Abstract
Site-specific analysis of protein glycosylation is important for biochemical and clinical research efforts. Glycopeptide analysis using liquid chromatography - collision induced dissociation/electron transfer dissociation - mass spectrometry (LC-CID/ETD-MS) allows simultaneous characterization of glycan structure and attached peptide site. However, due to the low ionization efficiency of glycopeptides during electrospray ionization (ESI), 200–500 fmol of sample per injection is needed for a single LC-MS run, which makes it challenging for the analysis of limited amounts of glycoprotein purified from biological matrices. To improve the sensitivity of LC-MS analysis for glycopeptides, an ultra-narrow porous layer open tubular (PLOT) LC column (2.5 m × 10 μm i.d.) was coupled to a linear ion trap mass spectrometer (LTQ-CID/ETD-MS) to provide sensitive analysis of N-linked protein glycosylation heterogeneity. The potential of the developed method is demonstrated by the characterization of site-specific glycosylation using haptoglobin (Hpt) as a model protein. To limit the amount of haptoglobin to low pmole amounts of protein, we affinity purified it from 1 μL of pooled lung cancer patients plasma. A total of 26 glycoforms/glycan compositions on three Hpt tryptic glycopeptides were identified and quantified from 10 LC-MS runs with a consumption of 100 fmol Hpt digest (13 ng protein, 10 fmol per injection). Included in this analysis was the determination of the glycan occupancy level. At this sample consumption level, the high sensitivity of the PLOT LC-LTQ-CID/ETD-MS allowed glycopeptide identification and structure determination, along with relative quantitation of glycans presented on the same peptide backbone, even for low abundant glycopeptides at the ~100 attomole level. The PLOT LC-MS is shown to have sufficient sensitivity to allow characterization of site-specific protein glycosylation from trace levels of glycosylated proteins.
INTRODUCTION
Protein glycosylation, a biologically significant and complex co- and post-translational modification, involves the attachment of glycans to the protein backbone through asparagine (N-linked) and/or serine/threonine (O-linked)1. Glycosylation plays a key role in, among others, protein folding, subcellular localization, turnover and activity, cell proliferation and cell-cell interaction2–4. Cancer has a profound effect on the glycosylation machinery of a cell, leading to glycans that are markedly altered (either in abundance or structure) from that produced by a normal cell2. Glycosylation changes have been reported for many cancers5–8, as well as other diseases9. One of the observed alterations in cancer is site specific glycosylation, and various studies have reported on this modification. For example, N251 monosialylated alpha-fetoprotein (AFP) for hepatocellular carcinoma5, N45 fucosylated prostate specific antigen (PSA) for prostate cancer6, N134 sialylated kallikrein 6 for ovarian cancer7, N211 difucosylated tetraantennary glycan of haptoglobin for pancreatic cancer8 and N630 under-glycosylated transferrin for congenital disorder of glycosylations (CDGs)9.
Site-specific glycan characterization can be important not only for diagnostic purposes, but also for fundamental structural biology. Site-specific glycosylation on epidermal growth factor receptor (EGFR), for example, has been shown to modulate its function10. A point mutation of EGFR (N579 to Q579), while maintaining glycosylation at other extra-cellular sites of the receptor, can produce a functionally distinct receptor 10. These previous studies 5–10 clearly indicate the necessity of characterization of protein glycosylation in a site-specific manner. The LC-MS analysis of glycopeptides can be more difficult than analysis of non-modified peptides or oligosaccharides, as the chemical properties can be quite different between the glycan (hydrophilic) and peptide (hydrophobic) components11. Since the glycosidic linkage in the glycopeptide is weaker than the peptide backbone, collision induced dissociation (CID)-MS will mainly produce glycosidic fragmentation, thus limiting information on the peptide sequence and linkage site12. The addition of electron transfer dissociation (ETD) has allowed the direct LC-MS analysis of glycopeptides to characterize, in the same run, both glycan structure (CID-MS/MS) and peptide sequence/site attachment (ETD-MS/MS)13.
Comprehensive characterization of specific glycoproteins from biological matrices (enriched, for example, by immunoprecipitation) present significant challenges from both the limited amount of material generally available and the overall complexity of the protein. A single glycosylation site can contain a number of glycans (micro-heterogeneity) and a specific glycopeptide can be a minor constituent in a peptide/glycopeptide digest mixture1. Furthermore, glycopeptides will generally have poorer electrospray ionization efficiency than peptides; thus, co-eluting peptides may suppress glycopeptide signal14. On top of these factors, due to the complexity of glycosylation, multiple LC-MS runs are necessary (fragmentation of different ions) to achieve comprehensive glycosylation characterization. A standard 75 μm i.d. capillary LC column routinely requires 200 to 500 fmol (or higher) sample for single LC-MS run15, and multiple runs could easily result in up to 10 pmol of protein sample consumption. This level may simply not be available, e.g. from tissue or low level plasma circulating glycoproteins. For characterization of protein glycosylation, where only limited amounts of a glycosylated protein may be available, ultrasensitive LC-MS methods are required.
It is well-known that the ionization efficiency and thus the MS response will increase if the LC flow rate is decreased16–18. Up to 100 times increase in response factor has been observed for glycan analysis, when the flow rate for ESI was decreased from 1 μL/min to ~ 30 nL/min19. Such a low flow rate is best utilized with ultra-narrow i.d. LC columns for optimal separation, which is challenging. We have previously introduced 10 μm i.d. porous layer open tubular (PLOT) LC columns for highly sensitive LC-MS analysis featuring high resolving power20–23. The low level sample consumption of PLOT LC column should be especially important for multiple runs, necessary for comprehensive glycosylation analysis.
In this paper, the performance of combining the 10 μm i.d. PLOT LC with LTQ-CID/ETD-MS is demonstrated in the comprehensive site specific glycan characterization of immunoprecipitated Hpt from grade 4 non-small cell lung cancer patient plasma at the sample consumption level of 100 fmol or lower. Haptoglobin is a well-known acute phase plasma glycoprotein having an (αβ)2 tetramer structure with 4 N-linked glycosylation sites on its β chain8. The concentration of the glycoprotein in plasma increases during inflammation-associated conditions, including cancer. Recently, several studies have reported on cancer-induced N-linked glycosylation changes on Hpt8, 24, 25, including prostate25, pancreatic8 and lung cancer24. Thus, Hpt is a relevant glycoprotein to select in order to demonstrate the power of the approach. Using affinity purified Hpt from pooled lung cancer patient plasma, the site-specific glycan identification, structure determination including core/antenna fucosylation isomers, relative quantitation and site occupancy, was accomplished from a total of 10 LC-MS runs, each with 10 fmol injection (total consumption of 100 fmol), allowing identification and quantitation down to 100 attomole of specific glycopeptide isoforms. Multiple injections were necessary for glycopeptide characterization in the data dependent mode to obtain high quality MS/MS spectra (especially for low abundant glycopeptides) and to determine the site-occupancy for partially occupied glycosylation sites. The developed method is demonstrated to have sufficient sensitivity to allow characterization of site-specific protein glycosylation from trace amounts of glycosylated proteins.
