Abstract
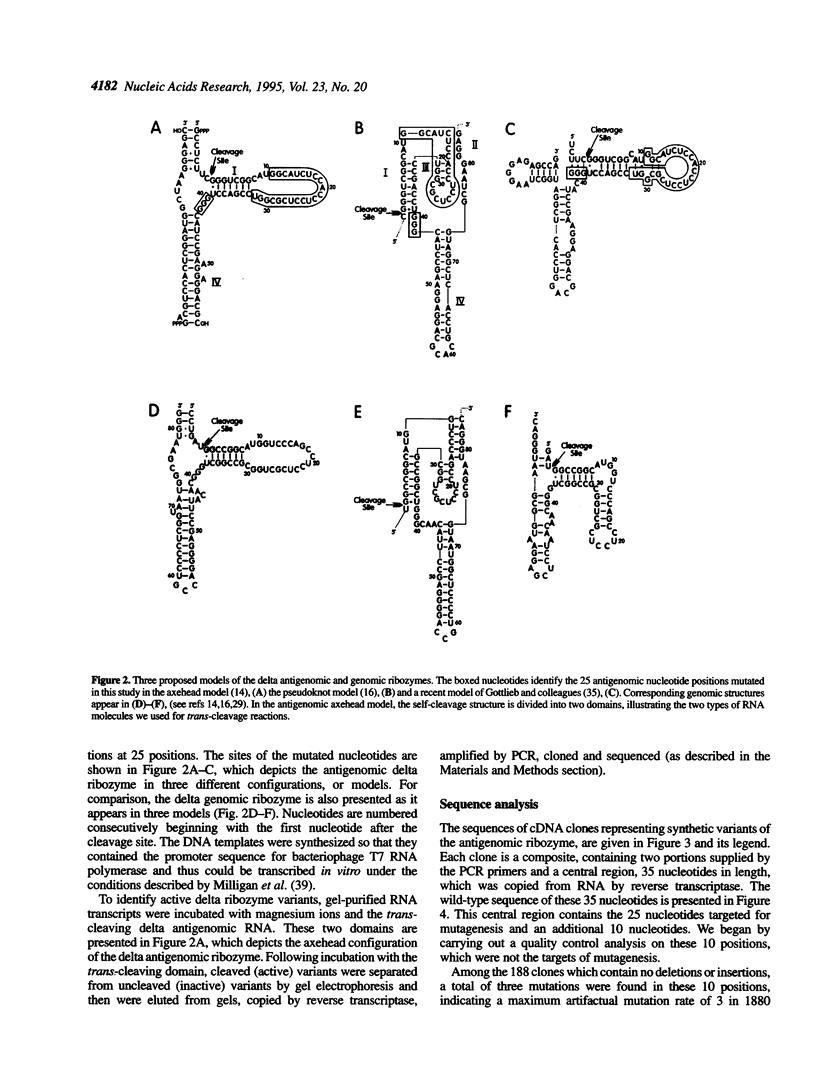
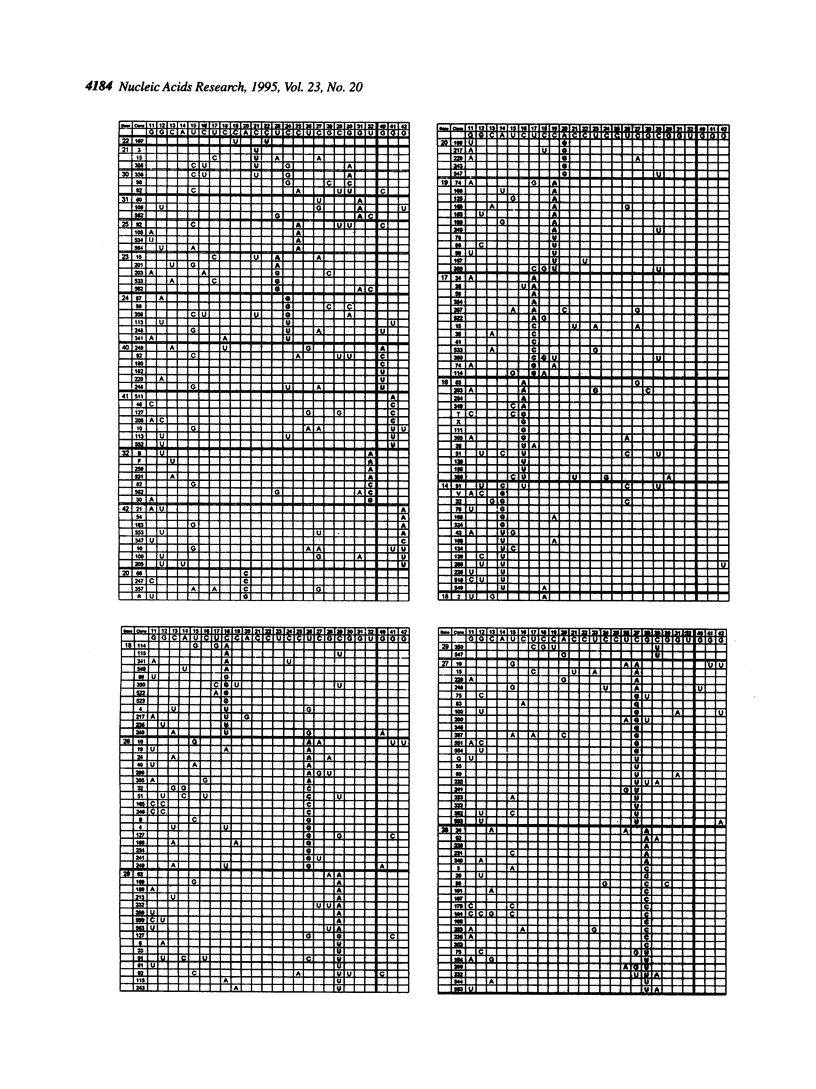
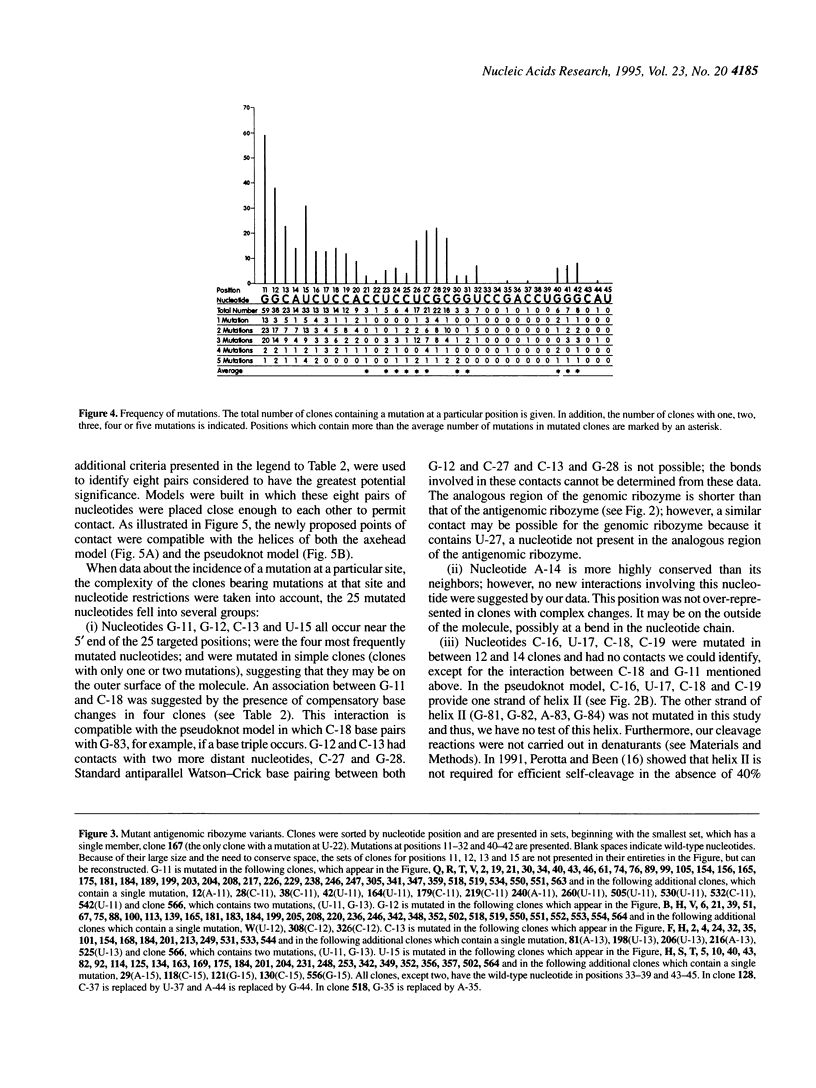
We mapped 359 mutations at 25 positions in synthetic variants of the antigenomic ribozyme of the hepatitis delta agent by analyzing the sequences of 188 cDNA clones. These data were used to identify three features of the ribozyme: highly conserved nucleotides, positions with restricted nucleotide substitutions and three-dimensional relationships between nucleotides. The distribution of mutations at the 25 positions was as follows: G-11 (the eleventh nucleotide from the cleavage site) was mutated in 56 clones; G-12 in 36; U-15 in 33; C-13 in 26; G-28 in 23; C-27 in 21; C-29 in 19; U-26 in 17; C-18 in 14; A-14 in 13; C-16 in 13; C-19 in 12; U-17 in 11; A-20 in 10; G-42 in 9; G-40 in 7; G-41 in 7; C-24 in 6; U-32 in 6; U-23 in 5; C-25 in 4; C-21 in 3; G-30 in 3; G-31 in 3; C-22 in 1. All clones containing a mutation at C-25 had an A at this position, suggesting that the extra cyclic amino group present in adenine and cytosine may function during the cleavage event. Mutations at certain positions were common in simple clones (containing only one or two mutations), while mutations at other positions were over-represented in more complex clones. Both compensatory base changes and co-mutational frequencies were used to identify eight pairs of nucleotides which may interact with each other: G-11 and C-18, G-12 and C-27, C-13 and G-28, C-21 and U-23/C-24, C-21 and G-30, U-23 and G-31/U-32, C24 and G-30, C-27 and G-42. These pairs, which involve some of the most conserved positions in the molecule, suggest interactions among nucleotides previously depicted in open-loop structures. The newly proposed points of contact between pairs of nucleotides are compatible with both the axehead and pseudoknot secondary structural models and were combined with previously proposed Watson-Crick base paired helices to produce two three dimensional models. In both of these, C-25 and C-76 are placed near the cleavage site.
