Abstract
The principal role of the placenta is the maintenance of pregnancy and promotion of fetal growth and viability. The use of transgenic rodents has greatly enhanced our understanding of placental development and function. However, embryonic lethality is often a confounding variable in determining whether a genetic modification adversely affected placental development. In these cases, it is beneficial to specifically manipulate the placental genome. The purpose of this review is to summarize available methodologies for specific genetic modification of the rodent placenta. By restricting genetic alterations to the trophoblast lineage, it is possible to gain a deeper understanding of placental development that perhaps will lead to gene-targeted therapies to rescue irregular placentation in transgenic animals or in women at high-risk for placenta-associated pregnancy complications.
Keywords: Placenta, trophoblast, genetic manipulation, In vivo techniques
1. Introduction
The placenta functions to facilitate the exchange of molecules between maternal and fetal circulations and to provide endocrinological and immunological support for pregnancy. Aberrant placental development is implicated in many pregnancy complications, impacting both fetal development and maternal health [1]. Therefore, gaining a deeper understanding of placental development, both normal and irregular, is of significant clinical importance.
The placenta contains anatomically distinct zones wherein architecture and function is dependent on the progressive differentiation of trophoblast cells. Trophoblast cells comprise a heterogeneous population of cells derived from progenitor trophoblast stem (TS) cells [2]. Trophectoderm, the eventual source of TS cells, is the first lineage-committed cell-type to appear during embryogenesis, emerging as polarized cells segregated to the periphery of the blastocyst. The remaining internal cells form an inner cell mass from which two populations of lineage-committed cells have been derived: extraembryonic endoderm (XEN) and embryonic stem (ES) cells [3]. While XEN and ES cells are fated to form yolk sac endoderm and all germ layers of the fetus, respectively, TS cells undergo a multilineage differentiation pathway resulting in distinct subpopulations of specialized trophoblast cells. Characterization of these trophoblast subtypes has revealed unique ontogeny, morphology, and function [4].
In rodents, the placenta can be divided into two regions: the junctional zone and the labyrinth zone. The junctional zone is an active endocrinological and invasive tissue situated at the uterine-trophoblastic border, and is comprised of trophoblast giant cells, spongiotrophoblast cells, and glycogen cells. Later in gestation, the invasive trophoblast lineage develops within the junctional zone, and actively migrates into the maternal decidua [5]. The labyrinth zone is the site at which transfer of nutrients between maternal and fetal circulations occurs, and is comprised of mesenchymal villous structures bordered by fused syncytiotrophoblast and a discontinuous cytotrophoblast layer bathing in maternal blood. Although some aspects of placentation differ between humans and rodents, there are fundamental similarities between these species in terms of placental structure, development, and function [6–8]. Thus, the capacity to genetically modify rodent trophoblast subpopulations, or the placenta in general, provides valuable information on processes integral for placental development and function.
The purpose of this review is to discuss available methodologies for genetic modification of the rodent placenta. Whole-body gene manipulation has enabled researchers to identify roles for genes in many tissues; however placental function is inherently vital for fetal viability; thus embryonic lethality is often a confounding variable in deciphering whether a genetic modification affected placental or fetal development. By restricting the genetic manipulation to the placenta, it is possible to more clearly define the effects on placental development and function. Techniques for genetic modification of the placenta, which are represented schematically in Figure 1, will be categorized as follows: i) transgene expression using trophoblast-specific promoters; ii) cell-based modulation of placental gene expression, and iii) viral strategies for genetic manipulation of the placenta.
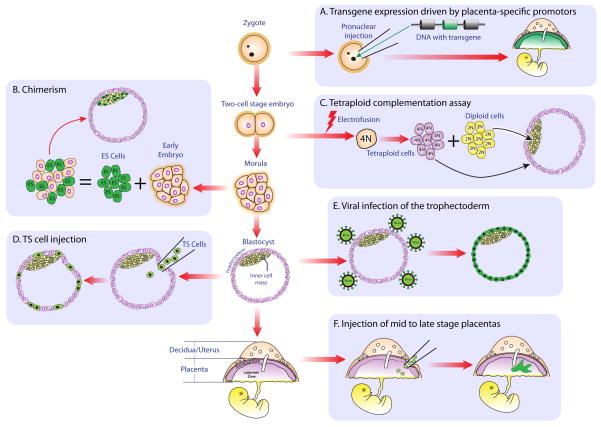
Figure 1. Schematic representation of techniques to facilitate genetic modification of the rodent placenta.
Unboxed flow chart in middle of diagram depicts progression from zygote through to mature conceptus. A) Pronuclear injection of transgene-containing DNA driven by placenta-specific promoter. Note that only the mature placenta expresses the transgene (green). B) Generating a chimera by aggregating an early embryo and transgenic ES cells. Please observe that ES cells are only capable of contributing to the inner cell mass. C) Tetraploid complementation. Electrofusion of a two-cell stage embryo results in the generation of a single tetraploid cell. Tetraploid cells continue to proliferate and develop normally up until the blastocyst stage, but rarely contribute to embryonic structures. Therefore, aggregation of tetraploid and diploid embryos results in lineage exclusivity, with tetraploid cells wholly contributing to extraembryonic tissue. D) Injection of TS cells into the blastocoel. Injected TS cells contribute exclusively to the trophectoderm. E) Viral infection of a blastocyst results in transduction of the peripheral trophectoderm, whereas inner cells are shielded from viral exposure. F) Intraplacental engraftment of cells or injection of virus to mature placentae, resulting in regions of placenta that express foreign cellular proteins or are transduced with virus, respectively.