EXPERIMENTAL
Samples and Reagents
Plasma from 20 patients diagnosed with advanced non-small cell lung cancer (NSCLC, all grade 4 squamous cell carcinoma) was obtained from Proteogenex (Culver City, CA). The individual plasma samples were pooled on an equal volume basis. Anti-haptoglobin (Hpt) polyclonal antibody (pAb) and human Hpt ELISA kit were acquired from Genway (San Diego, CA). Protein G coated magnetic beads were purchased from Invitrogen (Carlsbad, CA). Bis(sulfosuccinimidyl) suberate (BS3) was from Thermo Fisher Scientific (Waltham, MA). Sequencing grade trypsin was obtained from Promega (Madison, WI). Phosphate buffered saline (PBS, 10 times concentrated) was from MP Biomedicals (Solon, OH). All other reagents were from Sigma-Aldrich (St. Louis, MO).
Haptoglobin Immunoaffinity Purification
Anti-Hpt pAb was crosslinked to the magnetic protein G beads using BS3, according to the protocol supplied by the manufacturer. Briefly, 10 μg anti-Hpt pAbs in 40 μL PBS with 0.05% Tween-20 (PBST) was incubated with 0.3 mg protein G coated magnetic beads for 30 min at room temperature (RT). After a PBST wash, 250 μL of freshly-prepared 5 mM BS3 (in PBST) was added to the pAb-beads, and the crosslinking reaction was carried out at RT for 30 min. The reaction was quenched by addition of 12.5 μL of 1 M tris(hydroxymethyl)aminomethane buffer (Tris, pH 7.5). The beads were washed with PBST 3 times and were then ready for use.
For Hpt purification, 1 μL of the pooled lung cancer plasma was diluted to 50 μL with PBST and then incubated with protein G beads to deplete human immunoglobulin. The resulting IgG-depleted plasma was incubated with pAb-beads for 1 hr at RT. After extensive washing (6 times with PBST, followed by 3 times with PBS), the beads were resuspended in 100 μL PBS and transferred to a clean tube. The bound Hpt was eluted using 50 mM glycine (pH 2.8) and neutralized with 1M Tris. The Hpt concentration in the pooled plasma and in the affinity purified sample was determined by the Hpt ELISA kit, according to the protocol provided by the manufacturer.
Trypsin and PNGase-F Digestion
For trypsin digestion, immunoaffinity purified Hpt (1.4 μg, ~ 10 pmol) was buffer exchanged with 6 M guanidine hydrochloride in 50 mM ammonium bicarbonate by ultracentrifugation (10,000 × g, 10 min per cycle, 5 cycles) using a Microcon filter device with a 10 kDa MWCO membrane. The exchanged protein solution was reduced with dithiothreitol (DTT, final concentration 2 mM) for 30 min at 37 °C and alkylated with iodoacetamide (IAA, final concentration 10 mM) in the dark at RT for 30 min. The reduced and alkylated Hpt solution was then transferred to a 10 kDa MWCO filter device to remove guanidine hydrochloride and excess DTT and IAA and buffer exchanged to digestion buffer (50 mM ammonium bicarbonate, pH 7.5). Trypsin (enzyme: substrate = 1: 50) was added to digest the protein for 4 hr at 37 °C. For trypsin plus PNGase F digestion, the tryptic digest (0.3 μg, 2 pmol) was incubated with PNGase F(enzyme: substrate = 1: 50 ) at 37 °C for 4 hr. The digestion was stopped by addition of 1% formic acid. The resulting Hpt digest was aliquoted at 1 μL per vial (~ 100 fmol/vial) and stored at −80 °C. The Hpt digest was diluted 10 times with 0.1% formic acid to ~ 10 fmol/μL immediately before LC-MS analysis, and 1 μL sample (10 fmol Hpt, 1.4 ng Hpt) was injected.
PLOT LC-LTQ-CID/ETD-MS
The polystyrene-divinylbenzene (PS-DVB) PLOT LC column (2.5 m × 10 μm i.d.) was prepared according to procedures previously described [Yue, G. 2007]. Briefly, a degassed solution containing 200 μL styrene, 200 μL divinylbenzene, 600 μL ethanol and 5 mg azobisisobutyronitrile was filled into a 10 μm i.d. capillary pretreated with 3-(trimethoxysilyl)propyl methacrylate. Both ends of the capillary were sealed with septa, and the capillary was heated at 74 °C for ~16 h in a water bath. The column was washed with acetonitrile (> 50 column volumes) and evaluated using digested bovine serum albumin standard peptides (Michrom Bioresources, Auburn, CA), according to published protocols21, 22. A solid phase extraction (SPE) PS-DVB monolithic column (5 cm × 50 μm) was used for sample loading22. A diagram of the overall PLOT LC system can be found in Supplementary Material, Figure S1. The mobile phase flow was split immediately before the SPE-PLOT assembly to minimize the gradient delay, and the flow rate was maintained at 20 nL/min after the split. Gradient elution was performed using an Ultimate 3000 pump system (Dionex, Sunnyvale, CA) with mobile phase A as 0.1% (v/v) formic acid in water and mobile phase B as 0.1% (v/v) formic acid in acetonitrile. The gradient consisted of the following steps: (i) 10 min at 0% B for sample loading/washing; (ii) linear from 0 to 35% B over 35 min; (iii) linear from 35 to 80% B over 10 min; and finally (iv) isocratic at 80% B for 10 min.
An LTQ-CID/ETD-MS (Thermo Fisher Scientific, San Jose, CA) was used in all experiments. The mass spectrometer was operated in the data-dependent mode to switch automatically between full-scan MS, CID-MS2 and ETD-MS2. Briefly, after a survey full-scan MS spectrum from m/z 400 to 2000 (at a target value of 30,000 ions), a subsequent CID-MS2 (at a target value of 20,000 ions and 35% normalized collision energy) and ETD-MS2 (at a target value of 20,000 ions) were performed on the same precursor ion. The precursor ion was isolated with a ± 2 m/z width, starting from the most intense ion in the survey scan. CID-MS2 and ETD-MS2 were repeated four additional times to fragment the second to the fifth highest precursor ion. For ETD-MS2, the ion-ion reaction time between the isolated precursor and reagent anion (fluoranthene) was 100 ms, with the reagent ion (at a target value of 200,000) 10 fold higher than the precursor ion13, 26. The total cycle time (11 scans) of 1.6 s was continuously repeated for the entire LC-MS run under data-dependent conditions with dynamic exclusion (30 seconds for 2 repeated spectra).
