Abstract
MicroRNA (miRNA) expression was assessed in human cerebral cortical gray matter (GM) and white matter (WM) in order to provide the first insights into the difference between GM and WM miRNA repertoires across a range of Alzheimer's disease (AD) pathology. RNA was isolated separately from GM and WM portions of superior and middle temporal cerebral cortex (N = 10 elderly females, postmortem interval < 4 h). miRNA profiling experiments were performed using state-of-the-art Exiqon© LNA-microarrays. A subset of miRNAs that appeared to be strongly expressed according to the microarrays did not appear to be conventional miRNAs according to Northern blot analyses. Some well-characterized miRNAs were substantially enriched in WM as expected. However, most of the miRNA expression variability that correlated with the presence of early AD-related pathology was seen in GM. We confirm that downregulation of a set of miRNAs in GM (including several miR-15/107 genes and miR-29 paralogs) correlated strongly with the density of diffuse amyloid plaques detected in adjacent tissue. A few miRNAs were differentially expressed in WM, including miR-212 that is downregulated in AD and miR-424 which is upregulated in AD. The expression of certain miRNAs correlates with other miRNAs across different cases, and particular subsets of miRNAs are coordinately expressed in relation to AD-related pathology. These data support the hypothesis that patterns of miRNA expression in cortical GM may contribute to AD pathogenetically, because the aggregate change in miRNA expression observed early in the disease would be predicted to cause profound changes in gene expression.
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs that participate in gene expression regulation, mostly at the level of altering mRNA translation. In mammalian brain, miRNAs have been implicated in many fundamental functions including neurodevelopment, plasticity, and apoptosis (see reviews [12, 33]). Dysregulation of miRNAs has previously been reported in multiple diseases, including Alzheimer's disease (AD) [6, 11, 16, 24, 27, 35].
Research on the role(s) of miRNAs in AD must include the evaluation of human material, because the disease is essentially human-specific. One of the key experimental paradigms for evaluating human brain miRNAs involves gene expression profiling. These experiments may entail high-throughput assessment of the levels of multiple miRNAs across different tissue samples. In human brain miRNA expression profiling, technical details are critically important. For example, it is important to know clinical details of the human subjects, tissue sampling methods, pathological evaluation of adjacent tissue, RNA isolation methods used, miRNA profiling platform, and downstream data analyses.
Another important technical factor in brain miRNA expression profiling relates to the heterogeneity of the human cerebral cortex [36]. The peripheral ∼4 mm thick cortical cell layer (“gray matter”) overlies a deeper zone (“white matter”) that is traversed by myelinated axons. In terms of cell composition, gray matter comprises mostly neurons, astrocytes, endothelial cells, microglia, and relatively few oligodendrocytes. By contrast, white matter has a greater concentration of oligodendrocytes with markedly lower densities of neurons or blood vessels. Tissue sampling protocols that do not segregate white matter from gray matter will have increased variability that reflects differently sampled cell populations in addition to variance that is due to intrinsic factors (for example, disease state). It is a practical fact that individual samples of “cerebral cortex” can have dramatically different proportions of gray matter, white matter, and meningeal tissues.
Neuropathological hallmark lesions of AD—neuritic amyloid plaques (NPs) and neurofibrillary tangles (NFTs) [1]—are predominantly found in gray matter. Our prior work in AD miRNA profiling assessed RNA from gray matter exclusively [35] although we found differences in miRNA repertoires in a focused sampling of gray and white matter [36]. It has been suggested that white matter perturbations, and changes in axon myelination, could be fundamentally important to AD pathogenesis [3]. There have also been a number of miRNAs recently shown to be enriched in white matter and/or oligodendrocytes [8, 15, 38], and there have been miRNAs newly annotated since our prior study. For these reasons, we assessed separately the miRNA repertoire in cerebral cortical gray and white matter across a spectrum of early AD pathology, using state-of-the-art locked nucleic acid (LNA) microarrays.
Materials and methods
RNA isolation from a human cerebral cortex
Cases were selected on the basis of representing the spectrum of early progression of AD pathology, and of their being available more recently than our prior study [36] on miRNAs in AD brain. Premortem clinical evaluations and pathological assessments were as described previously [21, 31, 36]. The inclusion criteria that were applied: postmortem interval (PMI) < 4 h; no argyrophilic grains; no cortical Lewy bodies (LB); no evidence of frontotemporal dementia; no cancer in the brain parenchyma; and no large infarctions in the brain, or microinfarcts found within 3 cm of the brain tissue samples. These neuropathological confounds were assessed using standard neuropathological procedures as described in detail elsewhere [21].
RNA was isolated as previously described in detail [35, 36]. RNA was extracted from snap-frozen brain tissue in the superior and middle temporal gyri (SMT; Brodmann Areas 21/22) from the UK ADC under a University of Kentucky IRB protocol. All biochemical analyses were performed blind with respect to patient information. Briefly, prior to RNA extraction, gray matter was dissected away from white matter and meninges and large blood vessels were removed. Tissue (1–3 g) that had been snap-frozen in liquid nitrogen and then transferred to a −80°C freezer was thawed in isotonic lysis buffer with RNAsin (Promega, Madison, WI; 250 U/ml) and complete protease inhibitor pills (Roche, Basel, Switzerland). Trizol LS (Invitrogen, Carlsbad, CA) was used according to the manufacturer's instructions, except for an additional overnight −20°C precipitation step during isopropanol precipitation. RNA quality was confirmed using A260/A280 readings and also the Agilent Bioanalyzer for RNA Integrity Number (RIN).
Most aspects of pathological assessments were as described in detail previously [17, 21]. Briefly, sections were obtained from SMT (immediately adjacent to the tissues sampled for RNA studies), immersion-fixed in formalin and evaluated for AD-type pathology using conventional methods [18]. Amyloid plaques were separated into DPs (plaques without neurites) and NPs (plaques with degenerating neurites) in each region as described previously [21]. An arithmetic mean was calculated from the count of the five most involved fields for DPs (number of DPs per 2.35 mm2), NPs (number of NPs per 2.35 mm2), and NFTs (number of NFTs per 0.586 mm2) for each region.
Patient characteristics, along with pathological lesion counts in the same cases, are shown in Table 1. Cases 3, 4, 5, 6, 8, and 9 were evaluated separately in a different study that involved RT-qPCR [23] but none of these cases has ever been evaluated previously using a microarray or any other high-throughput profiling method.
