 10−52 |
0.38 10−21 |
0.45 10−30 |
0.15 0.001 |
−0.04 NS |
0.47 10−33 |
−0.24 10−8 |
−0.12 0.003 |
0.35 10−17 |
0.15 0.001 |
0.30 10−13 |
−0.06 NS |
0.06 NS |
| InMaj | 0.57 10−51 |
0.26 10−9 |
0.66 10−76 |
- | 0.60 10−59 |
0.72 10−95 |
0.77 10−117 |
0.54 10−45 |
0.40 10−24 |
0.65 10−71 |
−0.26 10−9 |
−0.01 NS |
0.47 10−34 |
0.28 10−11 |
0.50 10−38 |
0.20 10−6 |
0.36 10−19 |
| OutMin | 0.55 10−48 |
0.37 10−20 |
0.68 10−82 |
0.63 10−67 |
- | 0.68 10−81 |
0.79 10−130 |
0.75 10−108 |
−0.04 NS |
0.96 0 |
−0.26 10−9 |
−0.08 NS |
0.75 10−109 |
0.51 10−39 |
0.76 10−113 |
0.27 10−10 |
0.54 10−46 |
| OutMaj | 0.68 10−81 |
0.37 10−20 |
0.59 10−57 |
0.80 10−134 |
0.71 10−93 |
- | 0.95 10−311 |
0.88 10−197 |
0.71 10−92 |
0.84 10−161 |
−0.13 0.001 |
−0.08 0.048 |
0.69 10−85 |
0.55 10−49 |
0.75 10−107 |
0.27 10−10 |
0.60 10−58 |
| Perim | 0.70 10−88 |
0.39 10−22 |
0.63 10−68 |
0.83 10−153 |
0.80 10−135 |
0.97 0 |
- | 0.90 10−217 |
0.53 10−44 |
0.91 10−224 |
−0.20 10−6 |
−0.07 NS |
0.73 10−102 |
0.55 10−47 |
0.79 10−128 |
0.30 10−13 |
0.62 10−64 |
| CSA | 0.67 10−77 |
0.44 10−28 |
0.37 10−19 |
0.65 10−72 |
0.77 10−118 |
0.90 10−217 |
0.92 10−239 |
- | 0.48 10−35 |
0.89 10−201 |
−0.13 0.002 |
−0.05 NS |
0.76 10−112 |
0.62 10−63 |
0.84 10−157 |
0.38 10−20 |
0.74 10−102 |
| SF | 0.49 10−36 |
0.21 10−6 |
0.25 10−9 |
0.59 10−57 |
0.15 0.001 |
0.80 10−134 |
0.68 10−82 |
0.61 10−62 |
- | 0.23 10−7 |
0.07 NS |
−0.04 NS |
0.22 10−7 |
0.27 10−10 |
0.29 10−12 |
0.10 NS |
0.29 10−12 |
| CSMI | 0.63 10−68 |
0.41 10−25 |
0.63 10−67 |
0.71 10−91 |
0.96 0 |
0.87 10−181 |
0.92 10−238 |
0.90 10−215 |
0.40 10−23 |
- | −0.22 10−7 |
−0.08 NS |
0.80 10−132 |
0.58 10−54 |
0.83 10−152 |
0.31 10−14 |
0.63 10−67 |
| ED | −0.07 NS |
−0.15 0.001 |
−0.20 10−5 |
−0.20 10−5 |
−0.24 10−8 |
−0.09 NS |
−0.15 0.001 |
−0.10 NS |
0.08 NS |
−0.20 10−5 |
- | −0.12 0.003 |
−0.25 10−9 |
0.30 10−13 |
−0.10 NS |
−0.03 NS |
−0.07 NS |
| PYD | −0.05 NS |
0.04 NS |
−0.14 0.001 |
−0.02 NS |
−0.10 NS |
−0.10 NS |
−0.09 NS |
−0.08 NS |
−0.06 NS |
−0.11 0.001 |
−0.16 10−4 |
- | −0.18 10−4 |
−0.15 0.001 |
−0.16 10−4 |
0.58 10−54 |
−0.02 NS |
| Stiff | 0.41 10−25 |
0.36 10−19 |
0.42 10−25 |
0.40 10−23 |
0.75 10−111 |
0.57 10−52 |
0.63 10−65 |
0.67 10−79 |
0.16 10−4 |
0.75 10−111 |
−0.26 10−9 |
−0.18 10−5 |
- | 0.73 1099 |
0.89 10209 |
0.30 10−13 |
0.68 10−83 |
| YLoad | 0.32 10−15 |
0.26 10−9 |
0.27 10−10 |
0.25 10−9 |
0.55 10−48 |
0.48 10−34 |
0.49 10−36 |
0.56 1049 |
0.19 10−5 |
0.58 10−54 |
0.25 10−9 |
−0.16 10−4 |
0.77 10−116 |
- | 0.74 10104 |
0.23 10−8 |
0.61 10−62 |
| Mload | 0.52 10−42 |
0.38 10−20 |
0.44 10−29 |
0.47 10−34 |
0.79 10−129 |
0.68 10−80 |
0.72 10−97 |
0.78 10−120 |
0.28 10−11 |
0.82 10−146 |
−0.12 0.004 |
−0.17 10−4 |
0.90 10−222 |
0.78 10−120 |
- | 0.36 10−19 |
0.77 10−119 |
| Energy | 0.19 10−5 |
0.22 10−7 |
−0.07 NS |
0.17 10−4 |
0.22 10−7 |
0.20 10−6 |
0.23 10−7 |
0.33 10−15 |
0.09 NS |
0.25 10−9 |
−0.02 NS |
0.58 10−54 |
0.25 10−9 |
0.19 10−5 |
0.30 10−12 |
- | 0.35 10−18 |
| fBMD | 0.10 NS |
0.26 10−9 |
−0.09 NS |
0.10 NS |
0.37 10−20 |
0.22 10−7 |
0.27 10−10 |
0.45 10−30 |
0 NS |
0.38 10−21 |
−0.09 0.026 |
0 NS |
0.62 10−63 |
0.56 10−49 |
0.64 10−69 |
0.31 10−14 |
- |
Phenotypes are Mass = body mass, Length = rostro-anal length, InMin = inner minor axis, InMaj = inner major axis, OutMin = outer minor axis, OutMaj = outer major axis, Perim = periosteal perimeter, CSA = cortical cross-sectional area, SF = shape factor, CSMI = cross-sectional moment of inertia, ED = elastic deflection, PYD = post-yield deflection, Stiff = stiffness, YLoad = yield load, MLoad = maximum load, Energy = energy to failure, fBMD = BMD by ex vivo DXA. Top line of each cell shows R. Bottom line shows p value. The a = 0.05 significance threshold is p < 0.00725, corresponding to an estimated 6.9 independent tests. NS = not significant.