Data Processing
For peptide identification, the mass spectrometric raw data was database searched against the Hpt sequence (SwissProt P00738) using SEQUEST (Thermo Fisher Scientific). Peptide identification was based on the following criteria: ΔCn ≥ 0.1, peptide probability < 0.001, Xcorr ≥ 1.9, 2.5 and 3.8 for singly, doubly and triply charged ions, respectively. For glycopeptide characterization including glycan structure, peptide sequence and site of attachment, CID-MS/MS and ETD-MS/MS were manually annotated, as described below.
RESULTS AND DISCUSSION
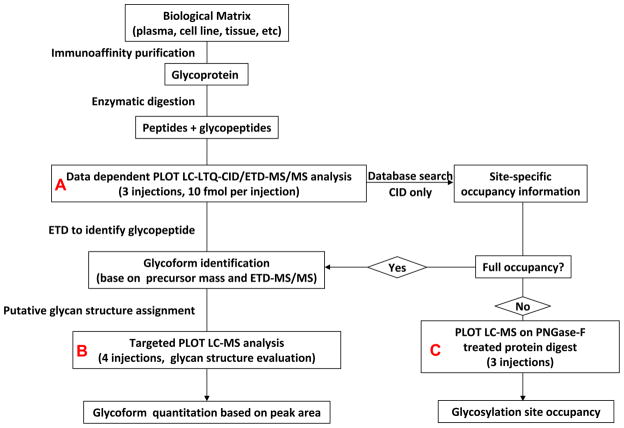
The general strategy for protein glycosylation analysis using 10 μm i.d. PLOT LC-LTQ-CID/ETD-MS is shown in Figure 1. The target glycoprotein is first affinity purified from a biological mixture. A polyclonal antibody, rather than monoclonal antibody, is used to pull down as many target molecule isoforms as possible. The purified glycoprotein (Hpt) is digested with a suitable enzyme to generate a mixture of peptides and glycopeptides, which are then analyzed by PLOT LC-LTQ-CID/ETD-MS. As noted, a total of 10 LC-MS runs (Figure 1, Panels A-C, Table S1) were performed for comprehensive characterization of Hpt glycosylation. For each individual LC-MS run, only 10 fmol of protein digest was injected. For this sample amount, the PLOT LC-MS had sufficient sensitivity to characterize and quantitate lower level glycans present down to the 100 attomole level (dynamic range of 100, see below). Compared to a C18 column, the PS-DVB based column has lower hydrophobicity20, and its open tubular structure improves the recovery of large glycopeptides21. Operating at the flow rate of 20 nL/min, the high-resolution PLOT LC column results in improved analyte ionization and decreased ion suppression compared to conventional size (75 μm i.d.) columns23. All of these features together contribute to improved sensitivity for glycopeptide analysis.
Figure 1.
General strategy for site-specific characterization of protein glycosylation using the 10 μm i.d. PLOT LC-LTQ-CID/ETD-MS system.
Haptoglobin Affinity Purification
Haptoglobin is a high abundant plasma glycoprotein having an (αβ)2 tetramer structure (M.W. ~ 130 kDa). Even though its concentration is relatively high, we chose Hpt as a model glycoprotein to demonstrate the performance of our strategy, because aberrant glycosylation changes have been reported for Hpt in many cancers8, 25 including lung cancer24. A pool of 20 individual advanced non-small cell lung cancer patient plasmas (stage 4) was selected. The concentration of haptoglobin was measured before and after affinity purification using an anti-human haptoglobin ELISA. and determined to be 2.7 μg/μL Hpt, (~ 20 pmol Hpt) and 1.4 μg Hpt (~ 10 pmol, respectively, indicating a 50 % recovery. The purity of the affinity purified haptoglobin was determined from by SDS-PAGE and estimated to be approximately 80 % (data not shown).
Haptoglobin contains 4 N-linked glycosylation sites on its β-chain (N184, N207, N211 and N241). Hpt, known to be readily digested27 with trypsin, was proteolytically cleaved with this enzyme. For other glycoproteins, a strategy of multiple enzymes may be necessary28. Trypsin digestion generates 3 glycopeptides, namely T1, T2 and T3, with T2 having two glycosylation sites (Table 1). The two glycans on T2 are very close and thus difficult to digest into single site glycopeptides. Although it has been reported that these two sites can be separated into single glycosylated peptides8, in this work, for simplicity, we treat the glycopeptide T2 as a single entity since our goal is to demonstrate the ability of the strategy to achieve low level glycosylation characterization. Further treatment for single site analysis of T2 would require more material, but will likely not affect significantly the amount of sample consumed.
Table 1.