Table 1. Cases included in current study.
| Case | Age | Final MMSE score | ApoE alleles | PMI (hours) | Braak NFTs stage | CERAD plaque stage | Diffuse plaques | Neuritic plaques | NFTs | RIN-GM | RIN-WM |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 85 | 28 | 3/3 | 2.50 | 0 | No | 0 | 0 | 0 | 5.7 | 3.4 |
| 2 | 79 | 29 | 3/3 | 1.75 | I | No | 0 | 0 | 0 | 6.0 | 6.1 |
| 3 | 84 | 29 | 3/3 | 2.50 | I | No | 0 | 0 | 0 | 6.6 | 6.6 |
| 4 | 92 | 29 | 3/3 | 3.00 | II | No | 0 | 0 | 0 | 7.3 | 6.9 |
| 5 | 86 | 26 | – | 3.25 | III | B | 5 | 5.8 | 0 | 7.2 | 7.9 |
| 6 | 92 | 30 | 2/3 | 3.25 | III | B | 36.6 | 8.2 | 1.2 | 7.1 | 6.6 |
| 7-D | 84 | 13.5 | 3/3 | 2.60 | IV | C | 50 | 12.8 | 0 | 2.3 | 2.3 |
| 8 | 88 | 28 | 3/3 | 3.00 | III | C | 41 | 16.4 | 0 | 5.1 | 5.5 |
| 9 | 99 | 16 | 3/4 | 2.10 | V | C | 50 | 20 | 23.6 | 5.9 | 6.2 |
| 10 | 91 | 13 | 3/3 | 3.00 | V | C | 46.2 | 35 | 42.6 | 5.5 | 6.1 |
List of cases with data about clinical, neuropathological, and RNA quality parameters. The focus of this study comprises the spectrum of disease from “no pathology” to “early AD” pathology. All ten individuals were Caucasian females. Final MMSE examinations were administered an average 10.9 months before death (median 7.2 months before death). Diffuse plaques, neuritic plaques, and neurofibrillary tangles (NFTs) were counted on adjacent tissue sections of superior and mid-temporal cortical tissue as described in “Methods”. Cases are arranged according to density of neuritic plaques in these tissues. Note that case #7-D has low RNA integrity numbers (RIN), which indicates substantial RNA degradation. White matter of case 1 (italic values) has marginal RNA quality. “Plaque stage” refers to CERAD scores, with B indicating moderate density of neuritic plaques and C indicating high density of neuritic plaques
Microarray methods
The quality of the total RNA was verified by an Agilent 2100 Bioanalyzer profile. 1 μg total RNA from sample and reference was labeled with Hy3™ and Hy5™ fluorescent label, respectively, using the miRCURY™ LNA Array power labeling kit (Exiqon, Denmark) following the procedure described by the manufacturer. The Hy3™-labeled samples and a Hy5™-labeled reference RNA samples (hereafter referred as Cy3 and Cy5) were mixed pair-wise and hybridized to the miRCURY™ LNA array version 11.0 (Exiqon, Denmark), which contains capture probes targeting all miRNAs for human, mouse or rat registered in the miRBASE version 14.0 at the Sanger Institute. The hybridization was performed according to the miRCURY™ LNA array manual using a Tecan HS4800 hybridization station. After hybridization the microarray slides were scanned and stored in an ozone-free environment (ozone level below 2.0 ppb) to prevent potential bleaching of the fluorescent dyes. The miRCURY™ LNA array microarray slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies, Inc., USA) and the image analysis was carried out using the ImaGene 8.0 software (BioDiscovery, Inc., USA). The quantified signals were background corrected (Normexp with offset value 10 as described previously by Ritchie et al. [30]) and normalized using the global Lowess (LOcally WEighted Scatterplot Smoothing) regression algorithm.
Northern blots on selected miRNAs
Northern blots were performed as described previously [35].
Data analyses
Data were obtained and normalized as above and exported to a Microsoft Excel™ spreadsheet that served as the basis for additional analyses. The first level of analyses was performed on the “single-color” raw data obtained from each data point that expressed the absolute expression values for each miRNA. This was used to obtain hierarchical clustering based on miRNA expression. Data were converted to a .txt file and imported to Partek Genomics Suite™ software. Hierarchical clustering was performed using both Euclidean and Pearson coefficient-based algorithms to demonstrate how cases clustered based on miRNA expression. Next, we focused on the 170 miRNAs that had positive signals in all samples and we used data from the “expression matrix” that show miRNA expression in relation to the relative ratio of hybridization signal for the particular miRNA being assessed (Cy3 on the microarray) versus a sample comprising pooled RNA from all samples (Cy5 on the microarray). Complete data from all the microarrays are provided in Supplemental Table 1. In the expression matrix, all capture probes with both Hy3 and Hy5 signals lower than 1.5× of the median signal intensity of the given slide were excluded. With these data we determined whether miRNAs were enriched in gray matter versus white matter using paired Student's t tests. We also tested correlations (including R values) between the miRNA expression levels and the densities of counted AD-type neuropathological lesions (DPs, NPs, and NFTs), using statistical functions of Microsoft Excel™. Next we performed linear regression-based tests of specific groups of miRNAs in comparison to the counted AD-type pathological lesions; here, P values of the regression were determined unlike the other tests that we just reported the correlation coefficient. Also, using the Partek Genomics Suite™, we obtained a similarity matrix (using the Pearson coefficient algorithm) and then hierarchically clustered this matrix to visualize the patterns of similarity of the miRNA expression patterns across the different cases. This clustered similarity matrix was the basis for separating the different miRNAs into groups related to their expression patterns.
Results
Cases evaluated, parameters of interest, and RNA quality
Data about individuals and various relevant clinical and pathological parameters are shown in Table 1. Cases for this study were chosen to represent no-pathology controls and the spectrum of early changes in AD. None of the cases had “end-stage” disease clinically (final MMSE scores all were ≥13 and most were considerably higher), or pathologically (there was no Braak stage VI case included). The histopathology (Fig. 1) was read in a quantitative method as routinely performed at the University of Kentucky Alzheimer's Disease Center [18, 21]. This enabled us to correlate in a quantitative manner the AD-type pathologies (DPs, NPs, and NFTs) with the miRNA profiling obtained from immediately adjacent tissue samples. Note that RNA quality was poor for one sample (case 7-degraded) and marginal for another (case 1-WM). We present the data from this sample in the Supplemental Table 1, although we did not include this sample in downstream analyses as described below. The rationale for not completely eliding these data is that we wanted to reveal all the data we obtained, and the effect of RNA degradation on miRNA repertoire will be of interest to some researchers.
Fig. 1.