Above the diagonal are sex- and cross-adjusted correlations
Below the diagonal are unadjusted correlations
We performed the primary linkage analysis by interval mapping [14] as implemented in R/qtl [15], using the Haley-Knott regression method [16] We performed the linkage analysis using sex and cross arm as additive covariates (.i.e. phenotypes adjusted by linear regression for sex and cross direction). We performed secondary analyses by composite interval mapping analysis [17] and multiple trait linkage analysis [18] as implemented in QTL Cartographer [19, 20]. For tests of QTL interactions with sex, cross direction, or both, we explicitly compared the covariate in question as an interactive covariate in the full linkage model and compared it to a model in which both sex and cross direction were treated as additive covariates. The interaction LOD score is the difference between full model LOD score and additive model LOD score [15, 21]. We established significance thresholds empirically by permutation tests [22]. For the interaction tests, we used the same seed value to ensure that the permutation tests examined the same simulated data sets. The additive model includes 2 degrees of freedom (additive and dominance effects), and the interactive models include an additional 2 degrees of freedom for each included interactive factor (factor x QTL additive effect and factor x QTL dominance effect). Thus, the full model including interactions for both sex and cross direction has 6 degrees of freedom. The significance threshold for the interaction test was experiment-wide p < 0.05.
Other statistical analyses
Correlations between pairs of phenotypes were measured on the transformed, normalized data. Comparisons of intercross subgroups were by 2-way ANOVA, with sex and cross direction as factors, with post hoc evaluation of significant differences between groups by the Holm-Sidak test. If necessary, we transformed the data to satisfy the assumptions of parametric statistical testing or used the non-parametric rank sum test or ANOVA on ranks when we were unable to normalize the data. To determine the appropriate correction for multiple comparisons in the correlation analysis, we adapted a regression method originally developed for linkage mapping [23]. Statistical analyses were performed with SigmaStat 3.0 (SPSS). Summary data are shown as mean + SEM.
RESULTS
Basic characteristics of the cross and correlations among directly measured phenotypes
We studied a total F2 progeny of 603 animals, including 124 8×23 females, 169 8×23 males, 163 23 × 8 females, and 147 23×8 males. The disparities in the numbers of males and females within the cross arms do not reflect differential survival, but simply pragmatic animal husbandry considerations. Phenotypic measures used for principal components analysis included geometric, biomechanical, and body size measurements. These were chosen to match phenotypes included in our published whole bone phenotype linkage mapping in the same cross [6]. Geometric measurements of the plane of fracture in the femoral mid-diaphysis included inner and outer axis lengths, shape factor, perimeter, cross-sectional area, and cross-sectional moment of inertia. Biomechanical measurements obtained from 3 point bending of the femora included elastic and post-yield deflection during 3 point bending, stiffness, yield load, maximum load, and energy to failure. Other measurements included body mass, rostro-anal length, and femoral bone mineral density by DXA.
Table 1 shows the correlations between each pair of phenotypes. The high correlation among them is unsurprising, as the phenotypes are related to each other. To the extent that they are correlated, the phenotypes reflect redundant information. Indeed, application of a regression-based method for estimating the number of independent tests in correlated data sets reveals that our data contain 6.9 independent tests. The Bonferroni-adjusted significance threshold for the correlation analysis is therefore p < 0.0725.
Principal components analysis
PC analysis expresses the experimental data as a linear function of orthogonal (i.e. statistically independent) principal components (PCs). We achieved a substantial dimensional reduction of our data set, with the first 4 PCs accounting for nearly 80% of the total variance of the 17 phenotype set. These 4 PCs each had an Eigenvalue > 1, the standard criterion for retention in subsequent analyses. The results are summarized in table 2, while the Eigenvectors are given in table 3.
Table 2.
