Abstract
In cells the de novo nucleation of actin filaments from monomers requires actin-nucleating proteins. These fall into three main families – the Arp2/ 3 complex and its nucleation promoting factors (NPFs), formins, and tandem-monomer-binding nucleators. In this review, we highlight recent advances in understanding the molecular mechanism of actin nucleation by both well-characterized and newly-identified nucleators, and explore current insights into their cellular functions in membrane trafficking, cell migration and division. The mechanisms and functions of actin nucleators are proving to be more complex than previously considered, with extensive cooperation and overlap in their cellular roles.
Introduction
Actin is one of the most highly conserved and abundant proteins in eukaryotic cells and is a major constituent of the cytoskeleton. Actin monomers (G-actin) assemble to form polarized filaments (F-actin) that have a fast-growing barbed end and a slower-growing pointed end. The first step in the assembly of actin filaments is nucleation, which is defined as the formation of a stable multimer of actin monomers. This is the rate-limiting step in polymerization due to the instability of actin dimer intermediates and the activity of actin monomer-sequestering proteins that suppress spontaneous nucleation in cells. To overcome the kinetic hurdle for nucleation, cells use a diverse set of actin-nucleating proteins, including the actin-related protein 2/ 3 (Arp2/ 3) complex, formins and tandem-monomer-binding nucleators. These proteins play important roles in many essential cellular processes.
In this review, we first compare the biochemical mechanisms of actin nucleation, focusing on recent advances and newly-discovered nucleators. We next describe progress in our understanding of the function of these nucleators in key actin-dependent processes including membrane trafficking, cell migration and division, examining how the characterization of known nucleators and the identification of new nucleators has revealed new ways in which actin polymerization contributes to cell function.
Actin nucleators and their mechanism of action – old news and recent developments
The Arp2/3 complex
The first major actin nucleator to be discovered was the Arp2/ 3 complex, which is composed of evolutionarily-conserved subunits including the actin-related proteins Arp2 and Arp3 and five additional subunits ARPC1–5 (reviewed in [1]). The Arp2/ 3 complex by itself is an inefficient nucleator, and its activation requires binding to the sides of actin filaments and to proteins called nucleation promoting factors (NPFs) that have WCA domains consisting of G-actin binding WH2 (W) domains and Arp2/ 3-binding central/ acidic (CA) sequences. Once activated, the Arp2/ 3 complex nucleates the formation of new filaments that extend from the sides of existing filaments at a 70° angle to form a Y-branched network (Figure 1, left panel).
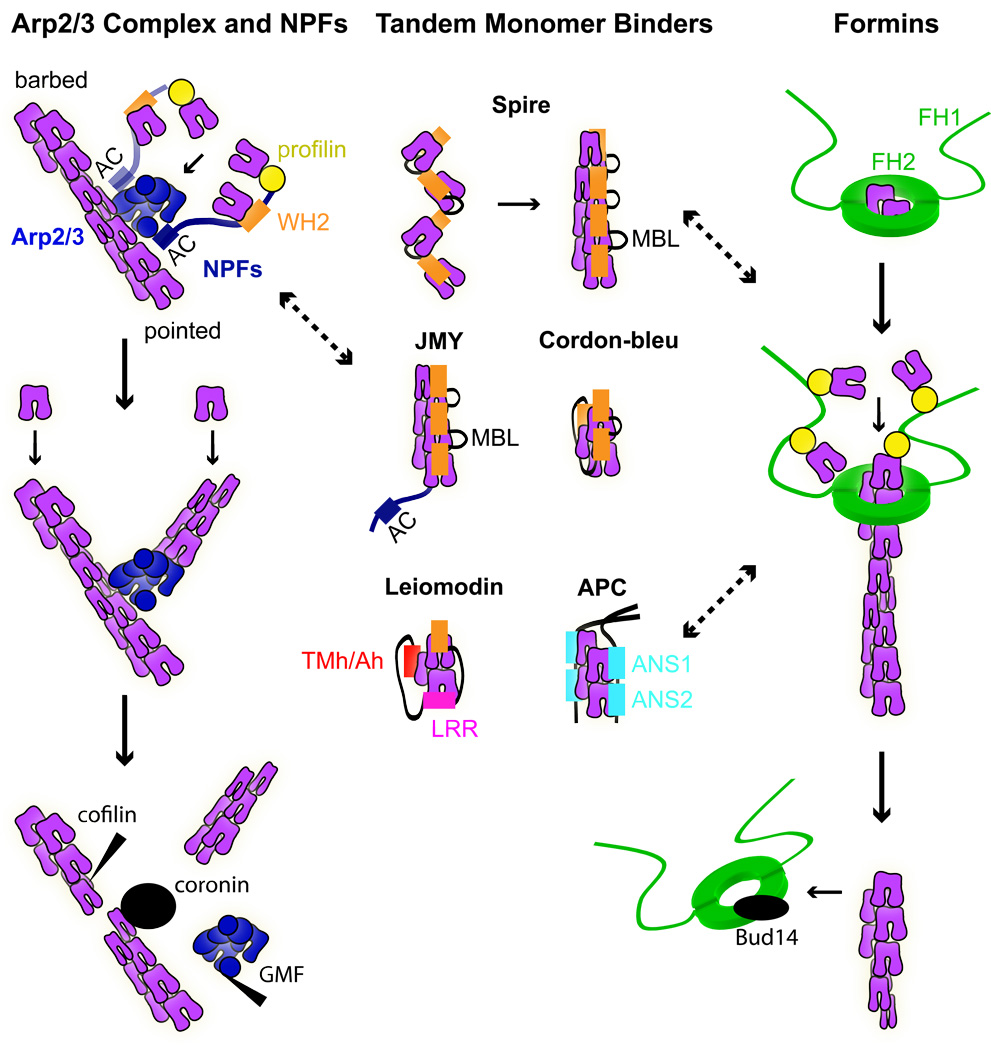
Figure 1. Models of actin nucleation.