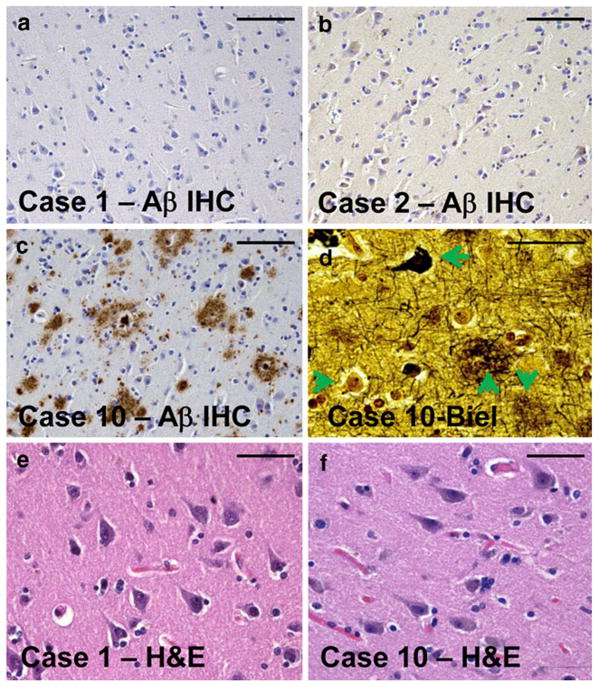
Representative photomicrographs from the superior and middle temporal cortical gyri of individuals sampled for the current study. Tissue sections for histopathology (Table 1) were immediately adjacent to the tissue samples used for RNA isolation. Amyloid plaques are stained using Aβ immunohistochemistry (IHC) as shown in a–c. Note that in cases 1 and 2 (a, b), there is no observable Aβ plaques. By contrast, case 10 (c, d, f) had the highest densities of Alzheimer's disease (AD)-related pathology. Bielschowsky (Biel) silver stain (d) confirms the presence in case 10 of diffuse amyloid plaques (downward arrow), neuritic amyloid plaques (upward arrow), neurofibrillary tangles (leftward arrow), and also viable-appearing neurons (rightward arrow). Hematoxylin and eosin (H&E) stains of both cases 1 and 10 demonstrate viable-appearing pyramidal neurons in the temporal cortex. Even in the most severely affected AD brain evaluated in the present study, the disease had not progressed to a stage of widespread neuronal cell death. Scale bars 100 μm (a–c) and 50 μm (d–f)
Initial assessment of miRNA microarray data and Northern blots
Raw data from all the miRNA microarrays are presented in Supplemental Table 1. In order to assess the quality of the data, we performed two separate profiling microarrays on the same sample, months apart. Case 1 was chosen arbitrarily. These results were very encouraging, as shown in Supplemental Fig. 1: R2 correlation coefficient for all the miRNAs on the microarray was 0.978.
Although the results of the microarray appeared robust in a sample-to-sample comparison, we were concerned because some of the miRNAs that were more recently annotated had unexpectedly high apparent expression signals. However, we and others had previously shown that some of these miRNAs are probably not actually miRNAs by a conventional definition because their northern blot signatures fail to show the conventional “mature” ∼22 nts form and ∼70 nts pre-miRNA [25, 34]. For this reason, we selected ten different miRNAs for northern blotting that had high evident expression on the microarrays from all the sampled microarrays. These miRNAs, along with the sequences of the probes used for northern blotting, are shown in Fig. 2. Most of these RNA species were either not detected (5/10) or had quite non-conventional northern blotting band patterns (5/10). Full northern blot results are shown in Supplemental Fig. 2.
Fig. 2.
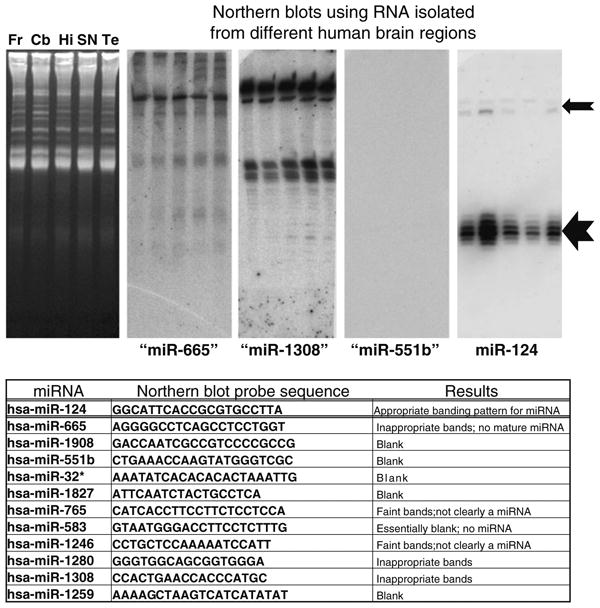
Northern blots were performed on ten different miRNAs with relatively high expression according to the microarrays used in the current studies despite lack of previous evidence of brain expression. 20 μg of RNA was isolated from human frontal cortex (Fr), cerebellum (Cb), hippocampus CA1 (Hi), substantia nigra (SN), and superior and middle temporal gyri (Te), and these samples were run on 15% urea-polyacrylamide gel electrophoresis. Representative ethidium-bromide stained gel is shown in left. Results are shown for miR-665, miR-1308, mir-551b, and miR-124 with the latter being the only one that has been firmly documented in human brain. Note that the northern blotting staining pattern for miR-124 is exactly as expected with a ∼22 nts band (large arrow) and a slower-migrating ∼70 nts band (smaller arrow). However, none of the other putative miRNAs stained as expected for a conventional miRNA (see table). These results, along with prior studies, confirm that some annotated miRNAs are not necessarily functioning as conventional miRNAs in human brain
Microarray data analyses
Since all ten candidates evaluated by northern blots did not appear to be conventional miRNAs, we presumed that the microarray data referent to more recently annotated miRNAs were problematic. For the sake of further clustering analyses (Figs. 3, 4, 5), we decided to rely on the best-characterized miRNAs, which we had to delineate with arbitrary criteria unless we performed northern blots on multiple samples for each candidate miRNA—a practical impossibility. Our criteria for inclusion in downstream analyses and clustering were that we only assessed the miRNAs that were expressed in all samples, and which had an annotated number <miR-600. These criteria were adopted with the caveat that neither all false negatives (true miRNAs excluded) nor all false positives (non-miRNAs included) were completely eliminated. Although downstream data incorporated these criteria to narrow further analyses, we underscore that the primary data from all the detected miRNAs are available in Supplemental Table 1.
Fig. 3.
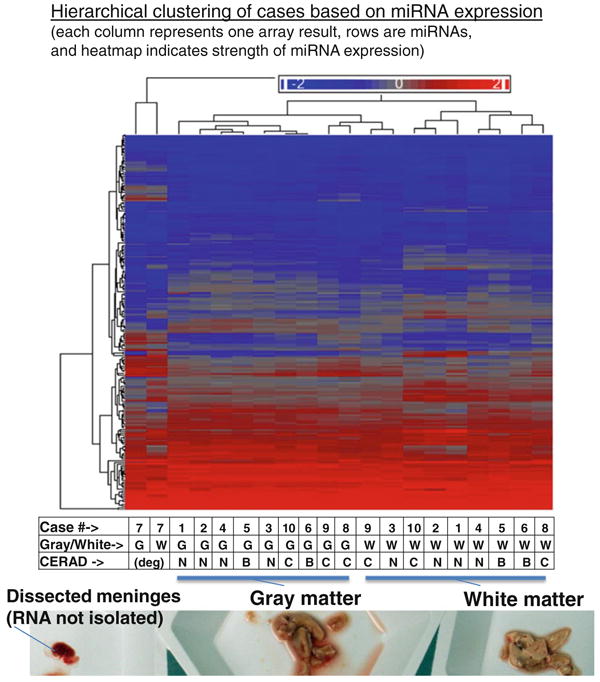
Hierarchial clustering of microarray results from gray matter and white matter of all ten brain samples evaluated in the current study. A representative photograph of gray and white matter tissue that had been freshly dissected for this study, prior to RNA isolation, is shown at the bottom of the figure. Each column of the heatmap figure represents a particular microarray sample. Each row is a miRNA (total N = 170 presumed conventional miRNAs). Heatmap coloring is based on miRNA expression levels. Both rows and columns are hierarchically clustered according to the Euclidean (default) method. The table at the bottom indicates which case the samples were derived from, whether the sample was gray or white matter, and the CERAD scores that indicate the densities of Alzheimer's disease-type neuritic amyloid plaques (B indicating moderate density of neuritic plaques and C indicating high density of neuritic plaques). Both samples from case 7, with degraded RNA, clustered separately from all the others. Note that among the non-degraded samples, gray matter-derived samples cluster separately from all the white matter samples. Further, there is a tendency of cases to cluster together that have similar CERAD scores
Fig. 4.