Principal Components Analysis
| PC1 | PC2 | PC3 | PC4 | |
|---|---|---|---|---|
| Eigenvalue | 2.91 | 1.44 | 1.29 | 1.17 |
| R2 | 0.496 | 0.122 | 0.098 | 0.081 |
| cumulative r2 | 0.496 | 0.619 | 0.717 | 0.797 |
Table 3.
Eigenvectors of the first 4 PCs
| PC1 | PC2 | PC3 | PC4 | |
|---|---|---|---|---|
| Body Mass | 0.248 | 0.179 | 0.109 | −0.065 |
| Rostro-Anal Length | 0.170 | −0.051 | 0.187 | 0.027 |
| Inner Minor Axis | 0.214 | 0.265 | −0.156 | 0.314 |
| Inner Major Axis | 0.261 | 0.295 | 0.086 | 0.071 |
| Outer Minor Axis | 0.302 | −0.067 | −0.080 | 0.259 |
| Outer Major Axis | 0.316 | 0.212 | 0.037 | −0.140 |
| Perimeter | 0.327 | 0.169 | 0.034 | −0.042 |
| Cross-Sectional Area | 0.318 | −0.002 | 0.069 | −0.144 |
| Shape Factor | 0.186 | 0.357 | 0.120 | −0.418 |
| Cross-Sectional Moment of Inertia | 0.329 | −0.003 | −0.032 | 0.102 |
| Elastic Deflection | −0.058 | −0.023 | −0.142 | −0.702 |
| Post-Yield Deflection | −0.037 | −0.088 | 0.672 | 0.077 |
| Yield Load | 0.224 | −0.334 | −0.209 | −0.265 |
| Maximum Load | 0.300 | −0.261 | −0.118 | −0.0016 |
| Stiffness | 0.275 | −0.310 | −0.148 | 0.129 |
| Energy to Failure | 0.101 | −0.252 | 0.585 | −0.089 |
| Femoral BMD | 0.153 | −0.506 | −0.002 | −0.062 |
Because the PCs were calculated from standardized phenotypes, inspection of the rotations in each Eigenvector provides some insight into the heuristic interpretation of what the PCs represent (table 3). PC1 represents a combination of size and stiffness, with approximately equal contributions from outer minor axis, outer major axis, perimeter, cross-sectional area, stiffness, and cross-sectional moment of inertia. PC2 represents size without mechanical performance, including large negative contributions from yield load, maximum load, stiffness, energy to failure, and BMD and large positive contributions from inner minor axis, inner major axis, outer major axis, and shape factor. PC3 is roughly analogous to ductility, with its large positive contributions from post-yield deflection and energy to failure, but negative contributions from yield load, maximum load, and stiffness. PC4 has its largest positive contributions from inner minor axis and outer minor axis, with large negative contributions from elastic deflection and shape factor.
The parental strains in this experiment, HcB-8 and HcB-23, were chosen because previous study of biomechanical performance and bone geometry had revealed significant differences between them. It is therefore useful to determine the extent to which they differ in their PC phenotypes, which are summarized in table 4. All 4 PCs differ between the parental strains, with p < 0.001 for PCs 1, 2, and 3, and p = 0.020 for PC4. PCs 1 (p = 0.015), 2 (p <0.001), and 3 (p = 0.014) also display differences between females and males. None of the PCs demonstrate a significant strain x sex interaction.
Table 4.
PC Phenotypes of HcB-8 and HcB-23
| HcB-8 | HcB-23 | |||
|---|---|---|---|---|
| Females (N = 5) | Males (N = 7) | Females (N = 14) | Males (N = 16) | |
| PC11,2 | −2.49 + 0.84 | −1.98 + 0.71 | −0.21 + 0.50 | 2.60 + 0.47 |
| PC21,2 | −1.66 + 0.39 | 1.35 + 0.33 | −0.16 + 0.23 | 2.15 + 0.22 |
| PC31,2 | 0.94 + 0.40 | −0.44 + 0.33 | −2.73 + 0.24 | −2.91 + 0.22 |
| PC41 | −2.05+ 0.66 | −2.20 + 0.56 | −2.71 + 0.40 | −4.03 + 0.37 |
Significant difference (p between 0.020 and < 0.001) between HcB-8 and HcB-23.
Significant difference (p between 0.015 and < 0.001) between females and males.
PC Linkage Mapping
We performed linkage analysis of the first 4 principal components. No QTLs for PC3 were detected in either the entire F2 progeny or in most of the subgroups, while the other 3 principal components all yielded several significant QTLs. Figure 1, table 5, and supplementary figure 1 summarize the results, and show the linkage signals on chromosomes 1, 2, 4, 10, and 19 in the full F2 population and the various sex and cross direction defined subgroups. The calculated effect sizes are summarized in table 6. The mapped QTLs, sex, and cross direction account for 40% of the PC1 variance, 51% of the PC2 variance, 6% of the PC3 variance, and 18% of the PC4 variance. Table 7 and supplementary figure 1 summarize the QTL x sex and QTL x cross direction interactions. A QTL x sex interaction exists when a QTL has a significantly more powerful effect in one sex than the other. A QTL x cross direction may indicate the existence of a maternal effect or genomic imprinting.