Full text
PDF
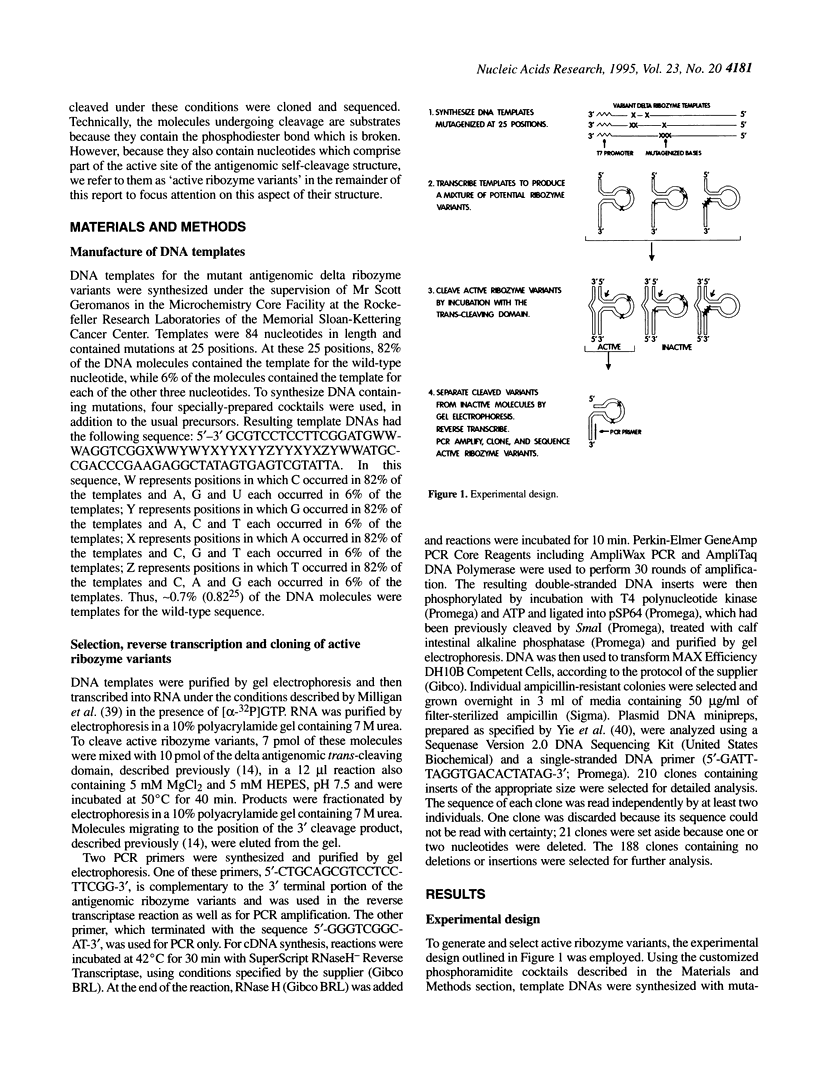





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belinsky M. G., Britton E., Dinter-Gottlieb G. Modification interference analysis of a self-cleaving RNA from hepatitis delta virus. FASEB J. 1993 Jan;7(1):130–136. doi: 10.1096/fasebj.7.1.8422959. [DOI] [PubMed] [Google Scholar]
- Branch A. D., Benenfeld B. J., Baroudy B. M., Wells F. V., Gerin J. L., Robertson H. D. An ultraviolet-sensitive RNA structural element in a viroid-like domain of the hepatitis delta virus. Science. 1989 Feb 3;243(4891):649–652. doi: 10.1126/science.2492676. [DOI] [PubMed] [Google Scholar]
- Branch A. D., Lee S. E., Neel O. D., Robertson H. D. Prominent polypurine and polypyrimidine tracts in plant viroids and in RNA of the human hepatitis delta agent. Nucleic Acids Res. 1993 Jul 25;21(15):3529–3535. doi: 10.1093/nar/21.15.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch A. D., Levine B. J., Baroudy B. M., Buckler-White A., Gerin J. L., Robertson H. D. The novel tertiary structure in delta RNA may function as a ribozyme control element. Prog Clin Biol Res. 1991;364:257–264. [PubMed] [Google Scholar]
- Branch A. D., Levine B. J., Polaskova J. A. An RNA tertiary structure of the hepatitis delta agent contains UV-sensitive bases U-712 and U-865 and can form in a bimolecular complex. Nucleic Acids Res. 1995 Feb 11;23(3):491–499. doi: 10.1093/nar/23.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch A. D., Robertson H. D. A replication cycle for viroids and other small infectious RNA's. Science. 1984 Feb 3;223(4635):450–455. doi: 10.1126/science.6197756. [DOI] [PubMed] [Google Scholar]