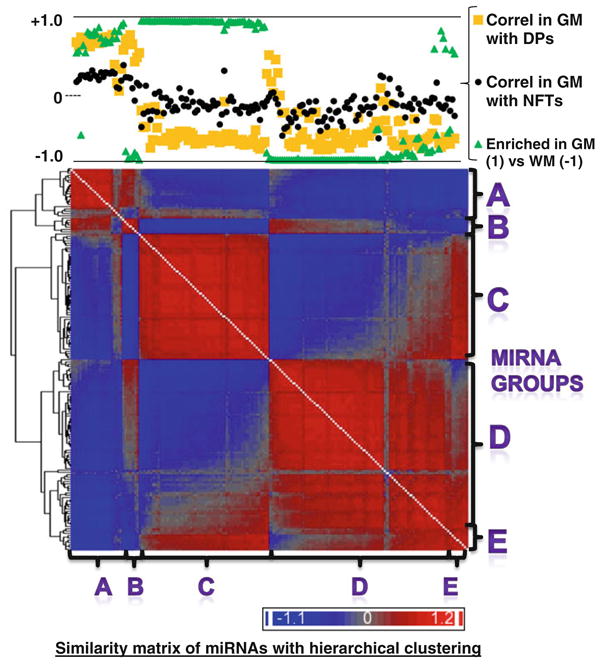
Hierarchial clustering of gene expression similarity matrix that allowed us to visualize the similarity of the patterns of miRNA expression across different cases including both gray matter and white matter samples. In this heatmap, all the rows and columns are miRNAs [each miRNA (N = 170, corresponding to the presumed conventional miRNAs shown in Supplemental Table 1) occupies one column and one row] and the heatmap represents the degree of similarity of expression for the various different miRNAs. This diagram allows us to visualize the phenomenon that groups of miRNAs appear to have similar expression patterns that contrast sharply with other groups of miRNAs. The chart at the top of the figure shows how these same miRNAs are correlated with the densities of Alzheimer's disease (AD)-type pathology [diffuse plaques (DPs) and neurofibrillary tangles (NFTs)] with correlation being indicated by the R value of correlation across cases 1–10. The degree of enrichment in gray matter (GM) or white matter (WM) is also charted. All these values are available in Supplemental Table 1. Based solely on visual inspection of this diagram, we segregated the miRNAs into five different groups (A–E) and sought to test the hypothesis that miRNAs in the different groups may have systematically different tendencies to be enriched in gray or white matter, and/or different correlative relationship with the density of Alzheimer's disease-type pathological lesions
Fig. 5.
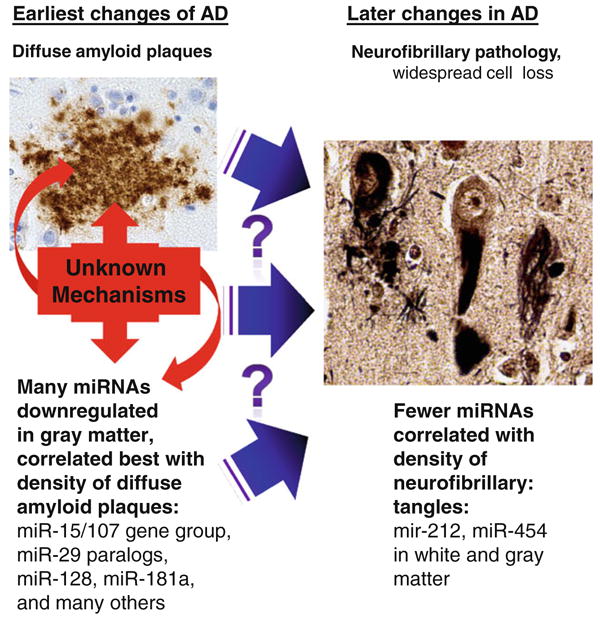
Results indicate a shift in gray matter miRNA expression early in the pathological progression of Alzheimer's disease (AD). It is not known what causes some individuals but not others to have extensive diffuse amyloid deposition, or the pathogenetic that cause a subset of those persons to progress to the later stages of AD that correlate with the presence of neurofibrillary pathology. However, the current study indicates that many miRNA changes occur, predominantly in gray matter, and in relatively strong correlation with the early changes and thus may also contribute to the additional downstream pathology. Only a handful of miRNAs, such as miR-212 and miR-454, appear to change in correlation to the presence of neurofibrillary pathology, and these changes are seen in both white matter and gray matter. Photomicrograph of diffuse plaques shows Ab immunohistochemistry; neurofibrillary pathology is stained using the Gallyas silver impregnation method
Using the above criteria for inclusion of miRNAs, hierarchical clustering was performed to test how the different cases corresponded in terms of the miRNA expression patterns (Fig. 3). The result of Euclidean-based hierarchical clustering is shown, but the Pearson coefficient-based analyses (more based on the correlation of the patterns rather than the strength of expression) showed essentially the same pattern of clustering. Note that the expression of miRNAs from case 7 (with degraded RNA) causes these samples to cluster separately from the others on the heatmap. This confirms prior scholarship that miRNA degeneration is an important consideration in miRNA profiling [32], and this is an important caveat in our study. Aside from those samples, there was an apparent clustering between samples derived from gray matter relative to those derived from white matter, and also cases with more AD pathology tended to cluster together relative to no-pathology cases.
Since the clustering data indicated that we could discriminate between the miRNA repertoire of gray matter and white matter-derived RNA samples, we sought to determine which miRNAs were most enriched in those compartments. Paired Student's t tests were run to evaluate the miRNAs with expression most different between gray matter and white matter. These results are shown in Table 2. Note that some of the miRNAs we find enriched in white matter are known to be highly expressed in oligodendrocytes [8, 15, 38], which is a preliminary indication of biological relevance of our results.