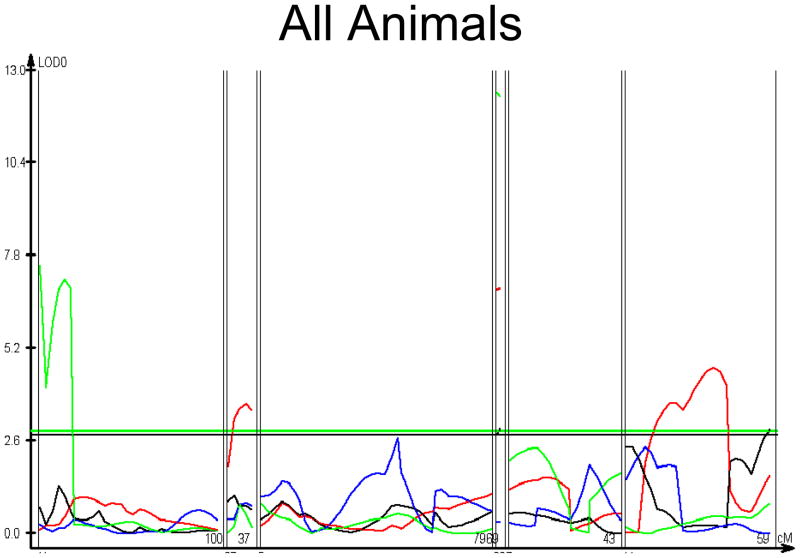
Figure 1. Linkage Map of the PCs.
Location in cM, limited to informative chromosome regions is shown on the X axis. LOD score is shown on the Y axis. PC 1 = red, PC 2 = black, PC3 = blue, PC 4 = green. Chromosome 19 is omitted because it contains only a single informative locus; its linkage data are given in tables 4 and 5. Linkage maps for the subgroups are provided as supplementary figure 1.
Table 5.
Summary of Linkage Data1
| Trait (5% threshold) | All F2 | All Female | All Male | All 8×23 | 8×23 Female | 8×23 Male | All 23×8 | 23×8 Female | 23×8 Male |
|---|---|---|---|---|---|---|---|---|---|
| PC1 (2.8) |
2, 33, 3.7 4, 66, 6.9 10, 39, 4.7 |
1, 67, 3.2 2, 27, 3.3 4, 66, 3.0 |
4, 66, 5.1 6, 15, 4.1 10, 39, 4.1 |
1, 49, 5.0 10, 30, 3.5 |
1, 84, 3.3 10, 27, 2.8 |
4, 66, 3.2 6, 14, 3.1 |
4, 66, 5.1 | 4, 66, 4.9 | 6, 18, 3.3 10, 37, 3.1 |
| PC2 (2.8) | 4, 67, 2.9 10, 40, 2.9 19, 16, 4.7 |
1, 47, 4.0 19, 16, 3.1 |
3, 19, 3.2 10, 57, 5.7 |
4, 66, 5.2 19, 16, 3.1 |
1, 45, 3.1 | 4, 66, 3.8 | 10, 43, 4.6 | 10, 46, 5.9 | |
| PC3 (2.9) | 10, 19, 3.9 | ||||||||
| PC4 (2.9) |
1, 41, 7.5 4, 66, 12.4 |
1, 41, 3.5 4, 66, 4.8 6, 7, 3.5 |
1, 41, 3.4 4, 66, 8.1 |
1, 49, 5.0 4, 66, 8.1 |
1, 45, 4.4 | 4, 66, 4.8 | 4, 66, 7.2 | 4, 66, 4.2 |
Cells show chromosome, map position in cM, and LOD score. Bold entries are significant at an experiment-wide confidence level of p<0.01
Table 6.
Principal Component QTL Effect Sizes
| Chromosome | Position | HcB-8 Phenotype1 (SEM) | Heterozygote Phenotype1 (SEM) | HcB-23 Phenotype1 (SEM) | Additive Effect2 | Dominance Effect3 | % F2 Variance | |
|---|---|---|---|---|---|---|---|---|
| PC1 | 2 | 33 | −0.62 (0.20) | 0.04 (0.13) | 0.66 (0.21) | 0.64 | 0.02 | 3.4 |
| 4 | 66 | −0.96 (0.19) | 0.33 (0.14) | 0.29 (0.18) | 0.63 | 0.67 | 5.1 | |
| 10 | 39 | −0.26 (0.21) | −0.12 (0.13) | 0.51 (0.21) | 0.39 | −0.25 | 1.4 | |
| PC2 | 4 | 66 | 0.31 (0.09) | −0.02 (0.07) | −0.25 (0.08) | −0.29 | −0.05 | 3.6 |
| 19 | 16 | 0.32 (0.08) | −0.04 (0.07) | −0.21 (0.08) | −0.27 | −0.10 | 3.5 | |
| PC4 | 1 | 41 | 0.33 (0.09) | 0.02 (0.07) | −0.36 (0.09) | −0.40 | 0.04 | 4.5 |
| 4 | 66 | 0.56 (0.09) | −0.11 (0.07) | −0.31 (0.08) | −0.44 | −0.24 | 8.5 |
Phenotype values are age and sex adjusted.
Additive effect = ½[average HcB-23 phenotype-average HcB-8 phenotype]
Dominance effect = average heterozygote phenotype-1/2[average HcB-8 phenotype+average HcB-23 phenotype]
Table 7.