- Casey J. L., Brown T. L., Colan E. J., Wignall F. S., Gerin J. L. A genotype of hepatitis D virus that occurs in northern South America. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9016–9020. doi: 10.1073/pnas.90.19.9016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. J., Kalpana G., Goldberg J., Mason W., Werner B., Gerin J., Taylor J. Structure and replication of the genome of the hepatitis delta virus. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8774–8778. doi: 10.1073/pnas.83.22.8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb P. A., Prasad Y., Smith J. B., Williams A. P., Dinter-Gottlieb G. Evidence that alternate foldings of the hepatitis delta RNA confer varying rates of self-cleavage. Biochemistry. 1994 Mar 15;33(10):2802–2808. doi: 10.1021/bi00176a008. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Kawakami J., Kumar P. K., Suh Y. A., Nishikawa F., Kawakami K., Taira K., Ohtsuka E., Nishikawa S. Identification of important bases in a single-stranded region (SSrC) of the hepatitis delta (delta) virus ribozyme. Eur J Biochem. 1993 Oct 1;217(1):29–36. doi: 10.1111/j.1432-1033.1993.tb18214.x. [DOI] [PubMed] [Google Scholar]
- Kos A., Dijkema R., Arnberg A. C., van der Meide P. H., Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986 Oct 9;323(6088):558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- Kumar P. K., Suh Y. A., Miyashiro H., Nishikawa F., Kawakami J., Taira K., Nishikawa S. Random mutations to evaluate the role of bases at two important single-stranded regions of genomic HDV ribozyme. Nucleic Acids Res. 1992 Aug 11;20(15):3919–3924. doi: 10.1093/nar/20.15.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P. K., Suh Y. A., Taira K., Nishikawa S. Point and compensation mutations to evaluate essential stem structures of genomic HDV ribozyme. FASEB J. 1993 Jan;7(1):124–129. doi: 10.1096/fasebj.7.1.8422958. [DOI] [PubMed] [Google Scholar]
- Kumar P. K., Taira K., Nishikawa S. Chemical probing studies of variants of the genomic hepatitis delta virus ribozyme by primer extension analysis. Biochemistry. 1994 Jan 18;33(2):583–592. doi: 10.1021/bi00168a025. [DOI] [PubMed] [Google Scholar]