Table 2. Gray matter- and white matter-enriched miRNAs: the top 15.
| miRNAs enriched in white matter | Microarray signal (Log 2) | P value | miRNAs enriched in gray matter | Microarray signal (Log 2) | P value | |
|---|---|---|---|---|---|---|
| 1 | hsa-miR-338-3p | 11.57 | <0.0001 | hsa-miR-485-3p | 8.90 | <0.0001 |
| 2 | hsa-miR-219-2-3p | 10.09 | <0.0001 | hsa-miR-129-5p | 9.90 | <0.0001 |
| 3 | hsa-miR-20a | 9.76 | <0.0001 | hsa-miR-143 | 11.81 | 0.0004 |
| 4 | hsa-miR-17 | 10.04 | <0.0001 | hsa-miR-34a | 10.51 | 0.0004 |
| 5 | hsa-miR-106a | 9.56 | <0.0001 | hsa-miR-124 | 15.41 | 0.0007 |
| 6 | hsa-miR-19a | 10.73 | <0.0001 | hsa-miR-149 | 9.77 | 0.0007 |
| 7 | hsa-miR-584 | 8.60 | <0.0001 | hsa-miR-136 | 8.96 | 0.0009 |
| 8 | hsa-miR-338-5p | 9.74 | <0.0001 | hsa-miR-138 | 12.43 | 0.0009 |
| 9 | hsa-miR-219-5p | 12.61 | <0.0001 | hsa-miR-145 | 8.94 | 0.0011 |
| 10 | hsa-miR-32 | 8.87 | <0.0001 | hsa-miR-129-3p | 11.38 | 0.0013 |
| 11 | hsa-miR-34c-5p | 7.79 | <0.0001 | hsa-miR-381 | 8.81 | 0.0013 |
| 12 | hsa-miR-16 | 12.61 | <0.0001 | hsa-miR-128 | 11.85 | 0.0016 |
| 13 | hsa-miR-151-5p | 10.76 | <0.0001 | hsa-miR-432 | 8.08 | 0.0016 |
| 14 | hsa-miR-181a | 11.83 | <0.0001 | hsa-miR-378 | 7.91 | 0.0017 |
| 15 | hsa-miR-181b | 9.26 | <0.0001 | hsa-miR-29b | 13.17 | 0.0017 |
Individual miRNAs that were most enriched in white matter (left) or gray matter (right). Enrichment was determined using paired Student's t tests comparing the samples across cases between gray and white matter. The top miRNAs listed had the lowest P values. Note that some of the miRNAs we find to be enriched in white matter (for example, miR-219 and miR-338) have been shown to be highly enriched in oligodendrocytes. Microarray log signal is shown (referent to white matter samples on the left, gray matter samples on the right) to convey the expression levels for these miRNAs
We next sought to correlate the miRNA expression data seen across the different cases with the severity of AD-type pathology that was counted in immediately adjacent tissue sections (Table 1; Fig. 1). We queried the data for gray matter samples separately from white matter samples, and did separate analyses for all three type of AD pathology: DPs, NPs, and NFTs. Simple correlation coefficients were obtained comparing the miRNA expression across the cases, relative to the pathology counts. Table 3 shows the results for the top 5 miRNAs in each group. Note that some of the miRNAs have a positive correlation with pathology counts, and some negative.
Table 3. Top miRNAs with expression correlated to Alzheimer's-type pathology.
| Correlation with AD pathology | AD pathology subtype | White matter or gray matter | Five miRNAs correlated best (according to linear regression coefficient) versus amount of AD pathology: diffuse plaques (DPs), neuritic plaques (NPs), and neurofibrillary tangles (NFTs) | ||||
|---|---|---|---|---|---|---|---|
| Negative | DPs | Gray | miR-185 | miR-27b | miR-151-5p | miR-382 | miR-361-3p |
| NPs | miR-212 | miR-27b | miR-382 | miR-15a | miR-151-5p | ||
| NFTs | miR-212 | miR-338-3p | miR-194 | miR-219-2-3p | miR-338-5p | ||
| Positive | DPs | miR-214 | miR-576-5p | miR-583 | miR-298 | miR-516b | |
| NPs | miR-214 | miR-583 | miR-184 | miR-300 | miR-516a-5p | ||
| NFTs | let-7e | miR-583 | miR-125b | miR-214 | miR-184 | ||
| Negative | DPs | White | miR-425 | miR-330-5p | miR-326 | miR-98 | miR-301a |
| NPs | miR-212 | miR-194 | miR-98 | miR-425 | miR-330-5p | ||
| NFTs | miR-212 | miR-194 | miR-551b | let-7f | miR-98 | ||
| Positive | DPs | miR-214 | miR-298 | miR-424 | miR-525-5p | miR-184 | |
| NPs | miR-424 | miR-525-5p | miR-214 | miR-509-5p | miR-142-5p | ||
| NFTs | miR-424 | miR-142-5p | miR-142-3p | miR-223 | miR-145 | ||
The top five miRNAs whose expression across different cases most closely correlated with the density of Alzheimer's disease (AD)-type pathological lesions. Results are stratified by whether the correlation was positive or negative, the subtype of AD-type lesion (diffuse plaques DPs, neuritic plaques NP, and neurofibrillary tangles NFTs), and whether the samples being compared derived from gray matter or white matter. The individual miRNAs were ordered according to the correlation coefficient that resulted from comparison between the miRNA expression in different cases versus the density of the AD-type lesions. The miRNA with the highest expression is shown on the left. Complete data with all the correlation coefficients for the various miRNAs are provided in Supplemental Table 1
Prior studies have implicated a handful of miRNAs as being downregulated in AD and which may regulate AD-related genes, including miR-15, miR-107, miR-29, and miR-128 [6, 11, 16, 24, 28, 35]. MiR-15 and miR-107 belong to a collection of genes referred to as the miR-15/107 group because of sequence similarity and overlap of function [9]. The miR-15/107 group in humans comprises miR-15a, miR-15b, miR-16, miR-103, miR-107, miR-195, miR-424, miR-497, and miR-503. Those expressed in our microarray are shown in Table 4. Note that indeed multiple members of the miR-15/107 group systematically tend to change in expression in correlation with the presence of AD-type pathology—mostly in correlation with the numbers of counted DPs. As noted above, miR-424 is increased in white matter in AD apparently, in contrast to other members of the miR-15/107 group which are downregulated in gray matter.