Haptoglobin Tryptic Glycopeptides
| Glycopeptide | Amino acid (AA) sequence | AA number |
|---|---|---|
| T1 | MVSHHN184LTTGATLINEQWLLTTAK | 24 |
| T2 | NLFLN207HSEN211ATAK | 13 |
| T3 | VVLHPN241YSQVDIGLIK | 16 |
Glycopeptides T1 and T3
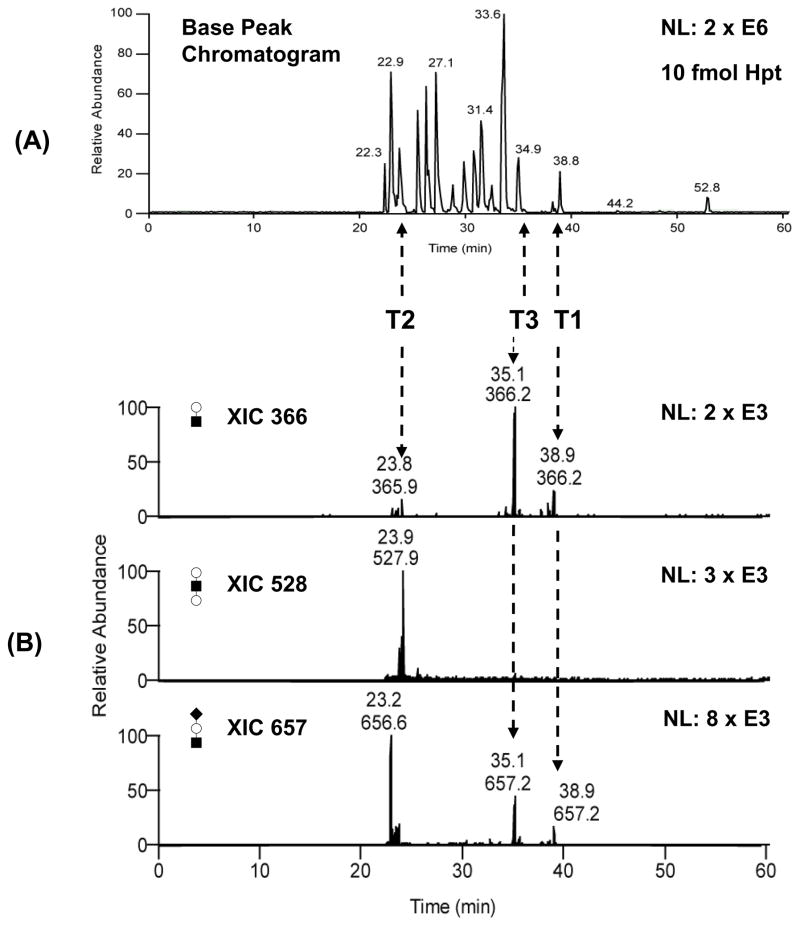
Glycopeptides, sharing the same peptide sequence but differing in glycan structure, will closely elute in reversed phase LC separation14. When subjected to CID fragmentation, glycopeptides will produce low molecular weight glycan oxonium ions such as m/z 366 (Hex-HexNAc+), 528 (Hex-HexNAc-Hex+) and 657 (NeuAc-Hex-HexNAc+), etc9, 29. These oxonium ions can be used to identify the chromatographic peaks of glycopeptides in a complex LC-MS chromatogram. Three data-dependent PLOT LC-LTQ-CID/ETD-MS analyses were initially performed consuming a total of 30 fmol of the Hpt tryptic digest. Figure 2–A shows the base peak chromatogram for one of the LC-MS runs. A signal intensity of 2E6 was obtained at this low sample level; this intensity is comparable to that obtained for ~ 200 fmol Hpt, or 20 fold higher, using a 75 μm packed C18 column for LC/MS (data not shown). To locate glycopeptides from the base peak chromatogram, the above three glycan oxonium ions at m/z of 366, 528 and 657 were extracted from the MS/MS spectra (Figure 2–B). Together with ETD-MS/MS (see below), the elution times for T1 and T3 were identified to be approximately 39 and 34.5 min, respectively (Figure 2–B). In addition, the retention time of T2 (to be discussed later) was estimated to be 23.5 min; the shorter time is expected, given the two glycosylation sites on T2.
Figure 2.
(A) Base peak chromatogram of PLOT LC-MS analysis for 10 fmol Hpt tryptic digest. (B) Oxonium ions were extracted to locate the glycopeptides in the complex LC-MS chromatogram. NL: normalized level. Key to symbols: Empty circle = Hexose, filled square = HexNAc, filled diamond = NeuAc.
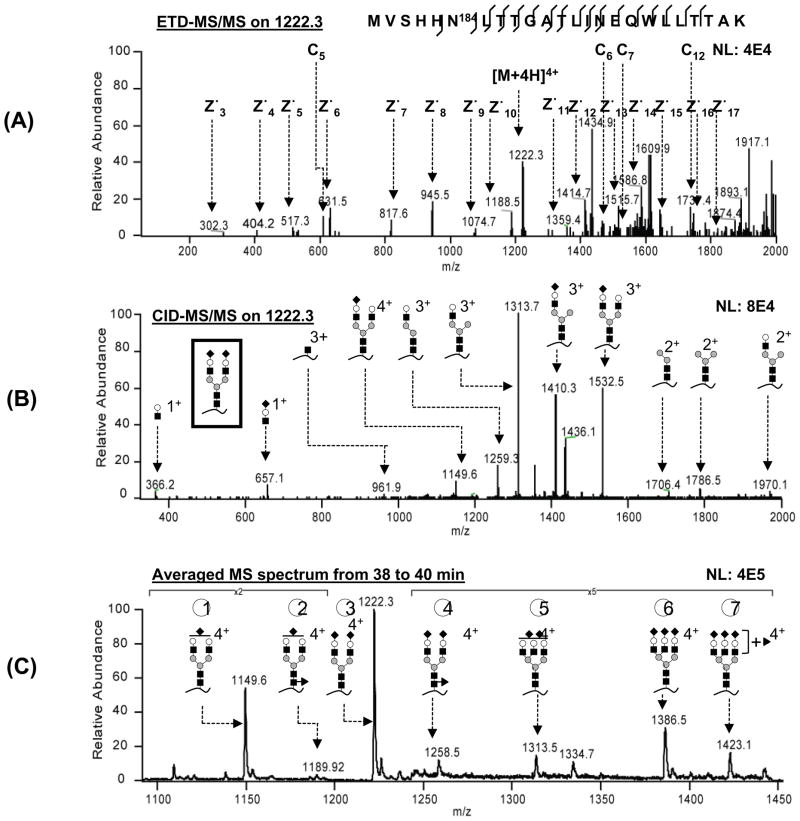
ETD-MS/MS was used to confirm on-line the peptide sequence of the moderate to high intensity glycopeptides for T1 and T3. An example is shown in Figure 3–A, where a T1 glycopeptide (m/z 1222.3), eluting at 39.2 min, was fragmented by ETD. Nine c and 15 z• ions could be identified, with the intact glycan structure attached to c6–9, c11–12 and c14–15. The complete z• ion series and 4 c ions are annotated in Figure 3–A. This ETD-MS/MS spectrum identified the peptide sequence to be MVSHHN184LTTGATLINEQWLLTTAK and confirmed the glycosylation site (N184).
Figure 3.
Glycopeptide analysis using ETD and CID-MS/MS. (A) ETD-MS/MS cleaves the peptide backbone, generating c and z• ions. (B) CID-MS/MS cleaves the glycan linkage, generating oxonium ions and glycopeptide fragments. (C) Averaged mass spectra contain all the detectable glycans on this glycopeptide. NL: normalized level. Key to symbols: empty circle = galactose, filled circle = mannose, filled square = HexNAc, filled diamond = NeuAc, filled triangle = fucose.