- Kuo M. Y., Sharmeen L., Dinter-Gottlieb G., Taylor J. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J Virol. 1988 Dec;62(12):4439–4444. doi: 10.1128/jvi.62.12.4439-4444.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. S., Wu H. N., Huang T. H. The catalytic domain of human hepatitis delta virus RNA. A proton nuclear magnetic resonance study. FEBS Lett. 1993 Jun 21;324(3):296–300. doi: 10.1016/0014-5793(93)80138-k. [DOI] [PubMed] [Google Scholar]
- Macnaughton T. B., Wang Y. J., Lai M. M. Replication of hepatitis delta virus RNA: effect of mutations of the autocatalytic cleavage sites. J Virol. 1993 Apr;67(4):2228–2234. doi: 10.1128/jvi.67.4.2228-2234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S., Kumar P. K., Jeoung Y. H., Kawakami J., Nishikawa F., Suh Y., Ohtsuka E., Taira K. Chemical probing studies of the hepatitis delta virus (HDV) genomic ribozyme. Nucleic Acids Symp Ser. 1993;(29):119–120. [PubMed] [Google Scholar]
- Perrotta A. T., Been M. D. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature. 1991 Apr 4;350(6317):434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- Perrotta A. T., Been M. D. Assessment of disparate structural features in three models of the hepatitis delta virus ribozyme. Nucleic Acids Res. 1993 Aug 25;21(17):3959–3965. doi: 10.1093/nar/21.17.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta A. T., Been M. D. Cleavage of oligoribonucleotides by a ribozyme derived from the hepatitis delta virus RNA sequence. Biochemistry. 1992 Jan 14;31(1):16–21. doi: 10.1021/bi00116a004. [DOI] [PubMed] [Google Scholar]
- Perrotta A. T., Been M. D. The self-cleaving domain from the genomic RNA of hepatitis delta virus: sequence requirements and the effects of denaturant. Nucleic Acids Res. 1990 Dec 11;18(23):6821–6827. doi: 10.1093/nar/18.23.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A., RajBhandary U. L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- Rizzetto M., Canese M. G., Aricò S., Crivelli O., Trepo C., Bonino F., Verme G. Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut. 1977 Dec;18(12):997–1003. doi: 10.1136/gut.18.12.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstein S. P., Been M. D. Evidence that genomic and antigenomic RNA self-cleaving elements from hepatitis delta virus have similar secondary structures. Nucleic Acids Res. 1991 Oct 11;19(19):5409–5416. doi: 10.1093/nar/19.19.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmeen L., Kuo M. Y., Dinter-Gottlieb G., Taylor J. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J Virol. 1988 Aug;62(8):2674–2679. doi: 10.1128/jvi.62.8.2674-2679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. B., Dinter-Gottlieb G. Antigenomic Hepatitis delta virus ribozymes self-cleave in 18 M formamide. Nucleic Acids Res. 1991 Mar 25;19(6):1285–1289. doi: 10.1093/nar/19.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y. A., Kumar P. K., Kawakami J., Nishikawa F., Taira K., Nishikawa S. Systematic substitution of individual bases in two important single-stranded regions of the HDV ribozyme for evaluation of the role of specific bases. FEBS Lett. 1993 Jul 12;326(1-3):158–162. doi: 10.1016/0014-5793(93)81782-u. [DOI] [PubMed] [Google Scholar]
- Suh Y. A., Kumar P. K., Nishikawa F., Kayano E., Nakai S., Odai O., Uesugi S., Taira K., Nishikawa S. Deletion of internal sequence on the HDV-ribozyme: elucidation of functionally important single-stranded loop regions. Nucleic Acids Res. 1992 Feb 25;20(4):747–753. doi: 10.1093/nar/20.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner N. K., Schaff S., Thill G., Petit-Koskas E., Crain-Denoyelle A. M., Westhof E. A three-dimensional model of hepatitis delta virus ribozyme based on biochemical and mutational analyses. Curr Biol. 1994 Jun 1;4(6):488–498. doi: 10.1016/s0960-9822(00)00109-3. [DOI] [PubMed] [Google Scholar]