Table 4. Correlation between miRNA expression and Alzheimer's-related changes (P values for linear regression test) for a selected subset of miRNAs.
| MiRNA | miR-15/107 group | Enriched in GM or WM | Gray matter by neuropathology | White matter by neuropathology | ||||
|---|---|---|---|---|---|---|---|---|
| DPs | NPs | NFTs | DPs | NPs | NFTs | |||
| miR-124 | N | GM | 0.152 | 0.463 | 0.864 | 0.871 | 0.908 | 0.863 |
| miR-125b | N | Neither | 0.678 | 0.832 | 0.460 | 0.876 | 0.780 | 0.794 |
| miR-103 | Y | GM | 0.014 | 0.082 | 0.399 | 0.214 | 0.344 | 0.627 |
| miR-107 | Y | GM | 0.034 | 0.114 | 0.465 | 0.374 | 0.419 | 0.631 |
| miR-15a | Y | WM | 0.005 | 0.041 | 0.269 | 0.240 | 0.618 | 0.932 |
| miR-15b | Y | WM | 0.081 | 0.184 | 0.526 | 0.213 | 0.509 | 0.869 |
| miR-16 | Y | WM | 0.036 | 0.045 | 0.227 | 0.112 | 0.296 | 0.476 |
| miR-195 | Y | WM | 0.033 | 0.070 | 0.361 | 0.824 | 0.820 | 0.733 |
| miR-424 | Y | Neither | 0.416 | 0.405 | 0.717 | 0.062 | 0.018 | 0.018 |
| miR-128 | N | GM | 0.042 | 0.229 | 0.782 | 0.775 | 0.735 | 0.845 |
| miR-29a | N | GM | 0.031 | 0.075 | 0.441 | 0.915 | 0.808 | 0.934 |
| miR-29b | N | GM | 0.058 | 0.182 | 0.702 | 0.770 | 0.882 | 0.898 |
| miR-29c | N | GM | 0.033 | 0.130 | 0.551 | 0.566 | 0.897 | 0.636 |
| miR-214 | N | Neither | 0.009 | 0.116 | 0.486 | 0.039 | 0.293 | 0.660 |
Testing specific miRNAs for correlation with the density of AD-type lesions. These miRNAs were chosen because they had been previously described to have altered expression in association with AD. Prior studies did not test white matter separately. Here, we used the P value for the linear regression to test for statistical significance (P < 0.05, bold values). Negative correlation between miRNA variability and AD pathology are in roman, positive correlation in italics. Note that most members of the miR-15/107 gene group (miR-15a, miR-15b, miR-16, miR-195, miR-424, miR-103, and miR-107) show a negative correlation to AD-type amyloid plaques, and only in gray matter, with the exception of miR-424, which shows a positive correlation to AD-type pathology that appears most specific to white matter samples. Complete gene expression data for the various miRNAs are provided in Supplemental Table 1
Since our data indicated that multiple different miRNAs were changed in correlation with the severity of AD pathology, we sought to better understand whether there were group patterns in miRNA expression in our dataset. To study this, a similarity matrix was created using the Partek Genomics Suite™ software, comparing all 170 conventional miRNAs that were included in our sample as described above. Figure 4 shows the hierarchical clustering of the similarity matrix, assessing the conventional miRNAs in the non-degraded samples including both gray matter and white matter. In this heatmap diagram, all the rows are miRNAs and all the columns miRNAs, and the heatmap intensity indicates the degree of similarity with blue being positively correlated and red being more negatively correlated. Note that this hierarchical clustering reveals groups of miRNAs with similar expression patterns across the different samples, and which tend to correlate negatively with other groups of miRNAs.
At the top of Fig. 4 is a graph that shows the results of analyses of three different parameters for each of the miRNAs in the columns below: correlation for each miRNA's expression with the number of counted DPs across the different cases; correlation for that miRNA's expression with the number of NFTs across the different cases; and the degree to which that miRNA was enriched in gray matter versus white matter. The simple correlation coefficients were calculated comparing the expression matrix values for each miRNA for each case versus the number of pathological lesions in the same cases. The enrichment in gray matter versus white matter shows the number of the P value of paired Student's t tests comparing all the cases expression values in gray matter versus white matter, with positive values being enriched in gray matter and negative in white matter.
Observation of Fig. 4 provided the basis of separating the 170 highly expressed miRNAs into groups based on similarity of expression, relative enrichment in gray matter or white matter, and down- or upregulation in correlation with AD pathology. Based on this pattern in the hierarchical clustering, we designated five different groups of miRNAs (A–E based solely on their positions in this heatmap). The characteristics of the groups, and the miRNAs that comprise each group, are provided in Table 5. The miRNAs in Table 5 are listed in the order in which they appeared in the similarity matrix. The most closely similarly expressed miRNAs across all the cases are listed most closely together in the table; for example, miR-219-5p, miR-338-3p, miR-219-2-3p, miR-338-5p had very similar expressions across all the tissue sampled.
Table 5. miRNAs in Alzheimer's disease.
| miRNA group | Gray matter or white matter enriched | Upregulated or downregulated in Alzheimer's disease | miRNAs in group |
|---|---|---|---|
| A | Gray matter | Upregulated | miR-519e, miR-574-5p, miR-498, miR-518a-5p/miR-527, miR-525-5p, miR-300, miR-576-3p, miR-583, miR-146b-3p, miR-490-3p, miR-549, miR-516a-5p, miR-510, miR-184, miR-516b, miR-298, miR-214, miR-198, miR-451, miR-144, miR-424, let-7e |
| B | White matter | Upregulated | miR-509-5p, miR-574-3p, miR-576-5p, miR-302e, miR-220b, miR-208a, miR-215 |
| C | Gray matter | Downregulated | miR-485-3p, miR-381, miR-124, miR-34a, miR-129-5p, miR-29a, miR-143, miR-136, miR-145, miR-138, miR-129-3p, miR-128, miR-379, miR-299-5p, miR-218, miR-149, miR-135a, miR-7, miR-126, miR-411, miR-335, miR-9, miR-378, miR-488, miR-432, miR-127-5p, miR-127-3p, miR-491-5p, miR-376c, miR-377, miR-95, miR-222, miR-29b, miR-329, miR-495, miR-551b, miR-195, miR-125b, miR-30b, miR-221, miR-139-5p, miR-487a, miR-487b, miR-107, miR-146b-5p, miR-29c, miR-30a, miR-582-5p, miR-103, miR-342-3p, miR-331-3p, miR-30c, miR-30d, miR-382, miR-22, miR-125a-5p |
| D | White matter | Downregulated | miR-491-3p, miR-423-5p, miR-34b, miR-422a, miR-34c-5p, miR-584, miR-219-5p, miR-338-3p, miR-219-2-3p, miR-338-5p, miR-181a, miR-181b, let-7b, miR-151-3p, miR-197, miR-19a, miR-20a, miR-17, miR-106a, miR-32, miR-340, miR-19b, miR-21, miR-151-5p, miR-194, let-7c, miR-330-3p, miR-27b, miR-93, miR-15a, miR-339-5p, miR-193b, miR-106b, miR-16, miR-23b, miR-15b, miR-320d, miR-320b, miR-320c, miR-320a, miR-557, miR-33a, let-7a, miR-374b, miR-140-3p, miR-374a, miR-24, miR-140-5p, miR-26a, miR-513a-5p, miR-212, miR-142-5p, miR-142-3p, miR-26b, miR-520d-5p, miR-193a-3p, miR-92b, miR-330-5p, miR-186, let-7f, miR-223, miR-412, miR-185, miR-148b, miR-101, miR-99b, miR-27a, miR-589, let-7i, miR-361-3p, miR-361-5p, miR-423-3p, miR-190, miR-301a, miR-365, miR-23a, miR-363, miR-326 |
| E | Gray matter | Downregulated | miR-425, miR-191, miR-519d, let-7g, miR-98, miR-99a, miR-30e |
Five groups of miRNAs (A–E) that were shown graphically on Fig. 4. miRNAs were grouped by the similarity of gene expression as detected across all nondegraded samples including both gray and white matter. As might be expected, some of the variability corresponded to whether the tissue was sampled from gray matter or white matter-Group D was almost exclusively white matter-enriched miRNAs, whereas Group C was mostly gray matter-enriched. MiRNAs that were positively correlated with the density of diffuse amyloid plaques (DPs) were in group A and B. By contrast, the negatively correlated miRNAs were in groups C–E. The complete gene expression data is provided in Supplemental Table 1
Discussion
Thoroughly characterized human brain samples and a state-of-the-art miRNA profiling platform were used to assess the absolute and relative expression levels of different miRNAs across a range of early AD pathology. These data provide the first systematic evidence of differential white matter and gray matter changes in miRNAs in human brain, with and without brain pathology. The number of miRNAs altered in AD was higher in gray than white matter, but a handful of miRNAs were altered specifically in white matter. These data indicate that while the expressions of individual miRNAs may be impactful alone, we will not understand the aggregate impact of miRNAs until we better understand the patterns of miRNA gene expression in human brain.