Left: Arp2/ 3 complex is activated by binding to the CA region of NPFs and to the side of actin filaments. In turn, NPFs bind actin monomers via their WH2 domains and profilin-actin monomers via their proline-rich regions, and deliver these to a nucleating complex. NPF dimerization enhances their activity, suggesting that dimers may bind to two sites on the Arp2/ 3 complex. After branch formation, cofilin and GMF stimulate debranching by binding to F-actin and Arp2/ 3 complex, respectively. Coronin binds both to F-actin and Arp2/ 3 complex, replaces Arp2/ 3 complex, and synergizes with cofilin to promote debranching. Middle: Tandem-monomer-binding nucleators bring together actin monomers through their clustered actin-binding motifs to form a nucleus. Spire and JMY stabilize actin monomers aligned along the long-pitch helix with their WH2 domains and monomer-binding linkers (MBL). Cordon-bleu, leiomodin and dimeric APC, with their combination of WH2 domains, leucine rich repeats (LRR), tropomyosin and actin-binding helices (Tmh/ Ah), and actin-nucleating sequences (ANS1–2), stabilize cross-filament interactions along the short-pitch helix of an actin filament. Right: Formins generate actin polymerization nuclei by stabilizing actin dimers through their homodimeric FH2 domains. The FH2 dimer stays processively attached to the barbed end of an actin filament as the flanking FH1 domains deliver profilin-actin to the barbed end for continued elongation. In yeast, Bud14p interacts with the FH2 domain and displaces formins from growing barbed ends. Dotted arrows point to the cross-talk between different actin nucleators.
Although the ability of the Arp2/ 3 complex to nucleate Y-branched arrays is well-characterized, the mechanism of nucleation and branching is not fully understood. The most recent model of the Y-branch was obtained by docking the atomic-resolution crystal structure of an inactive conformation of the Arp2/ 3 complex into a 3D reconstruction of the branch obtained by electron tomography [2]. This study suggested that Arp2 and Arp3 interact with the pointed end of the daughter filament while the remaining subunits, in particular ARPC2 and ARPC4, make substantial contacts with the mother filament. However, the functional importance of specific subunits and surfaces of the complex has only begun to be thoroughly investigated. Recent studies employing mutagenesis of conserved surface residues on ARPC1, ARPC2 and ARPC4 have defined features that are functionally important for activity. In particular, residues on a conserved surface on ARPC2 and ARPC4 that is predicted to lie close to the mother filament in the Y-branch [2] were shown to be required for efficient actin nucleation, as well as for high affinity actin filament binding and Y-branch stability [3,4]. Moreover, a conserved surface on ARPC1 was shown to be important for nucleation and binding to the WCA domain of the NPF Las17 (the Saccharomyces cerevisiae ortholog of the mammalian NPF WASP) [5]. Further mutational analyses combined with structural studies are needed to elucidate the detailed mechanism by which the Arp2/ 3 complex nucleates and branches filaments.
Because actin nucleation by the Arp2/ 3 complex requires the activity of NPFs, understanding NPF function and regulation is central to determining the mechanism of actin nucleation. Mammalian cells express several NPFs, including the well-characterized Wiskott-Aldrich Syndrome protein (WASP), neuronal WASP (N-WASP), three WASP and verprolin homologs (WAVEs), and the more recently identified WASP homolog associated with actin, membranes and microtubules (WHAMM), WASP and Scar homolog (WASH), and junction mediating regulatory (JMY) protein. The canonical mode of regulation of NPFs is via allosteric modulation of the accessibility of the WCA domain, either by autoinhibition through intramolecular interactions between the WCA and upstream domains of the NPF, as is the case for WASP and N-WASP (reviewed in [6]), or trans-regulation by interacting proteins, as was shown for the WAVEs [7,8]. Recent studies have also identified oligomerization as another layer of NPF regulation, based on the findings that dimerization of the WCA region increases the affinity of NPFs for the Arp2/ 3 complex and the efficiency of actin nucleation[9]. Thus, oligomerization can act together with allostery to enable NPFs to integrate a wide variety of cellular inputs that lead to activation of the Arp2/ 3 complex.
In addition to exploring the mechanisms of Arp2/ 3 complex activation, recent work has begun to elucidate the mechanisms of Arp2/ 3 complex inactivation and recycling through a process termed debranching. It has been known for some time that ATP hydrolysis by actin and the Arp2 subunit of the Arp2/ 3 complex [1], as well as binding of coronin [10], promote debranching and recycling of the Arp2/ 3 complex. More recent studies identified two new molecular players important for debranching: actin-depolymerizing factor (ADF)/ cofilin and the ADF/ cofilin superfamily protein glia maturation factor (GMF). These factors stimulate debranching through two distinct mechanisms (Figure 1, left panel). ADF/ cofilin, a filament severing protein, is proposed to stimulate debranching by directly competing with Arp2/ 3 for binding to F-actin as well as by causing a structural change in actin that reduces the affinity for Arp2/ 3 [11]. These experiments were performed with ADF/ cofilin purified from Schizosaccharomyces pombe, and it has not yet been reported whether one or more of the three ADF/ cofilin family proteins expressed in mammalian cells (ADF, cofilin-1, and cofilin-2), which have some differences in their biochemical activities [12], might also regulate debranching. In contrast to ADF/ cofilin, GMF binds to the Arp2/ 3 complex but not to F-actin, and it prunes daughter filaments at branch points and inhibits Arp2/ 3-mediated nucleation of new filaments [13,14]. In the future, it will be important to address how the activities of Arp2/ 3-activating and Arp2/3-debranching proteins are coordinated to control Y-branch dynamics in cells.
Formins
The second major class of actin nucleators to be identified was the formins (reviewed in [15]). In contrast to the Arp2/ 3 complex, they are multidomain proteins that function as dimers to assemble unbranched actin filaments. Formins both nucleate actin and act as elongation factors that processively associate with growing barbed ends (Figure 1, right panel), a phenomenon that has now been directly observed by tracking quantum dots coated with formins riding filament ends [16]. Processive association of formins with growing ends allows the addition of actin subunits while preventing capping proteins from terminating polymerization. The defining structural feature of formins is their conserved formin homology (FH) FH1 and FH2 domains. The homodimeric FH2 domain is thought to catalyze actin filament nucleation by stabilizing actin dimers (Figure 1, right panel), although the FH2 domains of different formins vary widely in their nucleation activity. Elongation is then stimulated by the proline-rich FH1 domain, which binds to and increases the local concentration of profilin-bound G-actin to enable its delivery to the barbed end.