Although LC-MS analysis on PNGase-F treated tryptic digests can also correlate the peptide moiety to its corresponding glycopeptide, the ETD-MS/MS used here can be conducted during the same run as CID-MS/MS, thus consuming less sample. Moreover, ETD-MS/MS provides direct evidence of the site of glycan attachment to the peptides, which is not available from the PNGase-F method30. While low signal intensity glycopeptides did not produce ETD fragmentation spectra (ETD is less sensitive than CID), since all glycopeptides from the same peptide eluted within a 2 minute window (data not shown), it was reasonable to associate the low intensity glycopeptides with T1, T2 or T3 based on retention time. Returning to Figure 3, the glycan mass was determined to be 2206.1 Da, based on the mass difference between the observed precursor glycopeptide (m/z 1222.3, z = 4) and the theoretical peptide mass (2683.1 Da). A glycan composition of NeuAc2 HexNAc4Hex5 (theoretical mass 2205.9 Da) was assigned, and the glycan structure was constructed based on the known Man3GlcNAc2 core1 and the CID-MS/MS spectra. CID fragmentation in Figure 3–B generates glycan oxonium ions (366 and 657) and glycopeptide fragments, the latter allowing identification of the glycan structure as a disialylated biantennary complex (see inset in Figure 3–B).
In order to determine the various glycoforms on glycopeptide T1, the MS spectra collected during the elution time window of 39 ± 1 min were averaged (Figure 3–C). GlycoMod31 was used to assign the possible glycoforms by adding the specific glycan masses to the corresponding peptide. The theoretical m/z values were then calculated and compared with the observed m/z values in the averaged spectra to identify the glycans. Table S2 lists all the identified site-specific glycans with their corresponding molecule mass and m/z. The glycan structures on glycopeptide T1, supported by CID-MS/MS in either the data-dependent or targeted MS/MS mode (see below), are presented in Figure 3–C. In a similar manner, 9 glycans were assigned on glycopeptide T3, ranging from biantennary to tetraantennary structures, with or without fucosylation (Supplementary Material, Figure S2–C). It should be noted that, for the initial 3 data-dependent PLOT LC-LTQ-CID/ETD-MS runs, high quality CID-MS/MS spectra were obtained for 5 and 6 glycans on glycopeptides T1 and T3, respectively. Four additional targeted LC-MS runs (either CID-MS/MS only or CID/ETD-MS/MS, Figure 1, panel B) were necessary, focusing on missed low abundant glycopeptide precursors (T1-02 and T1-04 on glycopeptide T1, Figure 1–C; T3-06, T3-07 and T3-09 on glycopeptide T3, Supplementary Material, Figure S2–C).
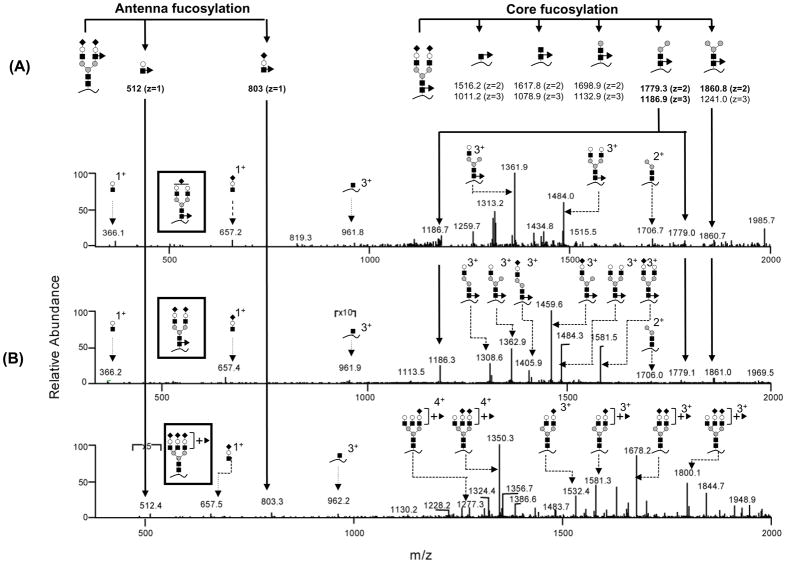
Core/Antenna Fucosylation Isomer Identification
Three glycoforms on glycopeptide T1 were found to be fucosylated (Figure 3–C, glycoforms 2, 4 and 7). In humans, the major sites of fucosylation of N-glycans are either antenna or core GlcNAc1. We attempted to differentiate the core from antenna fucosylated isomers. A theoretical fragmentation pattern was first determined to obtain the position-specific fragments. Five Y ions (for core-fucosylation) and 2 B ions (for antenna-fucosylation) (Figure 4–A, nomenclature according to ref32) were calculated to be sufficient to differentiate the position of the fucose attachment33. The CID-MS/MS spectra of glycoforms 2, 4 and 7 of T1 were manually surveyed for these theoretical fragments. The data suggested that glycoforms 2 and 4 are core-fucosylated, while glycoform 7 is antenna fucosylated (Figure 4–B). The fucosylated glycoforms on T3 were characterized in a similar manner, with all the 3 fucosylated glycoforms (glycoforms 3, 5 and 7) being found to be antenna-fucosylated (Supplementary Material Figure S2). These antenna fucosylated glycans are likely to be Lewis x type structures34. Exoglycosidase digestion could potentially be performed to determine the fucose linkage34, but with more sample consumption; however, this was not pursued in this study.
Figure 4.
Fucosylation position (core/antenna) differentiation based on CID-MS/MS. (A) Theoretical position-specific fragments which can differentiate core/antenna isomers. (B) CID-MS/MS spectra for 3 fucosylated glycoforms. The glycan structures are shown in the inset. Key to symbols can be found in Figure 3.
Glycopeptide T2
Glycopeptide T2 with two glycosylation sites eluted earlier than the other two glycopeptides. The observed glycan masses on T2 were also higher than those on T1 and T3, ranging from 4.1 to 5.8 kDa (Supplementary Material, Figure S3). We hypothesized that both sites are fully occupied, because, for the commonly encountered N-linked Hpt glycans8, 24, 25, if a single glycosylation site were occupied, the attached glycan will only contribute up to a maximum of 3.8 kDa additional mass to the peptide (assuming the largest glycan is sialylated tetraantennary complex glycan with core fucosylation and bisecting HexNAc). Ten major glycan compositions could be identified on T2, based on the observed m/z and MS/MS fragments, and their potential structures are shown in Figure S3. It was not possible to identify the exact glycan isoform structure, i.e. which site contained which specific glycan when the two glycans differed from each other. Moreover, electron transfer dissociation could not be used as it was unable to cleave the peptide backbone between the closely spaced glycosylation sites at N207 and N211.