Sex x QTL and Cross x QTL Interactions
| Covariate | Chromosome | Map Position | Trait (interaction p value) |
|---|---|---|---|
| Sex | 1 | 88 | PC2 (0.021) |
| 6 | 16 | PC1 (0.007) | |
| Cross Direction | 3 | 23 | PC2(0.027) |
| Both | 6 | 16 | PC1 (0.020) |
The chromosome 19 linkage signal was not apparent in the linkage mapping of the directly measured or calculated material property phenotypes, but those on the other 4 chromosomes coincide with QTLs for directly measured bone phenotypes that we previously reported [6, 24]. Interestingly, the chromosome 4 QTL that we found to affect bone geometry, BMD, and whole bone biomechanical performance shows significant linkage to PC1, PC2, and PC4. Thus, both at the level of whole bone biomechanical performance and structure and at the level of PCs, the chromosome 4 QTL is highly pleiotropic. Also paralleling the findings in direct phenotype linkage mapping, the PC1 QTL on proximal chromosome 6 is limited to males (tables 5 and 7), thus revealing a QTL x sex interaction.
DISCUSSION
We performed PC analysis of femoral anatomy, biomechanical performance, and body size phenotypes in a reciprocal F2 intercross of HcB-8 and HcB-23. The first 4 PCs account for 80% of the phenotypic variance, and PCs 1, 2, and 4 could be mapped genetically. Use of PC “virtual” phenotypes reveals a putative QTL on chromosome 19 that was not detected with either whole bone or material level phenotypes [6, 24]. The QTLs on chromosomes 1, 4, 6, and 10 coincide with those found in those analyses.
The QTL for PC2 on chromosome 19, however, was undetected in either the whole bone properties. The informative segment of chromosome 19 is short, with D19Mit41 the only known marker known to be informative between HcB-8 and HcB-23. The closest markers presently known not to be included within the informative segment are the SNPs rs68186 and rs3726735, located at 14,655,411 and 24,707,628 bp from the centromere, respectively. This 10 megabase candidate region includes 32 annotated genes and a similar number of uncharacterized transcripts [13]. The mouse SNP database identifies 22/84 SNP polymorphisms between C57BL/10ScSnJ and C3H/HeSnJ, the strains most closely related to the HcB’s progenitors that are included in the SNP database [25]. These strains have only been genotyped at low density, but the region includes 3508 imputed polymorphisms between C57BL/10J and C3H/HeJ [25]. Among the genes in the interval, Ostf1, encoding osteoclast stimulating factor 1 is an obvious positional candidate gene. Ostf1 was first identified as an intracellular signaling protein that promotes osteoclast maturation and activity [26], and originally isolated from mouse embryo cDNA on the basis of containing a SRC3 domain [27]. This gene is an attractive candidate because the PC2 loading is heavily weighted by inner minor and inner major axis length (table 3), phenotypes that reflect the size of the marrow space. This region might overlap two previously reported QTLs for tibial yield stress [28] and markers of anabolic response to mechanical loading [29]. The region may, but is unlikely to, coincide with a recently reported QTL for femoral robustness [30]. The robustness QTL is located distal to ours, and was detected in a different genetic background. In our prior work [6], we mapped “slenderness,” which is the inverse of robustness and did not detect any evidence of linkage to chromosome 19.
In any analysis of multiple related outcomes, determining which are most important poses a challenge. This is particularly true of the outcomes that are known to share some determinants. A starting point is to generate a correlation matrix in which each pair of outcomes is studied for covariation, as we have done in table 1. At this point, two basic strategies are possible. The first is to develop a multiple regression model in which one “primary” outcome is expressed as a function of several of the other “explanatory” outcomes. This approach has several fundamental problems. First, there is no convincing, a priori way to assign one outcome as being primary. Second, use of a stepwise heuristic, necessary to avoid model overfitting, may result in the explanatory outcomes being significantly correlated with each other, so that the final model expresses redundant information in the guise of distinct contributions to the “primary” outcome.
Principal component analysis takes a different approach. Each individual animal’s phenotypes can be considered as a vector with one dimension for each phenotype, seventeen dimensions in our case. The analysis finds the best 17-dimensional regression line to fit all the data by least squares, and defines this as the new X axis, or PC1. The best fitting line through the residuals is then chosen as PC2, with the constraint that it must be orthogonal (multidimensionally perpendicular) to PC1. This process is repeated iteratively until the number of principal components equals the number of phenotypes-1. The last PC is then the residual after the remaining PCs have been chosen as the best fitting orthogonal regression lines of the residuals. Downstream analyses are then limited to the subset of PCs that have larger than average contributions to the overall phenotypic variance, as embodied mathematically by an Eigenvalue > 1.
It is natural to wonder about the impact of including or omitting any particular input phenotype on the resulting PC analysis. This depends on the trait one chooses to omit. In general, omitting a trait that is highly correlated with multiple other traits will have little effect, while omitting a trait that is poorly correlated with the other traits has a large effect.
Several other groups have used PC analysis to study bone [31–39]. Most of these studies have used PCs to sample multiple sites with a single methodology, effectively using PCs as a tool to detect pleiotropic QTLs. Notable exceptions to this are [33], in which both DXA and quantitative ultrasound were used in a human population, and [31], in which biomechanical performance, bone anatomy, and BMD of the femur were studied. Other investigators performing a similar analysis observed a robust QTL on chromosome 4, but in their analysis, the linkage peak was concentrated within a single principal component [31], while in our experiment the linkage is distributed among 3 principal components.