Recent work aimed at understanding the mechanism of action of formins has focused on the processive association with elongating filaments, and in particular the source of energy for this process. Nevertheless, the mechanism remains controversial. One study proposed that the energy for processive movement is derived from ATP hydrolysis on actin that is coupled to addition of profilin-actin onto barbed ends [17]. However, a more recent study concluded that ATP hydrolysis on actin is not required for processivity, and instead postulated that the energy is derived from the binding of actin subunits to the barbed end [16]. Further investigation is required to resolve this controversy, and future efforts will focus on not only energetics but also on the structural basis for both nucleation and elongation, as well as potential mechanistic diversity within the formin family.
Apart from the mechanism of formin-mediated nucleation and elongation, recent studies have also characterized various modes of formin regulation. Formin activities can be regulated at multiple points, including initial activation, actin nucleation and elongation, and inactivation and recycling. The best understood mechanism of regulation is allosteric autoinhibition through intramolecular interactions between the Dia autoregulatory domain (DAD) and Dia inhibitory domain (DID) [15], similar to that discussed above for the NPFs WASP and N-WASP. Trans-regulation of formins by interacting proteins has since emerged as another mode of regulation. Previous studies had identified two proteins that inhibit formin activity in vitro, Dia-interacting protein/ WASP interacting SH3 protein (DIP/ WISH) [18] and Spire [19], although Spire is also thought to cooperate with formins to assemble actin filaments in Drosophila melanogaster oocytes [20]. More recently, S. cerevisiae Bud14p was identified as an inhibitor of the formin Bnr1p. Bud14p was shown to displace Bnr1p from growing barbed ends, and to restrict the length of actin filaments elongated by formins in vitro and in cells [21]. A second mechanism to attenuate formin-mediated elongation involves the actin-binding protein tropomyosin, which promotes annealing of actin filaments in a manner that can trap formins within the filament and promotes displacement of formins from barbed ends [22]. These studies suggest that formin displacement may be a critical point of regulation, and future studies will need to address how allosteric activation and formin displacement are coordinated in the formin regulatory cycle.
Tandem-monomer-binding nucleators
The third group of actin nucleators includes Spire, cordon-bleu (Cobl), leiomodin (Lmod) (reviewed in [23]) and the recently described JMY [24] and adenomatous polyposis coli (APC) [25]. These contain tandem G-actin-binding motifs, which bring together monomers to form a polymerization nucleus. In addition to nucleating actin assembly, Spire has also been reported to sever actin filaments and modulate barbed end polymerization [26]. While the WH2 domain is the most common actin-binding motif in these nucleators, they also have additional actin-binding elements. These include the monomer-binding linker (MBL) in Spire and JMY, tropomyosin and actin-binding helices (Tmh/ Ah) and the leucine-rich repeat (LRR) in leiomodin, and the actin-nucleating sequences (ANS1, 2) in APC. This heterogeneity implies that nucleators with distinct domain architecture may remain to be identified.
Despite their shared ability to nucleate actin by gathering monomers into a nucleation complex, members of this family have been proposed to form nuclei with distinct structural arrangements (Figure 1, middle panel). For example, Spire and JMY [24] have been proposed to stabilize monomers aligned along the long-pitch helix of the filament, whereas Cobl, Lmod [23] and APC [25] have been proposed to stabilize cross-filament interactions along the short-pitch helix. However, little structural information is available to verify these proposed mechanisms. An initial X-ray scattering analysis of a Spire-like hybrid WH2 cluster suggested that the nucleus consisted of actin subunits aligned along the long-pitch helix of the filament [27]. However, a more recent determination of the structure of a Spire-actin nucleus by X-ray crystallography suggested that actin monomers are initially organized in a compacted conformation that isomerizes to form a straight long-pitch configuration upon addition of monomers in cross-filament interactions [28]. The configuration of the APC-actin nucleus may be distinct, as APC differs from other members of this class in that its minimum nucleating domain has been shown to form a dimer in vitro [25]. Further structural and biochemical studies are needed to define the configuration of the actin nucleus and the mechanism of actin assembly by members of this class of nucleators.
Cooperation between different families of actin nucleators
Emerging evidence suggests that these nucleator families do not necessarily function individually, and that cross-talk occurs between different nucleators in cells. For example, Spire directly interacts with the formin Cappuccino (Capu) in vitro, and this interaction blocks Capu actin nucleation activity while enhancing Spire activity [19]. Spire and Capu also function together to organize a dynamic network of actin filaments in Drosophila oocytes [20]. APC synergizes with the formin mammalian homolog of Diaphanous 1 (mDia1) to promote actin nucleation in vitro and ectopic actin assembly in cells [25]. JMY both nucleates actin itself and cooperates with the Arp2/ 3 complex to nucleate actin in vitro, although how these activities are coordinated in cells is not known [24]. Finally, the NPF WASH was shown to function together with Spire and the formin Capu to regulate actin and microtubule organization during Drosophila oogenesis [29]. These findings suggest that an intricate interplay between different actin nucleators may enable increased spatial and temporal control over actin assembly in cells and allow greater flexibility in the overall architecture of actin filament networks.
Cellular functions of actin nucleators
The different classes of actin nucleators discussed above play important roles in a variety of cellular processes. Here, we focus on recent advances and controversies related to the function of actin nucleators in three key actin-dependent processes: membrane trafficking, leading edge protrusion during cell migration, and cell division.
Membrane trafficking
Actin polymerization plays a role in many membrane trafficking events including endocytic internalization, endocytic transport, and endoplasmic reticulum (ER)-to-Golgi transport. One function of actin nucleation common to each of these processes is the dynamic shaping and remodeling of membranes. The actin nucleators that contribute to these processes span all three classes and include the Arp2/ 3 complex and its NPFs, the inverted formin 2 (INF2), and the tandem -monomer-binding nucleator Spire.