Glycopeptide Isoform Quantitation
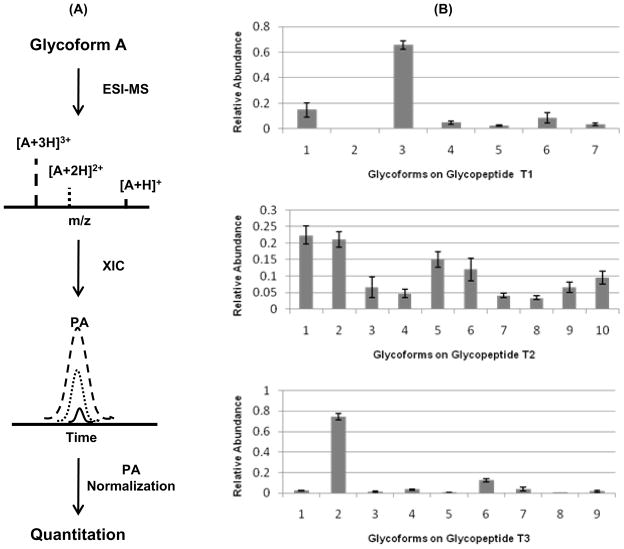
During positive ESI-MS analysis of glycopeptides, protonation occurs mainly on the peptide moiety; thus, the signal intensity of the glycopeptide is largely dependent on the peptide structure of the molecule. It is therefore reasonable to assume that the response factors of all glycoforms of a specific peptide are similar14. However, the ESI process typically generates multiply charged species from a single precursor35 which will complicate ESI-MS based quantitation. This is schematically shown in Figure 5–A. Therefore, peak areas (PAs) summed across all charge state ions were used to determine the relative abundance of a specific glycoform (Figure 5–B). The peak area of each individual glycoform was normalized to the total peak area of all glycoforms of the specific glycopeptide (relative quantitation)36.
Figure 5.
Glycopeptide quantitation. (A) Charge states result in different peak areas (PAs) for the same precursor; (B) Quantitation results using summed PA method. The glycan structures on glycopeptide T1, T2 and T3 can be found in Table S2.
The quantitation results, based on the 3 initial LC-MS runs (Figure 1, panel A), are shown in Figure 5–B. The X axis represents the specific glycoform, and the Y axis is the percentage abundance of different isoforms for a given glycopeptide in the glycosylated protein. For glycopeptide T1, the signal intensity ratio between the most abundant (T1-03, Figure 3–C) and least abundant glycoform (T1-02, Figure 3–C) was ~ 80 (Figure 5–B). This result suggests ~ 2 orders of magnitude difference in abundance for these 2 glycoforms or only ~ 100 attomoles of glycoform T1-02 with a 10 fmol sample injection. Importantly, the high sensitivity of the PLOT LC-MS allowed reproducible quantitation at this low level of sample amount. Similar dynamic ranges were observed for glycans on glycopeptide T3 (Supplementary Material, Figure S2–C, T3-02 and T3-08). For glycopeptide T2, the glycoforms appear to be more evenly distributed compared to T1 and T3. The PA results provide a basis for future comparison of glycosylation patterns across different samples, e.g. individual patient plasmas.
For non-small cell lung cancer, sialic acid and fucose expression on the Hpt-β chain has been reported to be elevated, albeit the glycosylation site and glycan structure contributing to the elevation was not obtained24. We have identified partially or fully sialylated glycans with attached fucose, e.g. T1-07, T3-05 and T3-07. Highly branched glycans were also observed in our study (tetra-antennary, T3-08 and T3-09, Table S3), and increased branching is typical for cancer1. In addition, core-fucosylated glycan was also observed (T1-02 and T1-04, Table S3). Core fucosylated glycoproteins have been reported to be involved in the development of many cancers including lung cancer37.
Glycosylation Site Occupancy
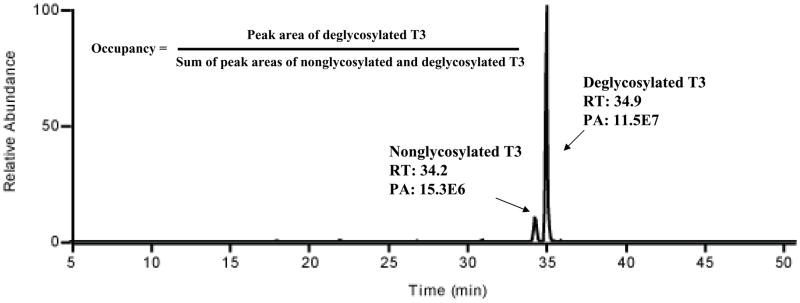
It is important to determine the degree of glycosylation occupancy at a specific site. Because of the limited amount of sample, we used a simpler approach to determine the site occupancy. Automatic database searching was employed to identify nonglycosylated glycopeptides from the Hpt tryptic digest. The three initial data-dependent LC-MS runs, performed for glycopeptide identification (Figure 1, panel A), were database searched against the known Hpt sequence using SEQUEST. The nonglycosylated form of T3 could be identified in the three runs, while the nonglycosylated forms of T1 and T2 were not found. Extracted ion chromatograms (corresponding to nonglycosylated T1 and T2) were also examined with no peak observed. This data suggests that T1 and T2 are 100% or close to 100% occupied while T3 is partially occupied. Next, we used PNGase F to remove the N-linked glycans from the tryptic glycopeptides and convert Asn into Asp. In order to confirm complete deglycosylation, the extracted ion chromatogram of the most intense glycoform on glycopeptide T3 (glycoform T3-03, m/z 1334.5, Supplemental Material Figure S2) was examined; the peak area of glycopeptide T3-03 after PNase F treatment was found to be ~ 3% of that without PNase F treatment, indicating that glycan removal was close to complete. Since the deglycosylated and nonglycosylated peptides have a small difference in hydrophobicity (Asp vs. Asn), we were able to separate the two peptide forms on the high resolution PLOT LC column, as shown in Figure 6. This separation allowed the calculation of the peak area and an estimate of the glycosylation occupancy. The occupancy was calculated to be 88 ± 2% (n = 3), assuming similar ESI-MS responses for the nonglycosylated and deglycosylated peptides. To our knowledge, this is the first report that glycopeptide T3 was partially occupied. Since we used a pooled sample in this work, 88% represents an average value, and it is thus possible that some patients will have even lower occupancy, possibly dependent on the extent of their disease. In a separate study, the occupancy of glycopeptide T3 of Hpt from pooled normal plasma sample was determined to be 93 ± 5 % (data not shown).
Figure 6.
Glycosylation site occupancy determination. Deglycosylated and nonglycosylated peptides T3 were baseline separated using the high-resolution PLOT column. The site occupancy was calculated from the equation shown in the figure.