The QTL on chromosome 4 is pleiotropic both with regard to PCs and with regard to directly measured phenotypes. It is linked to 10 directly measured phenotypes {Saless, 2009 #106}, yet is unlinked to any calculated material property phenotypes {Saless, 2010 #124}. While pleiotropy is an attractive explanation for covariation among traits, it is also indicative of the existence of a “physiological gap” between the pleiotropic gene’s primary function and the studied phenotypes. Pleiotropy is helpful insofar as positional candidate genes can be prioritized on the basis of known biology that relates the multiple pleiotropic phenotypes to a single mechanism. We have interpreted this pattern of findings as suggesting that the QTL’s primary phenotype is to modulate the amount of cell proliferation in response to loading and that the other phenotypes are all consequences of the difference in the bone modeling response [6, 24]. Should this interpretation be correct, then the effects of differential modeling should also be apparent at sites other than the femur.
Theoretical and simulation-based studies predict that incorporation of multiple phenotypes increases the statistical power to detect QTLs for complex traits [40]. Further, these authors emphasize that the greatest increase in power occurs when an allele’s effect is opposite in direction for the included traits. This condition is satisfied on chromosome 10, where the QTL for PC1 (HcB-23 allele increases) coincides with QTLs for BMD (HcB-23 allele increases) and toughness (HcB-8 allele increases) [6, 24]. On average, use of PCs improves the statistical power to find QTLs. However, this is not always sufficient to allow a QTL to be found. Our data illustrate both aspects of this. On the positive side, we were able to map a QTL on chromosome 19 that we did not detect when we mapped any of the original phenotypes. The remaining PC QTLs coincide with linkage peaks for one or more “natural” phenotypes. This validates the overall analysis at an empirical level. However, our failure to map any QTLs for PC3 shows that the additional statistical power is not necessarily sufficient to allow a QTL to be mapped. There are 2 alternative interpretations: PC3 is not heritable or measurements of the phenotypes that contribute most to PC3 are too imprecise to allow us to discern the underlying genetics. Our data do not allow us to distinguish between these alternatives.
The work reported here has several strengths. The choice of HcB recombinant congenic mice as parental strains reduced the effective genome size in our cross. This resulted in increased statistical power, as reflected by the lower significance thresholds for linkage than are encountered in whole-genome linkage scans. In a whole genome mouse intercross, the expected 5% experiment-wide significance threshold is a LOD score of 4.3 [41], while in our experiment permutation testing showed experiment-wide significance at LOD scores between 2.7 and 2.9, depending on the phenotype. This has the practical consequence of increasing the sensitivity of the experiment for finding QTLs. Recombination during the construction of the HcB strains also led to the shortening of individual donor chromosome segments by crossing over during the inbreeding process. The locations of the recombination events, in turn, constrained the possible locations of mapped QTLs.
Limitations of our study should also be noted. Using recombinant congenic strains as parental strains made it impossible for us to detect bone QTLs in genomic regions where HcB-8 and HcB-23 harbor the same allele. The endpoints of the informative chromosome regions have not been precisely localized. We have analyzed the bone properties of the F2 progeny only at a single age, so our results are unable to address developmental phenotypes either during growth or following maturity. The biological interpretation of the principal components is also potentially problematic, as they are synthetic phenotypes. We had hoped that the limited power of biomechanical testing [42], particularly in the context of measuring plasticity, energy absorption, and their material equivalents, strain and toughness, might be mitigated by the use of principal component analysis. However, PC3, the PC that corresponds best to these properties, proved to be unsuitable for linkage analysis.
In summary, we performed a PCs analysis of bone biomechanical, geometric, and body size phenotypes and found that 80% of the total phenotypic variation could be accounted for in the first 4 principal components. Three of the 4 PCs were successfully mapped, and the PC QTLs confirmed QTLs identified by linkage mapping of either the directly measured phenotypes or the calculated material properties on chromosomes 1, 2, 4, 6, and 10. A QTL for PC2 has been mapped to a 10 megabase region of chromosome 19, which was not detected using either whole bone or calculated material property phenotypes.
Supplementary Material
Acknowledgments
Funding Sources: This material is based upon work supported by the Office of Research and Development, Biomedical Laboratory R&D Service, Department of Veterans Affairs (RDB) and performed in the Geriatrics Research, Education, and Clinical Center at the William S. Middleton Memorial Veterans Hospital. This report is Madison GRECC manuscript 2010-09. This work is supported by NIH grant AR-54753 (RDB).
This material is based upon work supported by the Office of Research and Development, Biomedical Laboratory R&D Service, Department of Veterans Affairs (RDB) and performed in the Geriatrics Research, Education, and Clinical Center at the William S. Middleton Memorial Veterans Hospital. This report is Madison GRECC manuscript 2010-09. This work is supported by NIH grant AR-54753 (RDB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Neema Saless, Cellular and Molecular Biology Program, University of Wisconsin, Madison.
Suzanne J. Litscher, Department of Medicine, University of Wisconsin, Madison
Ray Vanderby, Department of Orthopedic Surgery, University of Wisconsin, Madison.
Peter Demant, Department of Genetics, Roswell Park Cancer Institute.