The functional importance of actin polymerization in the internalization step of clathrin-mediated endocytosis is well established and many of the molecular players have been identified, particularly in the yeast S. cerevisiae, where the process is intensively studied (reviewed in [30]). The Arp2/ 3 complex is the primary actin nucleator during this process, and it is activated by a number of different NPFs including Las17p/ WASp, Abp1p, Pan1p, Myo3p and Myo5p in S. cerevisiae, and N-WASP in mammalian cells. Despite the fact that many key molecules have been identified, the biochemical and biophysical contributions of actin polymerization are matters of active investigation. Our relatively detailed understanding of both the molecules and subprocesses involved in endocytic internalization has enabled the recent development of a theoretical model that describes how membrane shaping is coupled with the underlying biochemical reactions [31]. According to this model, actin nucleation and polymerization are important for the generation of an initial force that drives the shaping of membrane invaginations, resulting in the initiation of a positive feedback loop involving the recruitment of Bin/ Amphiphysin/ Rvs (BAR) domain proteins and lipid phosphatases, and the development of PI(4,5)P2 lipidphase segregation. This phase separation is thought to generate an interfacial force that constricts the membrane invagination and drives vesicle scission. Interestingly, actin polymerization by the Arp2/ 3 complex and activated N-WASP in the absence of other factors was recently shown to drive vesicle scission from tubulated membrane intermediates in a reconstituted system in vitro [32]. These studies support the emerging view that actin nucleation and polymerization induce membrane deformation and scission via a capacity to generate force and/ or promote lipid phase segregation.
In contrast to the case of endocytic internalization, the role of actin in later stages of endocytic trafficking is poorly understood and the molecular players are just now being identified (Figure 2a). A handful of nucleating proteins that function in early-to-late endosome transport have been identified, including the NPF WASH as well as Spire1 and annexin A2. WASH localizes to patches on early and recycling endosomes, where it promotes Arp2/ 3 complex-induced actin nucleation [33–35]. WASH has been implicated in modulating endosome shape and is hypothesized to function in receptor recycling, retromer-mediated endosome-to-Golgi transport, and endosome-to-lysosome trafficking. Although native WASH was recently shown to exist in a multiprotein complex, how the activity of the complex is regulated has not yet been determined [33,35,36]. In addition to WASH and the Arp2/ 3 complex, Spire1 has also been localized to early endosomes and shown to bind to and function with annexin A2 in nucleating F-actin patches [37]. Although these studies highlight the functional importance of actin nucleation in shaping the membranes of early endosomes and in enabling endosomal trafficking, the precise mechanistic contribution of actin remains unknown. Because actin-mediated membrane reorganization facilitates vesicle scission during internalization, it is tempting to speculate that actin polymerization might perform a similar function during later stages of endocytic trafficking.
Figure 2. Models of the cellular localization and function of actin nucleators.
(A) A depiction of the role of actin nucleators in membrane-trafficking events including endocytic internalization as well as various stages of endocytic and ER-to-Golgi trafficking. Abbreviations: EE, early endosomes; ECV/ MVB, endosomal carrier vesicles/ multivesicular bodies; LE, late endosomes; RE, recycling endosomes; Lys, lysosome; ER, Endoplasmic reticulum; ERGIC, endoplasmic reticulum-Golgi intermediate compartment. (B) Diagram of the role of actin nucleators in lamellipodia and filopodia. In the dendritic organization model of actin organization in lamellipodia, branched actin networks are nucleated by the Arp2/ 3 complex and the NPFs WAVE1/ 2 and JMY. In the linear organization model, filaments are nucleated by the Arp2/ 3 complex but are unbranched, or are nucleated by mDia2 or perhaps JMY. In the tip nucleation model of filopodium formation, bundled arrays of actin filaments are nucleated by mDia2, whereas in the convergent elongation model, filaments are nucleated by the Arp2/ 3 complex and WAVE1/ 2, and are elongated by mDia2. (C) Cartoons depicting the role of actin nucleators in cell division. During cytokinesis, formins (Cdc12p in yeast, mDia2 in mammalian cells) nucleate actin filaments from multiple nodes at the division site that then coalesce into the contractile ring in the search, capture, pull and release model. During centrosome separation, dynamic actin reorganization by Dia and Arp2/ 3 drives centrosome separation in the early syncytial Drosophila embryo. For asymmetric spindle positioning in mouse oocytes, FMN2 nucleates a dynamic actin network that moves the spindle to the cell cortex. During the segregation of protein aggregates in S. cerevisiae, Bni1p generates actin cables extending from the polarisome that are required for transport of protein aggregates from the daughter to the mother cell. In (A), (B), and (C) nucleators are color-coded as follows: Arp2/ 3 complex and NPFs (blue), formins (green), tandem-monomer-binding nucleators (yellow). Question marks indicate that the precise role of the nucleating protein is unclear.
In addition to playing a role in endocytosis, actin nucleation has also been implicated in ER-to-Golgi transport (Figure 2a). The NPF WHAMM localizes to the cis-Golgi apparatus, and functions in maintaining Golgi shape and facilitating ER-to-Golgi transport [38]. In addition, WHAMM localizes to tubulo-vesicular membrane transport intermediates and promotes membrane tubulation and tubule elongation by inducing Arp2/ 3-mediated actin assembly and interacting with microtubules. Although WHAMM-mediated Arp2/ 3 activation remains the best-characterized way of nucleating actin during ER-to-Golgi transport, another nucleator was also recently implicated in this process. The formin INF2 was localized to the ER and was implicated in actin assembly in cells based on the observation that an activated mutant caused actin assembly and ER collapse onto the nucleus [39]. Interestingly, INF2 was also implicated in basolateral-to-apical transcytosis in polarized hepatoma cells [40]. Future studies will address the role of actin nucleation in membrane trafficking by exploring more precisely the stages of transport that require actin, whether the role of actin is general or restricted to certain cargos, and what biophysical role actin nucleation plays in reshaping and remodeling membranes.
Leading edge protrusion during cell migration
In migrating cells, actin polymerization in flat membrane protrusions called lamellipodia and finger-like protrusions called filopodia provides the driving force for leading edge protrusion (reviewed in [41]). Widely-accepted models have proposed that lamellipodia are composed of Y-branched filament networks nucleated by Arp2/ 3 complex while filopodia are composed of linear bundles nucleated by formins. However, recent work has challenged some of these assumptions. These studies suggest that the molecular mechanisms of actin nucleation in lamellipodia and filopodia are likely to be complicated and involve cross-participation of diverse actin nucleators.