There are some limitations to our approach. For example, we dissected gray matter from white matter tissue, which took a few minutes during which some RNA degradation could occur. Sethi et al. [32] found that miRNAs degrade differentially postmortem and indeed we find that our most degraded samples (case 7) had different miRNA expression. This is an important caveat but we feel that the benefits of having gray matter separated from white matter, which we have pointed out before [36], far outweighs the cost in terms of RNA degradation.
We also note that our sample size is quite small (N = 10 cases with one gray matter and one white matter sample from each) and we only evaluated a duplicate sample from a single case (Supplemental Fig. 1). However, that duplicate revealed an almost perfect positive correlation (R2 ∼ 0.98), and the 20 overall samples (including gray matter and white matter) make this one of the largest high-throughput brain profiling experiments recently reported in the literature. Furthermore, where we could correlate our results with prior studies, our data are very compatible with previous results. For example, miRNAs that we find enriched in white matter are also much enriched in oligodendrocytes [8, 13, 38], as one would expect. Further, the miRNAs that have been reported to be aberrantly expressed in AD (miR-15, miR-103/7, and miR-29) [6, 11, 23, 27, 35] were indeed changed in the “appropriate” direction in these new data.
Although there were similarities with our prior study where we profiled miRNAs in the superior and middle temporal gyri, the prior study only found differential expression of one miRNA related to AD pathology—miR-107. Here, we found a much different result: multiple miRNAs changed expression in relation to the AD pathology. There were at least three differences in our methods that may help explain this difference. One difference in the current study relative to our prior microarray data [35] or RT-qPCR data [23] is that we only used samples from females. We hypothesized that this might decrease biological variation since males and females have some differences in neurodegenerative disease phenotypes [2, 4, 7, 22] and miRNA expression repertoire [37]. Further, microarray technology has improved in the past 4 years, including the addition of many newly annotated miRNAs. This increase in the number of annotated miRNAs comes at some expense because we found that all ten relatively newly annotated miRNAs that we evaluated using northern blots did not show the results expected for a conventional miRNA. These data indicate that the present annotation of miRNAs includes some sequences that are either not miRNAs or, at best, have molecular signatures on northern blots that are unlike conventional miRNAs. We previously had found that miR-720, which appears highly expressed on cDNA microarray, does not resemble other miRNAs according to a conventional northern blot on RNA isolated from human brain [36]. These data indicate that there is a need for more basic, “descriptive” work on human brain miRNAs. Finally, the normalization methods we used in the current study were quite distinct from our prior microarray analyses, being dependent on Cy3/Cy5 normalization relative to pooled RNA, rather than global normalization in the prior study [35].
One contribution of this study is to describe which miRNAs are enriched in gray matter versus white matter. The white matter-enriched miRNAs included, as expected, miRNAs known to be enriched in oligodendrocytes [8, 15, 38]. Since there are multiple cell populations enriched in gray matter (neurons and endothelial cells, for example), it remains to be seen exactly which of those cell populations express the specific miRNAs. It is important to complement tissue-level work with lower-throughput methods such as in situ hybridization [26].
These new data (in agreement with prior work) indicate that it is necessary to assess the aggregate impact of multiple miRNAs rather than focusing exclusively on a single gene. For example, miRNAs in the miR-15/107 gene group have overlapping lists of mRNA targets [9, 14]. It will not be adequate to merely explain the impact of (for example) miR-15a or miR-107 because we can now see that miR-15a, miR-15b, mR-16, miR-195, miR-103, and miR-107 are all downregulated in AD gray matter, whereas miR-424 from this gene group is actually upregulated in AD white matter! More work will be required in order to understand how these aggregate neurochemical changes impact the brain.
Although each miRNA had its own unique expression signature, biological variation in miRNA expression could be used to separate the miRNAs into five groups. These groups had systematic tendencies in terms of enrichment in gray or white matter, and whether they were up- or downregulated in relation to the severity of AD-type pathology. These data should be interpreted with caution because the group designations are somewhat tautological: much of the biological variation in our samples may relate to the presence or absence of AD pathology.
In the future we may learn how brain miRNAs are coordinately expressed and the molecular pathways involved in AD. A key feature of the “amyloid cascade hypothesis” [10] in AD is that various genetic and environmental cues can lead to the deposition of amyloid plaques, and these plaques initiate or potentiate downstream changes that culminate in neurofibrillary pathology (NPs and NFTs), cell death, synapse elimination and, ultimately cognitive impairment. Amidst all of that pathology it is extremely challenging to determine causality, and to separate impactful changes from epiphenomena. For example, it has been posited that both plaques and tangles may be at least partly adaptive responses [5], although these alternative hypotheses should themselves be evaluated with caution [20]. Also, the correlative impact of DPs on cognition is controversial—most but not all previous studies indicate that DPs are not directly correlated with cognitive impairment [19, 20, 29]. However, in the present study, we have at least provided new insights into the stage of AD progression where specific miRNAs' expression seems to change, and the compartment where that change may mostly occur. We found that relatively many different miRNAs change in the gray matter (including miR-15/107 genes, miR-29 paralogs, miR-181a, miR-128, and other) and the AD-related pathological subtype that most closely correlated with the miRNA changed expression was the DPs. These ideas are presented schematically in Fig. 5. In future studies we may be able to understand whether those miRNAs contributed to the development of plaques, or vice versa. In any case, since AD is a human-specific condition it is important to know about these human brain miRNA expression patterns, which may help inform other experimental models.