Robert D. Blank, Department of Medicine, University of Wisconsin, Madison and GRECC Service, William S. Middleton Veterans Hospital
LITERATURE CITED
- 1.Jepsen KJ, Hu B, Tommasini SM, Courtland HW, Price C, Cordova M, Nadeau JH. Phenotypic integration of skeletal traits during growth buffers genetic variants affecting the slenderness of femora in inbred mouse strains. Mamm Genome. 2009;20:21–33. doi: 10.1007/s00335-008-9158-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jepsen KJ, Hu B, Tommasini SM, Courtland HW, Price C, Terranova CJ, Nadeau JH. Genetic randomization reveals functional relationships among morphologic and tissue-quality traits that contribute to bone strength and fragility. Mamm Genome. 2007;18:492–507. doi: 10.1007/s00335-007-9017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ritchie RO, Koester KJ, Ionova S, Yao W, Lane NE, Ager JW. Measurement of the toughness of bone: a tutorial with special reference to small animal studies. Bone. 2008;43:798–812. doi: 10.1016/j.bone.2008.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearson K. On Lines and Planes of Closest Fit to Systems of Points in Space. Philosophical Magazine. 1901;2:559–572. [Google Scholar]
- 5.Demant P, Hart AA. Recombinant congenic strains--a new tool for analyzing genetic traits determined by more than one gene. Immunogenetics. 1986;24:416–22. doi: 10.1007/BF00377961. [DOI] [PubMed] [Google Scholar]
- 6.Saless N, Litscher SJ, Lopez Franco GE, Houlihan MJ, Sudhakaran S, Raheem KA, O’Neil TK, Vanderby R, Demant P, Blank RD. Quantitative trait loci for biomechanical performance and femoral geometry in an intercross of recombinant congenic mice: restriction of the Bmd7 candidate interval. Faseb J. 2009;23:2142–54. doi: 10.1096/fj.08-118679. [DOI] [PubMed] [Google Scholar]
- 7.Beamer WG, Donahue LR, Rosen CJ, Baylink DJ. Genetic variability in adult bone density among inbred strains of mice. Bone. 1996;18:397–403. doi: 10.1016/8756-3282(96)00047-6. [DOI] [PubMed] [Google Scholar]
- 8.Lopez Franco GE, O’Neil TK, Litscher SJ, Urban-Piette M, Blank RD. Accuracy and precision of PIXImus densitometry for ex vivo mouse long bones: comparison of technique and software version. J Clin Densitom. 2004;7:326–33. doi: 10.1385/jcd:7:3:326. [DOI] [PubMed] [Google Scholar]
- 9.Becker RA, Chambers JM, Wilks AR. The New S Language. Cole Computer Science Series. Monterey, CA: Wadsworth and Brooks/Cole Advanced Books & Software; 1988. [Google Scholar]
- 10.Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. London: Academic Press; 1979. [Google Scholar]
- 11.Venables WN, Ripley BD. Modern Applied Statistics with S. New York: Springer; 2002. [Google Scholar]
- 12.Stassen AP, Groot PC, Eppig JT, Demant P. Genetic composition of the recombinant congenic strains. Mamm Genome. 1996;7:55–8. doi: 10.1007/s003359900013. [DOI] [PubMed] [Google Scholar]
- 13.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The mouse genome database (MGD): new features facilitating a model system. Nucleic Acids Res. 2007;35:D630–7. doi: 10.1093/nar/gkl940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps [published erratum appears in Genetics 1994 Feb;136(2):705] Genetics. 1989;121:185–99. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–90. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 16.Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–24. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- 17.Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–68. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang C, Zeng ZB. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics. 1995;140:1111–27. doi: 10.1093/genetics/140.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basten CJ, Weir BS, Zeng Z-B. Zmap- a QTL Cartographer. In: Smith C, Gavora JS, Benkel B, Chesnais J, Fairfull W, Gibson JP, Kennedy BW, Burnside EB, editors. 5th World Conference on Genetics Applied to Livestock Production: Computing Strategies and Software. Vol. 22. Guelph, Ontario: 1994. pp. 65–66. [Google Scholar]
- 20.Basten CJ, Weir BS, Zeng Z-B. QTL Cartographer. Raleigh, NC: North Carolina State University; 1999. [Google Scholar]
- 21.Ishimori N, Stylianou IM, Korstanje R, Marion MA, Li R, Donahue LR, Rosen CJ, Beamer WG, Paigen B, Churchill GA. Quantitative Trait Loci for Bone Mineral Density in an SM/J by NZB/BlNJ Intercross Population and Identification of Trps1 as a Probable Candidate Gene. J Bone Miner Res. 2008 doi: 10.1359/JBMR.080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–71. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camp NJ, Farnham JM. Correcting for multiple analyses in genomewide linkage studies. Ann Hum Genet. 2001;65:577–82. doi: 10.1017/S0003480001008922. [DOI] [PubMed] [Google Scholar]