The dendritic nucleation model, which posits that the Arp2/ 3 complex and NPFs polymerize Y-branched filament networks, has long been the accepted model for actin nucleation in lamellipodia (Figure 2b) [41]. Evidence supporting this model comes from the observations that Y-branching is integral to Arp2/ 3 complex activity in vitro, that Arp2/ 3 is required for lamellipodium formation in cells, and that Y-branches were observed in lamellipodia by electron microscopy with Arp2/ 3 localized to branch points [1,41]. However, this model has been challenged by a recent study in which actin filament networks in lamellipodia of live vitreously frozen cells were visualized by electron tomography [42]. Using this method, individual filaments could be traced along the lamellipodium and were observed to be long and organized into doublets of X-links, but few Y-branches were seen (Figure 2b). This finding opens a new debate about the dendritic nucleation model, and suggests an alternative model that does not rule out the importance of Arp2/ 3 complex for actin nucleation, but implies that Arp2/ 3 may not always form Y-branches in cells. Given the strength of the evidence supporting the dendritic nucleation model, discarding this model is premature, and additional confirmatory work is needed to buttress the case that Y-branches are indeed absent from actin networks within lamellipodia and other structures known to be assembled by the Arp2/ 3 complex.
Activation of the Arp2/ 3 complex in lamellipodia is known to be mediated by the WAVE subfamily of NPFs [41]. More recently, the multifunctional actin nucleator JMY was also shown to localize to lamellipodia in migrating cells, and its depletion slowed migration while its overexpression enhanced migration [24]. JMY was originally discovered as a transcriptional coactivator, and its function in cell migration was recently postulated to be due in part to a role in controlling cadherin expression and cell adhesion [43]. Moreover, the nucleating and Arp2/ 3-activating activities of JMY both appear to be important for its function in enhancing cell migration [24,43]. The dual ability of JMY to directly nucleate unbranched actin filaments and activate the Arp2/ 3 complex to polymerize branched actin filaments might help explain the presence of both filament populations in lamellipodia. The presence of a substantial population of unbranched filaments in lamellipodia also suggests the possible involvement of formins. However, evidence addressing a possible role for formins is only beginning to emerge. A melanoma-derived cell line was shown to require mDia2 for lamellipodium formation [44], although mDia2 was not required for membrane ruffling in HeLa cells [45]. T cells [46] and neutrophils [47] from mDia1 knockout mice exhibited impaired polarization and chemotaxis, further supporting the functional importance of formins during cell migration. Future experimental studies are required to determine how different actin nucleators work together to polymerize actin in lamellipodia and similar protrusive structures.
In filopodia, which contain bundled arrays of actin filaments, the precise role of actin nucleators is also a matter of active investigation and debate, and two competing models have been proposed. According to the tip nucleation model, filopodia arise by formin-mediated nucleation and elongation of linear filaments at the extending filopodial tip (Figure 2b) (reviewed in [48]). In support of this model, depletion of mDia2 in cells inhibits filopodium formation while overexpression of a constitutively active mutant induces it [44,49,50]. Although a role for mDia2 is clear, whether it functions primarily to nucleate or to elongate existing filaments remains uncertain. The alternative model, termed convergent elongation, proposes that the bundle of filaments in filopodia originates from a branched network of filaments nucleated by Arp2/ 3 complex in lamellipodia (Figure 2b). Initial support for this model came from electron micrographs showing that filaments in filopodia are continuous with those in lamellipodia [51], and from the observation that inhibition of Arp2/ 3 complex activity suppressed filopodium formation in neurons [52]. However, others have found that silencing of the Arp2/ 3 complex in melanoma cells does not abolish filopodium formation, suggesting that the requirement for the Arp2/ 3 complex may be cell-type dependent [53]. To further complicate matters, it was recently shown that the Arp2/ 3 complex localizes to puncta within filopodia of spreading cells that may correspond to lamellipodia activity within filopodia [54], and to the heads of neuronal dendritic spines at the tips of dendritic filopodia [50,55]. Continued examination of the relationship between filopodia and lamellipodia, both at an ultrastructural and mechanistic level, is required to determine in what contexts actin nucleation in both structures is interdependent or separable.
Cell division
Actin nucleation also plays crucial and varied roles at several stages of cell division. The best-studied of these is the assembly of the contractile ring, which is essential for cytokinesis. Recent work has highlighted unexpected roles for actin in other facets of cell division, including centrosome separation and asymmetric positioning of the spindle and chromosomes in oocytes, as well as segregation of protein aggregates in yeast.
The molecular requirements for contractile ring assembly have been best characterized in the fission yeast Schizosaccharomyces pombe. The key actin nucleator in this system is the formin Cdc12p, which is essential for actin assembly in the contractile ring (reviewed in [56]). How actin nucleation and elongation by Cdc12p contributes to contractile ring assembly is under active investigation. Cdc12p expressed by its native promoter localizes to a broad band of multiple nodes at the division site [57]. Actin filaments are nucleated from these nodes and elongate in random directions (Figure 2c). Following actin filament nucleation, myosin-II activity is required for the nodes to coalesce into the contractile ring. Mathematical simulations of this process support a search, capture, pull and release model in which actin filaments nucleated by Cdc12p are transiently captured by myosin-II in neighboring nodes, which then pull nodes together [58]. Interestingly, ectopic expression of an activated mutant of Cdc12p results in contractile ring assembly in interphase, a process that requires not only actin assembly but also activation of other contractile ring factors, suggesting that Cdc12p may function as a regulator that drives ring assembly via multiple pathways [59]. Studies in animal cells also provide evidence for the functional importance of formins during contractile ring assembly [56]. Most recently, the mammalian formin mDia2 was shown to localize to the cleavage furrow and perform important functions in actin assembly and cytokinesis [60,61]. It remains to be seen whether formins in mammalian cells participate in contractile ring assembly via a mechanism similar to that seen in S. pombe, or by another pathway.