Conclusions
A 10 μm i.d. PLOT LC-LTQ-CID/ETD-MS strategy has been successfully implemented for the sensitive site-specific characterization of N-linked protein glycosylation. The open tubular structure of the PLOT LC column led to good glycopeptide recovery and, the long column provided high resolving power. Furthermore and importantly, the ultralow flow rate of 20 nL/min significantly enhanced glycopeptide ionization efficiency. As demonstrated in this work, the coupling of the PLOT column to an LTQ-CID/ETD-MS (CID for glycan, ETD for peptide) should have wide application for trace level glycoprotein characterization. Twenty-six site-specific glycoforms/glycan compositions on 3 glycopeptides of Hpt were characterized and quantitated from 10 LC-MS runs using a sample consumption level of 100 fmol protein digest. Among the 10 LC-MS runs, 3 data-dependent runs were initially employed for glycopeptide identification (ETD) and glycan structure evaluation and quantitation (CID), followed by 4 additional targeted LC-MS/MS analyses (CID-MS/MS) on additional glycoforms for further in-depth glycan structure determination. Finally, three additional runs were conducted for glycosylation occupancy determination.
The PLOT LC-LTQ-CID/ETD-MS strategy is straightforward, robust, highly resolving and sensitive, allowing the identification of minor glycans from trace amounts of biological sample. For plasma biomarker discovery, the tissue leakage protein concentration ranges from 1ng/mL (or below) to 1 μg/mL38. Assuming a M.W. of 50 KDa and a recovery of 50%, from 1 mL plasma sample, 10 fmol to10 pmol protein will be available for LC-MS analysis. Theoretically, our strategy should be applicable for characterization of middle to high level tissue leakage glycoproteins in 1 mL of plasma. Also, the method should be useful for elucidation of the structure-function relationship of biologically important molecules such as receptors and the role of glycosylation in their biological function.
Finally, it should be noted that, in this study, the MS spectra were interpreted manually; however, reliable bioinformatics software for glycopeptide identification/annotation will undoubtedly become available in the future. Indeed, significant progress has already been made in this direction39, 40. Thus, together with bioinformatics software, the 10 μm i.d. PLOT LC-LTQ-CID/ETD-MS will ultimately allow comprehensive characterization and quantitation of site-specific protein glycosylation from trace amounts of sample in an automatic manner. Candidate glycoforms such as those highly branched in disease can then be selected for further validation using targeted MS analysis, such as selected reaction monitoring.
Supplementary Material
Acknowledgments
The authors thank NIH GM 15847 and NIH/NCI U01-CA128427 for support of this work. Contribution number 975 from the Barnett Institute.
References
- 1.Varki A, Cummings R, Esko J, Freeze H, Stanley P, Bertozzi C, Hart GW, Etzler M. Essentials of glycobiology. 2. Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 2.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126(5):855–67. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 3.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129(1):123–34. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 4.Grewal PK, Uchiyama S, Ditto D, Varki N, Le DT, Nizet V, Marth JD. The Ashwell receptor mitigates the lethal coagulopathy of sepsis. Nat Med. 2008;14(6):648–55. doi: 10.1038/nm1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poon TC, Mok TS, Chan AT, Chan CM, Leong V, Tsui SH, Leung TW, Wong HT, Ho SK, Johnson PJ. Quantification and utility of monosialylated alpha-fetoprotein in the diagnosis of hepatocellular carcinoma with nondiagnostic serum total alpha-fetoprotein. Clin Chem. 2002;48(7):1021–7. [PubMed] [Google Scholar]
- 6.Peracaula R, Tabares G, Royle L, Harvey DJ, Dwek RA, Rudd PM, de Llorens R. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology. 2003;13(6):457–70. doi: 10.1093/glycob/cwg041. [DOI] [PubMed] [Google Scholar]
- 7.Kuzmanov U, Jiang N, Smith CR, Soosaipillai A, Diamandis EP. Differential N-glycosylation of kallikrein 6 derived from ovarian cancer cells or the central nervous system. Mol Cell Proteomics. 2009;8(4):791–8. doi: 10.1074/mcp.M800516-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakano M, Nakagawa T, Ito T, Kitada T, Hijioka T, Kasahara A, Tajiri M, Wada Y, Taniguchi N, Miyoshi E. Site-specific analysis of N-glycans on haptoglobin in sera of patients with pancreatic cancer: a novel approach for the development of tumor markers. Int J Cancer. 2008;122(10):2301–9. doi: 10.1002/ijc.23364. [DOI] [PubMed] [Google Scholar]
- 9.Huddleston MJ, Bean MF, Carr SA. Collisional fragmentation of glycopeptides by electrospray ionization LC/MS and LC/MS/MS: methods for selective detection of glycopeptides in protein digests. Anal Chem. 1993;65(7):877–84. doi: 10.1021/ac00055a009. [DOI] [PubMed] [Google Scholar]
- 10.Whitson KB, Whitson SR, Red-Brewer ML, McCoy AJ, Vitali AA, Walker F, Johns TG, Beth AH, Staros JV. Functional effects of glycosylation at Asn-579 of the epidermal growth factor receptor. Biochemistry. 2005;44(45):14920–31. doi: 10.1021/bi050751j. [DOI] [PubMed] [Google Scholar]
- 11.Wuhrer M, Catalina MI, Deelder AM, Hokke CH. Glycoproteomics based on tandem mass spectrometry of glycopeptides. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;849(1–2):115–28. doi: 10.1016/j.jchromb.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 12.Zaia J. Mass spectrometry and the emerging field of glycomics. Chem Biol. 2008;15(9):881–92. doi: 10.1016/j.chembiol.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu SL, Huhmer AF, Hao Z, Karger BL. On-line LC-MS approach combining collision-induced dissociation (CID), electron-transfer dissociation (ETD), and CID of an isolated charge-reduced species for the trace-level characterization of proteins with post-translational modifications. J Proteome Res. 2007;6(11):4230–44. doi: 10.1021/pr070313u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wada Y, Azadi P, Costello CE, Dell A, Dwek RA, Geyer H, Geyer R, Kakehi K, Karlsson NG, Kato K, Kawasaki N, Khoo KH, Kim S, Kondo A, Lattova E, Mechref Y, Miyoshi E, Nakamura K, Narimatsu H, Novotny MV, Packer NH, Perreault H, Peter-Katalinic J, Pohlentz G, Reinhold VN, Rudd PM, Suzuki A, Taniguchi N. Comparison of the methods for profiling glycoprotein glycans--HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology. 2007;17(4):411–22. doi: 10.1093/glycob/cwl086. [DOI] [PubMed] [Google Scholar]