Supplementary Material
Acknowledgments
We are deeply grateful to all of the study participants at the University of Kentucky Alzheimer's disease Center. We thank Ela Patel, Erin Abner and Sonya Anderson for technical and data support and our clinical colleagues (Drs. Gregory Jicha, Gregory Cooper, and Frederick Schmitt) for their thorough evaluations. This study was supported by Grants R01 NS061933, R21AG036875, and P01 NS058484 from NIH.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00401-010-0756-0) contains supplementary material, which is available to authorized users.
References
- 1.Consensus recommendations for the postmortem diagnosis of Alzheimer's disease. The National Institute on Aging, and Reagan Institute Working Group on Diagnostic Criteria for the Neuropathological Assessment of Alzheimer's Disease. Neurobiol Aging. 1997;18:S1–S2. [PubMed] [Google Scholar]
- 2.Barrett AM. Probable Alzheimer's disease: gender-related issues. J Gend Specif Med. 1999;2:55–60. [PubMed] [Google Scholar]
- 3.Bartzokis G. Alzheimer's disease as homeostatic responses to age-related myelin breakdown. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging/2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Candore G, Balistreri CR, Grimaldi MP, et al. Age-related inflammatory diseases: role of genetics and gender in the pathophysiology of Alzheimer's disease. Ann N Y Acad Sci. 2006;1089:472–486. doi: 10.1196/annals.1386.008. [DOI] [PubMed] [Google Scholar]
- 5.Castellani RJ, Lee HG, Zhu X, Perry G, Smith MA. Alzheimer disease pathology as a host response. J Neuropathol Exp Neurol. 2008;67:523–531. doi: 10.1097/NEN.0b013e318177eaf4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cogswell JP, Ward J, Taylor IA, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 7.Czlonkowska A, Ciesielska A, Gromadzka G, Kurkowska-Jastrzebska I. Gender differences in neurological disease: role of estrogens and cytokines. Endocrine. 2006;29:243–256. doi: 10.1385/ENDO:29:2:243. [DOI] [PubMed] [Google Scholar]
- 8.Dugas JC, Cuellar TL, Scholze A, et al. Dicer1 and miR-219 are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finnerty JR, Wang WX, Hebert SS, et al. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402(3):491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardy J. Alzheimer's disease: the amyloid cascade hypothesis: an update and reappraisal. J Alzheimers Dis. 2006;9:151–153. doi: 10.3233/jad-2006-9s317. [DOI] [PubMed] [Google Scholar]
- 11.Hebert SS, Horre K, Nicolai L, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci USA. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosik KS, Krichevsky AM. The elegance of the microRNAs: a neuronal perspective. Neuron. 2005;47:779–782. doi: 10.1016/j.neuron.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Lau P, Verrier JD, Nielsen JA, et al. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28:11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linsley PS, Schelter J, Burchard J, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Casaccia P. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 2010;33:193–201. doi: 10.1016/j.tins.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J Inorg Biochem. 2007;101:1265–1269. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markesbery WR, Schmitt FA, Kryscio RJ, et al. Neuropathologic substrate of mild cognitive impairment. Arch Neurol. 2006;63:38–46. doi: 10.1001/archneur.63.1.38. [DOI] [PubMed] [Google Scholar]
- 18.Nelson PT, Abner EL, Scheff SW, et al. Alzheimer's-type neuropathology in the precuneus is not increased relative to other areas of neocortex across a range of cognitive impairment. Neurosci Lett. 2009;450:336–339. doi: 10.1016/j.neulet.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson PT, Abner EL, Schmitt FA, et al. Modeling the association between 43 different clinical and pathological variables and the severity of cognitive impairment in a large autopsy cohort of elderly persons. Brain Pathol. 2008;67:555–564. doi: 10.1111/j.1750-3639.2008.00244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson PT, Braak H, Markesbery WR. Neuropathology and cognitive impairment in Alzheimer disease: a complex but coherent relationship. J Neuropathol Exp Neurol. 2009;68:1–14. doi: 10.1097/NEN.0b013e3181919a48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson PT, Jicha GA, Schmitt FA, et al. Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort: neuritic plaques and neurofibrillary tangles “do count” when staging disease severity. J Neuropathol Exp Neurol. 2007;66:1136–1146. doi: 10.1097/nen.0b013e31815c5efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson PT, Schmitt FA, Jicha GA, et al. Association between male gender and cortical Lewy body pathology in large autopsy series. J Neurol. 2010 doi: 10.1007/s00415-010-5630-4. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson PT, Wang WX. MiR-107 is reduced in Alzheimer's disease brain neocortex: validation study. J Alzheimers Dis. 2010;21:75–79. doi: 10.3233/JAD-2010-091603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson PT, Wang WX, Wilfred BR, Tang G. Technical variables in high-throughput miRNA expression profiling: much work remains to be done. Biochim Biophys Acta. 2008;1779:758–765. doi: 10.1016/j.bbagrm.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson PT, Wilfred BR. In situ hybridization is a necessary experimental complement to microRNA (miRNA) expression profiling in the human brain. Neurosci Lett. 2009;466:69–72. doi: 10.1016/j.neulet.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer's disease cortex reveals altered miRNA regulation. PLoS ONE. 2010;5:e8898. doi: 10.1371/journal.pone.0008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel N, Hoang D, Miller N, et al. MicroRNAs can regulate human APP levels. Mol Neurodegener. 2008;3:10. doi: 10.1186/1750-1326-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price JL, McKeel DW, Jr, Buckles VD, et al. Neuropathology of nondemented aging: presumptive evidence for preclinical Alzheimer disease. Neurobiol Aging. 2009;30:1026–1036. doi: 10.1016/j.neurobiolaging.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchie ME, Silver J, Oshlack A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 31.Schmitt FA, Davis DG, Wekstein DR, et al. “Preclinical” AD revisited: neuropathology of cognitively normal older adult. Neurology. 2000;55:370–376. doi: 10.1212/wnl.55.3.370. [DOI] [PubMed] [Google Scholar]
- 32.Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer's disease temporal lobe neocortex. Neurosci Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 33.Smalheiser NR, Lugli G. microRNA regulation of synaptic plasticity. Neuromol Med. 2009;11:200–207. doi: 10.1007/s12017-009-8065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang X, Gal J, Zhuang X, et al. A simple array platform for microRNA analysis and its application in mouse tissues. RNA. 2007;13:1803–1822. doi: 10.1261/rna.498607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang WX, Rajeev BW, Stromberg AJ, et al. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang WX, Wilfred BR, Baldwin DA, et al. Focus on RNA isolation: obtaining RNA for microRNA (miRNA) expression profiling analyses of neural tissue. Biochim Biophys Acta. 2008;1779:749–757. doi: 10.1016/j.bbagrm.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W, Huang RS, Duan S, Dolan ME. Gene set enrichment analyses revealed differences in gene expression patterns between males and females. In Silico Biol. 2009;9:55–63. [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X, He X, Han X, et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65:612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