In addition to having a role in contractile ring assembly, actin nucleation is important for other facets of cell division, including centrosome separation, spindle and chromosome movements, and segregation of damaged proteins (Figure 2c). During cell divisions in the early syncytial Drosophila melanogaster embryo, centrosome separation is driven in part by microtubules and the motor dynein. However, this process was also recently shown to require actin nucleation by the Arp2/ 3 complex and the formin Diaphanous (Dia) [62]. Centrosome separation in mammalian cells also depends in part on cortical actin and myosin [63], although a function for actin nucleators has not been investigated. Apart from its roles early in cell division, actin nucleation is also important later, particularly in mammalian oocytes during female asymmetric meiotic divisions. Asymmetric divisions involve off-center positioning of the spindle and chromosomes, a process that in mouse oocytes depends on a dynamic actin network but not on microtubules. Actin nucleation by formin 2 (FMN2) was shown to play a key role in spindle and chromosome movement during this process [64–66]. Whether the force for movement is derived from actin polymerization [65] or myosin activity [66] is a matter of debate, although both force-generating mechanisms could operate at different stages of the process. A third facet of cell division that requires actin nucleation is the establishment of cellular age asymmetry in S. cerevisiae. In dividing yeast cells subjected to heat stress, aggregated proteins are segregated from daughter cells to mother cells, a process that is proposed to be related to aging of the mother cell [67]. The formin Bni1p generates actin cables extending from the polarisome at the distal end of the daughter cell that are required for directional transport of protein aggregates to the mother cell. It is unclear whether actin polymerization itself drives transport or whether myosin motors also participate. Another formin, Bnr1p, also contributes to asymmetry by allowing protein aggregates in mother cells to merge into an inclusion body. It is an open question as to whether there is a similar protein quality control mechanism during division in other species. The involvement of actin nucleators in such a diverse array of processes is remarkable, and suggests that we are just scratching the surface in terms of our mechanistic understanding of the roles that these proteins play in cell division.
Conclusions
The past two years have seen key advances both in understanding the function of previously-recognized actin nucleators, and in identifying new ones. These advances have come from a combination of biochemical and structural studies aimed at deciphering the mechanisms of actin nucleation, and cell biological studies aimed at defining the functions of actin nucleators at a cellular and organismal level. However, many unanswered questions remain. For example, we know little about the structures of actin nucleators in their activated states, and advances in this area will be essential for understanding the mechanisms of nucleation in greater detail. Moreover, how upstream signal transduction pathways regulate the activity of nucleators needs to be determined at a biochemical level, especially for those that were only recently discovered. More detailed cell biological experiments are also required to explain the function and regulation of actin nucleation during distinct cellular processes. Lastly, emerging evidence has suggested that actin nucleators cooperate with and antagonize each other in vivo, overturning simplified models that assigned one nucleator to one cellular process. Systems-level approaches will be required to determine how the complex cast of molecular players and their interactions are integrated during cell function and behavior. Thus, future studies will allow us to better appreciate the complexity of cellular actin nucleation pathways.
Acknowledgements
We thank Erin Benanti, Ken Campellone, Steve Duleh, Cat Haglund and Taro Ohkawa for helpful discussions and comments on the manuscript. This work was supported by grants from the NIH/ NIGMS (GM059609) and the University of California Cancer Research Coordinating Committee to M.D.W, and a predoctoral fellowship from the American Heart Association (0715035Y) to E.N.F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 2.Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daugherty KM, Goode BL. Functional surfaces on the p35/ARPC2 subunit of Arp2/3 complex required for cell growth, actin nucleation, and endocytosis. J Biol Chem. 2008;283:16950–16959. doi: 10.1074/jbc.M800783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goley ED, Rammohan A, Znameroski EA, Firat-Karalar EN, Sept D, Welch MD. An actin-filament-binding interface on the Arp2/3 complex is critical for nucleation and branch stability. Proc Natl Acad Sci U S A. 2010;107:8159–8164. doi: 10.1073/pnas.0911668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balcer HI, Daugherty-Clarke K, Goode BL. The p40/ARPC1 subunit of Arp2/3 complex performs multiple essential roles in WASp-regulated actin nucleation. J Biol Chem. 2010;285:8481–8491. doi: 10.1074/jbc.M109.054957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derivery E, Gautreau A. Generation of branched actin networks: assembly and regulation of the N-WASP and WAVE molecular machines. Bioessays. 2010;32:119–131. doi: 10.1002/bies.200900123. [DOI] [PubMed] [Google Scholar]
- 7.Lebensohn AM, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell. 2009;36:512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ismail AM, Padrick SB, Chen B, Umetani J, Rosen MK. The WAVE regulatory complex is inhibited. Nat Struct Mol Biol. 2009;16:561–563. doi: 10.1038/nsmb.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Padrick SB, Cheng HC, Ismail AM, Panchal SC, Doolittle LK, Kim S, Skehan BM, Umetani J, Brautigam CA, Leong JM, et al. Hierarchical regulation of WASP/WAVE proteins. Mol Cell. 2008;32:426–438. doi: 10.1016/j.molcel.2008.10.012.. Dimerization of NPFs was demonstrated to increase their affinity for the Arp2/ 3 complex and to enhance their ability to stimulate actin nucleation. Based on these observations, dimerization was proposed be an additional layer of NPF regulation that functions along with allosteric regulation to control NPF activity and actin nucleation.
- 10.Cai L, Makhov AM, Schafer DA, Bear JE. Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell. 2008;134:828–842. doi: 10.1016/j.cell.2008.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan C, Beltzner CC, Pollard TD. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr Biol. 2009;19:537–545. doi: 10.1016/j.cub.2009.02.060.. Cofilin binding to actin filaments was shown to reduce the affinity for Arp2/ 3 complex binding and the stability of Y-branches. The effects were proposed to be due to both direct competition for binding sites and propagation of structural changes in the filament.
- 12.Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gandhi M, Smith BA, Bovellan M, Paavilainen V, Daugherty-Clarke K, Gelles J, Lappalainen P, Goode BL. GMF is a cofilin homolog that binds Arp2/3 complex to stimulate filament debranching and inhibit actin nucleation. Curr Biol. 2010;20:861–867. doi: 10.1016/j.cub.2010.03.026.. GMF was shown to inhibit actin nucleation by the Arp2/ 3 complex and stimulate filament debranching. Because GMF binds to the Arp2/ 3 complex but not actin, the mechanism of debranching was proposed to be distinct from that of cofilin and to involve pruning of filaments at branch points.