- Thill G., Blumenfeld M., Lescure F., Vasseur M. Self-cleavage of a 71 nucleotide-long ribozyme derived from hepatitis delta virus genomic RNA. Nucleic Acids Res. 1991 Dec 11;19(23):6519–6525. doi: 10.1093/nar/19.23.6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thill G., Vasseur M., Tanner N. K. Structural and sequence elements required for the self-cleaving activity of the hepatitis delta virus ribozyme. Biochemistry. 1993 Apr 27;32(16):4254–4262. doi: 10.1021/bi00067a013. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Wang K. S., Choo Q. L., Weiner A. J., Ou J. H., Najarian R. C., Thayer R. M., Mullenbach G. T., Denniston K. J., Gerin J. L., Houghton M. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature. 1986 Oct 9;323(6088):508–514. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- Wu H. N., Huang Z. S. Mutagenesis analysis of the self-cleavage domain of hepatitis delta virus antigenomic RNA. Nucleic Acids Res. 1992 Nov 25;20(22):5937–5941. doi: 10.1093/nar/20.22.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. N., Lee J. Y., Huang H. W., Huang Y. S., Hsueh T. G. Mutagenesis analysis of a hepatitis delta virus genomic ribozyme. Nucleic Acids Res. 1993 Sep 11;21(18):4193–4199. doi: 10.1093/nar/21.18.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. N., Lin Y. J., Lin F. P., Makino S., Chang M. F., Lai M. M. Human hepatitis delta virus RNA subfragments contain an autocleavage activity. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1831–1835. doi: 10.1073/pnas.86.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. N., Wang Y. J., Hung C. F., Lee H. J., Lai M. M. Sequence and structure of the catalytic RNA of hepatitis delta virus genomic RNA. J Mol Biol. 1992 Jan 5;223(1):233–245. doi: 10.1016/0022-2836(92)90728-3. [DOI] [PubMed] [Google Scholar]
- Yie Y., Wei Z., Tien P. A simplified and reliable protocol for plasmid DNA sequencing: fast miniprep and denaturation. Nucleic Acids Res. 1993 Jan 25;21(2):361–361. doi: 10.1093/nar/21.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]