- 24.Saless N, Lopez Franco GE, Litscher S, Kattappuram RS, Houlihan MJ, Vanderby R, Demant P, Blank RD. Linkage mapping of femoral material properties in a reciprocal intercross of HcB-8 and HcB-23 recombinant mouse strains. Bone. 2010;46:1251–9. doi: 10.1016/j.bone.2010.01.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mouse SNP Database. Bar Harbor, ME: The Jackson Laboratory; 2010. [Google Scholar]
- 26.Reddy S, Devlin R, Menaa C, Nishimura R, Choi SJ, Dallas M, Yoneda T, Roodman GD. Isolation and characterization of a cDNA clone encoding a novel peptide (OSF) that enhances osteoclast formation and bone resorption. J Cell Physiol. 1998;177:636–45. doi: 10.1002/(SICI)1097-4652(199812)177:4<636::AID-JCP14>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 27.Sparks AB, Hoffman NG, McConnell SJ, Fowlkes DM, Kay BK. Cloning of ligand targets: systematic isolation of SH3 domain-containing proteins. Nat Biotechnol. 1996;14:741–4. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- 28.Lang DH, Sharkey NA, Mack HA, Vogler GP, Vandenbergh DJ, Blizard DA, Stout JT, McClearn GE. Quantitative trait loci analysis of structural and material skeletal phenotypes in C57BL/6J and DBA/2 second-generation and recombinant inbred mice. J Bone Miner Res. 2005;20:88–99. doi: 10.1359/JBMR.041001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kesavan C, Baylink DJ, Kapoor S, Mohan S. Novel loci regulating bone anabolic response to loading: expression QTL analysis in C57BL/6JXC3H/HeJ mice cross. Bone. 2007;41:223–30. doi: 10.1016/j.bone.2007.04.185. [DOI] [PubMed] [Google Scholar]
- 30.Jepsen KJ, Courtland HW, Nadeau JH. Genetically determined phenotype covariation networks control bone strength. J Bone Miner Res. 2010;25:1581–93. doi: 10.1002/jbmr.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koller DL, Schriefer J, Sun Q, Shultz KL, Donahue LR, Rosen CJ, Foroud T, Beamer WG, Turner CH. Genetic effects for femoral biomechanics, structure, and density in C57BL/6J and C3H/HeJ inbred mouse strains. J Bone Miner Res. 2003;18:1758–65. doi: 10.1359/jbmr.2003.18.10.1758. [DOI] [PubMed] [Google Scholar]
- 32.Freedman BI, Bowden DW, Ziegler JT, Langefeld CD, Lehtinen AB, Rudock ME, Lenchik L, Hruska KA, Register TC, Carr JJ. Bone morphogenetic protein 7 (BMP7) gene polymorphisms are associated with inverse relationships between vascular calcification and BMD: the Diabetes Heart Study. J Bone Miner Res. 2009;24:1719–27. doi: 10.1359/JBMR.090501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karasik D, Cupples LA, Hannan MT, Kiel DP. Genome screen for a combined bone phenotype using principal component analysis: the Framingham study. Bone. 2004;34:547–56. doi: 10.1016/j.bone.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 34.Karasik D, Kiel DP. Genetics of the musculoskeletal system: a pleiotropic approach. J Bone Miner Res. 2008;23:788–802. doi: 10.1359/jbmr.080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korostishevsky M, Vistoropsky Y, Malkin I, Kobyliansky E, Livshits G. Anthropometric and bone-related biochemical factors are associated with different haplotypes of ANKH locus. Ann Hum Biol. 2008;35:535–46. doi: 10.1080/03014460802304588. [DOI] [PubMed] [Google Scholar]
- 36.Peacock M, Koller DL, Hui S, Johnston CC, Foroud T, Econs MJ. Peak bone mineral density at the hip is linked to chromosomes 14q and 15q. Osteoporos Int. 2004;15:489–96. doi: 10.1007/s00198-003-1560-7. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Liu YJ, Xiao P, Shen H, Deng HY, Papasian CJ, Drees BM, Hamilton JJ, Recker RR, Deng HW. Chromosome 2q32 may harbor a QTL affecting BMD variation at different skeletal sites. J Bone Miner Res. 2007;22:1672–8. doi: 10.1359/jbmr.070722. [DOI] [PubMed] [Google Scholar]
- 38.Xiao P, Liu PY, Lu Y, Guo YF, Xiong DH, Li LH, Recker RR, Deng HW. Association tests of interleukin-6 (IL-6) and type II tumor necrosis factor receptor (TNFR2) genes with bone mineral density in Caucasians using a re-sampling approach. Hum Genet. 2005;117:340–8. doi: 10.1007/s00439-005-1327-7. [DOI] [PubMed] [Google Scholar]
- 39.Xiong DH, Shen H, Xiao P, Guo YF, Long JR, Zhao LJ, Liu YZ, Deng HY, Li JL, Recker RR, Deng HW. Genome-wide scan identified QTLs underlying femoral neck cross-sectional geometry that are novel studied risk factors of osteoporosis. J Bone Miner Res. 2006;21:424–37. doi: 10.1359/JBMR.051202. [DOI] [PubMed] [Google Scholar]
- 40.Allison DB, Thiel B, St Jean P, Elston RC, Infante MC, Schork NJ. Multiple phenotype modeling in gene-mapping studies of quantitative traits: power advantages. Am J Hum Genet. 1998;63:1190–201. doi: 10.1086/302038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–7. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 42.Leppanen OV, Sievanen H, Jarvinen TL. Biomechanical testing in experimental bone interventions--May the power be with you. J Biomech. 2008;41:1623–31. doi: 10.1016/j.jbiomech.2008.03.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.