- 14.Nakano K, Kuwayama H, Kawasaki M, Numata O, Takaine M. GMF is an evolutionarily developed Adf/cofilin-super family protein involved in the Arp2/3 complex-mediated organization of the actin cytoskeleton. Cytoskeleton. 2010;67:373–382. doi: 10.1002/cm.20451. [DOI] [PubMed] [Google Scholar]
- 15.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 16.Paul AS, Pollard TD. Energetic requirements for processive elongation of actin filaments by FH1FH2-formins. J Biol Chem. 2009;284:12533–12540. doi: 10.1074/jbc.M808587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero S, Didry D, Larquet E, Boisset N, Pantaloni D, Carlier MF. How ATP hydrolysis controls filament assembly from profilin-actin: implication for formin processivity. J Biol Chem. 2007;282:8435–8445. doi: 10.1074/jbc.M609886200. [DOI] [PubMed] [Google Scholar]
- 18.Eisenmann KM, Harris ES, Kitchen SM, Holman HA, Higgs HN, Alberts AS. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr Biol. 2007;17:579–591. doi: 10.1016/j.cub.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 19.Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007;179:117–128. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahlgaard K, Raposo AA, Niccoli T, St Johnston D. Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev Cell. 2007;13:539–553. doi: 10.1016/j.devcel.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chesarone M, Gould CJ, Moseley JB, Goode BL. Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev Cell. 2009;16:292–302. doi: 10.1016/j.devcel.2008.12.001.. Bud14p was shown to be an inhibitor of the S. cerevisiae formin Bnr1p, that displaces the formin from growing barbed ends in vitro and is important for moderating actin filament length in cells.
- 22. Skau CT, Neidt EM, Kovar DR. Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol Biol Cell. 2009;20:2160–2173. doi: 10.1091/mbc.E08-12-1201.. Tropomyosin was found to modulate the activity of the formin Cdc12p. Tropomyosin both enhanced processive elongation and annealing, and terminated elongation by trapping the formin within annealed filaments or dissociating it from barbed ends.
- 23.Qualmann B, Kessels MM. New players in actin polymerization--WH2-domain-containing actin nucleators. Trends Cell Biol. 2009;19:276–285. doi: 10.1016/j.tcb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 24. Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852.. JMY was shown to both directly nucleate actin polymerization and act as an NPF for the Arp2/ 3 complex. It was further shown that JMY shuttles between the nucleus and the leading edge and plays a role in cell migration.
- 25. Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189:1087–1096. doi: 10.1083/jcb.201001016.. The C-terminal basic domain of APC was shown to promote actin nucleation, and to synergize with the formin mDia1, in vitro and in cells. The proposed mechanism of nucleation involves APC dimerization and the recruitment of four monomers to form a polymerization nucleus.
- 26.Bosch M, Le KH, Bugyi B, Correia JJ, Renault L, Carlier MF. Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol Cell. 2007;28:555–568. doi: 10.1016/j.molcel.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Rebowski G, Boczkowska M, Hayes DB, Guo L, Irving TC, Dominguez R. X-ray scattering study of actin polymerization nuclei assembled by tandem W domains. Proc Natl Acad Sci U S A. 2008;105:10785–10790. doi: 10.1073/pnas.0801650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ducka AM, Joel P, Popowicz GM, Trybus KM, Schleicher M, Noegel AA, Huber R, Holak TA, Sitar T. Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation. Proc Natl Acad Sci U S A. 2010;107:11757–11762. doi: 10.1073/pnas.1005347107.. Structures of the WH2 domains of Spire bound to actin were determined by x-ray crystallography, suggesting a stepwise model of nucleation involving a transition from a side-to-side to a longitudinal configuration of monomers.
- 29.Liu R, Abreu-Blanco MT, Barry KC, Linardopoulou EV, Osborn GE, Parkhurst SM. Wash functions downstream of Rho and links linear and branched actin nucleation factors. Dev elopment. 2009;136:2849–2860. doi: 10.1242/dev.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson AS, Smythe E, Ayscough KR. Functions of actin in endocytosis. Cell Mol Life Sci. 2009;66:2049–2065. doi: 10.1007/s00018-009-0001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu J, Sun Y, Drubin DG, Oster GF. The mechanochemistry of endocytosis. PLoS Biol. 2009;7:e1000204. doi: 10.1371/journal.pbio.1000204.. By analyzing existing experimental information and carrying out new mathematical simulations, the authors construct the first integrated theoretical model of vesicle formation during clathrin-mediated endocytosis. Their model suggests a key role for actin nucleation in initiating a positive feedback loop that results in vesicle scission.
- 32. Romer W, Pontani LL, Sorre B, Rentero C, Berland L, Chambon V, Lamaze C, Bassereau P, Sykes C, Gaus K, et al. Actin dynamics drive membrane reorganization and scission in clathrin-independent endocytosis. Cell. 2010;140:540–553. doi: 10.1016/j.cell.2010.01.010.. Actin nucleation is observed to contribute to membrane domain reorganization and scission in the absence of dynamin. This suggests that actin nucleation/ polymerization itself is a trigger for membrane scission.
- 33. Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010.. WASH was localized to subdomains of early endosomes, and functionally linked to transferrin recycling to the plasma membrane.
- 34. Duleh SN, Welch MD. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton. 2010;67:193–206. doi: 10.1002/cm.20437.. WASH was shown to be important for maintaining endosome shape, and implicated in endosomal trafficking to lysosomes.
- 35. Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009.. WASH was observed to be localized to actin-rich subdomains of early endosomes, and shown to be involved in retromer-mediated retrograde trafficking.
- 36.Jia D, Gomez TS, Metlagel Z, Umetani J, Otwinowski Z, Rosen MK, Billadeau DD. WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc Natl Acad Sci U S A. 2010;107:10442–10447. doi: 10.1073/pnas.0913293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morel E, Parton RG, Gruenberg J. Annexin A2-dependent polymerization of actin mediates endosome biogenesis. Dev Cell. 2009;16:445–457. doi: 10.1016/j.devcel.2009.01.007.. Actin was shown to play a role in maintaining early endosome morphology and in enabling early-to-late endosomal transport. Moreover, annexin A2 was implicated in actin nucleation on early endosomes, a process that was mediated by Spire and the Arp2/ 3 complex.