- 15.Alley WR, Jr, Mechref Y, Novotny MV. Characterization of glycopeptides by combining collision-induced dissociation and electron-transfer dissociation mass spectrometry data. Rapid Commun Mass Spectrom. 2009;23(1):161–70. doi: 10.1002/rcm.3850. [DOI] [PubMed] [Google Scholar]
- 16.Wilm M, Mann M. Analytical properties of the nanoelectrospray ion source. Anal Chem. 1996;68(1):1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 17.Wilm M, Mann M. Electrospray and taylor-cone theory, Dole’s beam of macromolecules at last? Int J Mass spectrom Ion Process. 1994;136:167–180. [Google Scholar]
- 18.Marginean I, Kelly RT, Page JS, Tang K, Smith RD. Electrospray characteristic curves: in pursuit of improved performance in the nanoflow regime. Anal Chem. 2007;79(21):8030–6. doi: 10.1021/ac0707986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahr U, Pfenninger A, Karas M, Stahl B. High-sensitivity analysis of neutral underivatized oligosaccharides by nanoelectrospray mass spectrometry. Anal Chem. 1997;69(22):4530–5. doi: 10.1021/ac970624w. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Wu SL, Kim J, Karger BL. Ultratrace liquid chromatography/mass spectrometry analysis of large peptides with post-translational modifications using narrow-bore poly(styrene-divinylbenzene) monolithic columns and extended range proteomic analysis. J Chromatogr A. 2007;1154(1–2):295–307. doi: 10.1016/j.chroma.2007.03.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yue G, Luo Q, Zhang J, Wu SL, Karger BL. Ultratrace LC/MS proteomic analysis using 10-microm-i.d, Porous layer open tubular poly(styrene-divinylbenzene) capillary columns. Anal Chem. 2007;79(3) doi: 10.1021/ac061411m. [DOI] [PubMed] [Google Scholar]
- 22.Luo Q, Gu Y, Wu SL, Rejtar T, Karger BL. Two-dimensional strong cation exchange/porous layer open tubular/mass spectrometry for ultratrace proteomic analysis using a 10 microm id poly(styrene- divinylbenzene) porous layer open tubular column with an on-line triphasic trapping column. Electrophoresis. 2008;29(8):1604–11. doi: 10.1002/elps.200700741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo Q, Rejtar T, Wu SL, Karger BL. Hydrophilic interaction 10 microm I. D. porous layer open tubular columns for ultratrace glycan analysis by liquid chromatography-mass spectrometry. J Chromatogr A. 2009;1216(8):1223–31. doi: 10.1016/j.chroma.2008.09.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoagland LFt, Campa MJ, Gottlin EB, Herndon JE, 2nd, Patz EF., Jr Haptoglobin and posttranslational glycan-modified derivatives as serum biomarkers for the diagnosis of nonsmall cell lung cancer. Cancer. 2007;110(10):2260–8. doi: 10.1002/cncr.23049. [DOI] [PubMed] [Google Scholar]
- 25.Fujimura T, Shinohara Y, Tissot B, Pang PC, Kurogochi M, Saito S, Arai Y, Sadilek M, Murayama K, Dell A, Nishimura S, Hakomori SI. Glycosylation status of haptoglobin in sera of patients with prostate cancer vs. benign prostate disease or normal subjects. Int J Cancer. 2008;122(1):39–49. doi: 10.1002/ijc.22958. [DOI] [PubMed] [Google Scholar]
- 26.Udeshi ND, Compton PD, Shabanowitz J, Hunt DF, Rose KL. Methods for analyzing peptides and proteins on a chromatographic timescale by electron-transfer dissociation mass spectrometry. Nat Protoc. 2008;3(11):1709–17. doi: 10.1038/nprot.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Proc J, Kuzyk M, Hardie D, Yang J, Smith D, Jackson A, Parker C, Borchers C. Glycoproteomics Analysis of Human Liver Tissue by Combination of Multiple Enzyme Digestion and Hydrazide Chemistry. Journal of Proteome Research. 2010;9:5422–5437. [Google Scholar]
- 28.Chen R, Jiang X, Sun D, Han G, Wang F, Ye M, Wang L, Zou H. Glycoproteomics Analysis of Human Liver Tissue by Combination of Multiple Enzyme Digestion and Hydrazide Chemistry. Journal of Proteome Research. 2009;8:651–661. doi: 10.1021/pr8008012. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan B, Addona TA, Carr SA. Selective detection of glycopeptides on ion trap mass spectrometers. Anal Chem. 2004;76(11):3112–8. doi: 10.1021/ac035427d. [DOI] [PubMed] [Google Scholar]
- 30.Mechref Y, Novotny MV. Structural investigations of glycoconjugates at high sensitivity. Chem Rev. 2002;102(2):321–69. doi: 10.1021/cr0103017. [DOI] [PubMed] [Google Scholar]
- 31.Cooper CA, Gasteiger E, Packer NH. GlycoMod--a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics. 2001;1(2):340–9. doi: 10.1002/1615-9861(200102)1:2<340::AID-PROT340>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 32.Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J. 1988;5:397–409. [Google Scholar]
- 33.Satomi Y, Shimonishi Y, Hase T, Takao T. Site-specific carbohydrate profiling of human transferrin by nano-flow liquid chromatography/electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2004;18(24):2983–8. doi: 10.1002/rcm.1718. [DOI] [PubMed] [Google Scholar]
- 34.Saldova R, Royle L, Radcliffe CM, Abd Hamid UM, Evans R, Arnold J, Banks R, Hutson R, Harvey DJ, Antrobus R, Petrescu S, Dwek RA, Rudd PM. Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and IgG. Glycobiology. 2007;17(12):1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen S, Fenn JB. Gas-phase ions of solute species from charged droplets of solutions. Proc Natl Acad Sci U S A. 2007;104(4):1111–7. doi: 10.1073/pnas.0609969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rebecchi KR, Wenke JL, Go EP, Desaire H. Label-free quantitation: a new glycoproteomics approach. J Am Soc Mass Spectrom. 2009;20(6):1048–59. doi: 10.1016/j.jasms.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Inoue S, Gu J, Miyoshi E, Noda K, Li W, Mizuno-Horikawa Y, Nakano M, Asahi M, Takahashi M, Uozumi N, Ihara S, Lee SH, Ikeda Y, Yamaguchi Y, Aze Y, Tomiyama Y, Fujii J, Suzuki K, Kondo A, Shapiro SD, Lopez-Otin C, Kuwaki T, Okabe M, Honke K, Taniguchi N. Dysregulation of TGF-beta1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc Natl Acad Sci U S A. 2005;102(44):15791–6. doi: 10.1073/pnas.0507375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1(11):845–67. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 39.Ceroni A, Maass K, Geyer H, Geyer R, Dell A, Haslam SM. GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J Proteome Res. 2008;7(4):1650–9. doi: 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- 40.Tissot B, North SJ, Ceroni A, Pang PC, Panico M, Rosati F, Capone A, Haslam SM, Dell A, Morris HR. Glycoproteomics: past, present and future. FEBS Lett. 2009;583(11):1728–35. doi: 10.1016/j.febslet.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.