- 38.Campellone KG, Webb NJ. Cdc42 and MAL2 is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chhabra ES, Ramabhadran V, Gerber SA, Higgs HN. INF2 is an endoplasmic reticulum-associated formin protein. J Cell Sci. 2009;122:1430–1440. doi: 10.1242/jcs.040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madrid R, Aranda JF, Rodriguez-Fraticelli AE, Ventimiglia L, Andres-Delgado L, Shehata M, Fanayan S, Shahheydari H, Gomez S, Jimenez A, et al. The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Dev Cell. 2010;18:814–827. doi: 10.1016/j.devcel.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 41.Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- 42. Urban E, Jacob S, Nemethova M, Resch GP, Small JV. Electron tomography reveals unbranched networks of actin filaments in lamellipodia. Nat Cell Biol. 2010;12:429–435. doi: 10.1038/ncb2044.. The organization of actin in the lamellipodia of vitreously frozen cells was observed using electron tomography, and was found to consist of unbranched filaments, which contradicts previously published results and challenges the dendritic nucleation hypothesis. These observations suggest a controversial model in which the Arp2/ 3 complex nucleates filaments in lamellipodia but does not form Y-branches, in contradiction with the well-characterized biochemical activity of the Arp2/ 3 complex in vitro.
- 43.Coutts AS, Weston L, La Thangue NB. A transcription co-factor integrates cell adhesion and motility with the p53 response. Proc Natl Acad Sci U S A. 2009;106:19872–19877. doi: 10.1073/pnas.0906785106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beli P, Mascheroni D, Xu D, Innocenti M. WAVE and Arp2/3 jointly inhibit filopodium formation by entering into a complex with mDia2. Nat Cell Biol. 2008;10:849–857. doi: 10.1038/ncb1745. [DOI] [PubMed] [Google Scholar]
- 46.Sakata D, Taniguchi H, Yasuda S, Adachi-Morishima A, Hamazaki Y, Nakayama R, Miki T, Minato N, Narumiya S. Impaired T lymphocyte trafficking in mice deficient in an actin-nucleating protein, mDia1. J Exp Med. 2007;204:2031–2038. doi: 10.1084/jem.20062647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Y, Zhang J, Mullin M, Dong B, Alberts AS, Siminovitch KA. The mDial formin is required for neutrophil polarization, migration, and activation of the LARG/RhoA/ROCK signaling axis during chemotaxis. J Immunol. 2009;182:3837–3845. doi: 10.4049/jimmunol.0803838. [DOI] [PubMed] [Google Scholar]
- 48.Mellor H. The role of formins in filopodia formation. Biochim Biophys Acta. 2010;1803:191–200. doi: 10.1016/j.bbamcr.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 49.Block J, Stradal TE, Hanisch J, Geffers R, Kostler SA, Urban E, Small JV, Rottner K, Faix J. Filopodia formation induced by active mDia2/Drf3. J Microsc. 2008;231:506–517. doi: 10.1111/j.1365-2818.2008.02063.x. [DOI] [PubMed] [Google Scholar]
- 50.Hotulainen P, Llano O, Smirnov S, Tanhuanpaa K, Faix J, Rivera C, Lappalainen P. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol. 2009;185:323–339. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korobova F, Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol Biol Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, Rottner K, Stradal TE. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. M ol Biol Cell. 2006;17:2581–2591. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnston SA, Bramble JP, Yeung CL, Mendes PM, Machesky LM. Arp2/3 complex activity in filopodia of spreading cells. BMC Cell Biol. 2008;9:65. doi: 10.1186/1471-2121-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pollard TD. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol. 2010;22:50–56. doi: 10.1016/j.ceb.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Coffman VC, Nile AH, Lee IJ, Liu H, Wu JQ. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol Biol Cell. 2009;20:5195–5210. doi: 10.1091/mbc.E09-05-0428.. The formin Cdc12p was localized to a band of dynamic nodes from which actin filaments were nucleated in random directions, and myosin function was required to condense nodes into a contractile ring. This provides support for the search, capture pull and release model proposedby Vavylonis et al [58].
- 58.Vavylonis D, Wu JQ, Hao S, O'Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 59. Yonetani A, Chang F. Regulation of cytokinesis by the formin cdc12p. Curr Biol. 2010;20:561–566. doi: 10.1016/j.cub.2010.01.061.. Overexpression of an activated mutant of the formin Cdc12p results in contractile ring formation during interphase, an effect that depends on actin nucleation and another undescribed activity. This suggests that Cdc12p functions dowstream of cell cycle regulators to drive the initiation of cytokinesis.
- 60.Watanabe S, Ando Y, Yasuda S, Hosoya H, Watanabe N, Ishizaki T, Narumiya S. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol Biol Cell. 2008;19:2328–2338. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe S, Okawa K, Miki T, Sakamoto S, Morinaga T, Segawa K, Arakawa T, Kinoshita M, Ishizaki T, Narumiya S. Rho and Anillin-dependent Control of mDia2 Localization and Function in Cytokinesis. Mol Biol Cell. 2010;18:3193–3204. doi: 10.1091/mbc.E10-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cao J, Crest J, Fasulo B, Sullivan W. Cortical Actin Dynamics Facilitate Early-Stage Centrosome Separation. Curr Biol. 2010;8:770–776. doi: 10.1016/j.cub.2010.02.060.. This paper shows that dynamic rearrangement of actin is requ ired for centrosome separation in Drosophila embryos. Actin reorganization during this process is mediated by the formin Dia and the Arp2/ 3 complex.
- 63.Rosenblatt J, Cramer LP, Baum B, McGee KM. Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell. 2004;117:361–372. doi: 10.1016/s0092-8674(04)00341-1. [DOI] [PubMed] [Google Scholar]
- 64.Azoury J, Lee KW, Georget V, Rassinier P, Leader B, Verlhac MH. Spindle positioning in mouse oocytes relies on a dynamic meshwork of actin filaments. Curr Biol. 2008;18:1514–1519. doi: 10.1016/j.cub.2008.08.044. [DOI] [PubMed] [Google Scholar]
- 65.Li H, Guo F, Rubinstein B, Li R. Actin-driven chromosomal motility leads to symmetry breaking in mammalian meiotic oocytes. Nat Cell Biol. 2008;10:1301–1308. doi: 10.1038/ncb1788. [DOI] [PubMed] [Google Scholar]
- 66.Schuh M, Ellenberg J. A new model for asymmetric spindle positioning in mouse oocytes. Curr Biol. 2008;18:1986–1992. doi: 10.1016/j.cub.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 67.Liu B, Larsson L, Caballero A, Hao X, Oling D, Grantham J, Nystrom T. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 2010;140:257–267. doi: 10.1016/j.cell.2009.12.031. [DOI] [PubMed] [Google Scholar]