2. Trophoblast-specific gene promoter mediated manipulations
Genetic modulation of the placenta eliminates the ambiguity associated with embryonic lethality in whole-body transgenics. This approach is made feasible by insertion of genes downstream of promoters that are active in specific trophoblast subpopulations. Inserted genes are then expressed only in trophoblast subtypes containing the active promoters, where they facilitate gain-of-function or loss-of-function effects. The critical limitation for this strategy is the identification of gene promoters that are active with relative exclusivity in trophoblast cell-types. Thus far, several such genes have been recognized (Table 1). In some cases, such as with the purine-catabolizing enzyme adenine deaminase (ADA), the gene is active in all trophoblast derivatives [9]. Trophoblast ADA expression, which commences at mid-gestation and increases until term, protects the embryo from purine cytotoxicity. Using transgenic mice, it was found that a 770-bp region situated 5.4-kb upstream of the ADA transcription start site is sufficient to drive expression of a reporter construct exclusively in the mouse placenta [10]. Therefore, transgene insertion downstream of the ADA promoter may be a putative means of expressing transgenes in all trophoblast derivatives.
Table 1.
Characterized gene promoters effective in targeting trophoblast lineages in vivo
| Gene | Function/Expression | Relevant promoter regions necessary for trophoblast-specific expression | Relevant References |
|---|---|---|---|
| Adenosine deaminase | Purine metabolizing enzyme; expressed in all mouse trophoblast subpopulations beginning at mid-gestation. | 770-bp region situated 5.4 kb upstream of transcription start site; contains binding site for trophoblast-specific element binding protein. | [9,10] |
| CYP19 | Aromatase complex; converts androgens to estrogens; expressed in human, but not mouse placenta. The human promoter is active in the mouse placenta. | 501-bp 5′-flanking region of placenta-specific Cyp19 exon I.1. | [30–32] |
| Eomesodermin | Regulation of trophoblast lineage specification; expressed in TS cells and early trophoblast cells, primitive streak, telencephalon. | 180-kb 5′-sequence on bacterial artificial chromosome sufficient to drive expression. | [25] |
| Growth hormone releasing hormone | Hypothalamic hormone that stimulates release of growth hormone; expressed by rodent placenta; expression commences at mid-gestation and continues increasing until term. | 7.5-kb-5′ flanking region and sequences downstream to exon 1. | [24] |
| Human chorionic somato-mammotropin | Human growth hormone gene cluster; expressed in human syncytiotrophoblast at mid-gestation. No mouse homologue, but transcriptionally active in mice. | Histone acetylation and differential protein binding within a remote locus control region 14–32 kb upstream of gene cluster. | [27–29] |
| Placental lactogen II (PL-II; Prl3b1) | Member of expanded rodent prolactin-like protein superfamily; expressed by rodent trophoblast giant cells and in mice, spongiotrophoblast cells beginning at mid-gestation. | 2019 to 1340 bp upstream of transcription start site; contains elements for GATA, Ets, and AP-2γ factor binding. | [12–14] |
| Renin | Involved in regulation of blood volume and blood pressure; expressed in fetal adrenal gland, Wolffian duct, and trophoblast giant cells during mid-late pregnancy. | 4-kb 5′-flanking region. | [23] |
| Tpbpa (4311) | Unknown function; expressed in junctional zone trophoblast. | 340-bp region situated 3740 bp upstream of transcription start site. Contains E-box binding elements for basic helix-loop-helix transcription factor binding. | [21,22] |
Many genes exhibit a temporal or cell-specific expression pattern in the placenta and may be useful for targeting trophoblast subpopulations during certain gestational periods. Such is the case with the rodent prolactin (PRL) superfamily. The genes that encode these proteins are organized contiguously on the same chromosome. Each gene shows a characteristic temporal and trophoblast cell-specific expression pattern [11]. Although the promoter motifs that drive in vivo expression of most PRL family members are unknown, the endogenous placental lactogen-II (PL-II; Prl3b1) promoter, which is activated in rodent trophoblast giant cells and, at least in mice, spongiotrophoblast cells during the latter half of pregnancy, has been effectively used to target gene expression to the mouse placenta [12,13]. In particular, a region spanning −2019 to −1340 bp upstream of the PL-II transcription start site drove reporter expression specifically in the placenta of transgenic mice [14]. The promoter regions of several other PRL family members, including PL-I/Prl3d1 [15–17], PLP-A/Prl4a1 [18], dPRP/Prl8a2 [19], and PLP-Cv/Prl8a3 [20], have been characterized in vitro using the Rcho-1 rat trophoblast cell line, but the efficiency of these promoter regions in driving trophoblast-specific gene expression in vivo is unknown. Characterization of the promoter regions of these and other PRL family genes would be invaluable in targeting transgenes to specific trophoblast subpopulations at precise gestational periods.
The promoter regions of other genes that are expressed in trophoblast cells have been used to promote transgene expression within the placenta in vivo. One example is the promoter region for Tpbpa (4311), which exhibits restricted expression within a subset of cells of the ectoplacental cone commencing at gestational day 7.5 in mice, and subsequently in trophoblast cells comprising the junctional zone for the duration of pregnancy. Promoter-reporter analyses identified a 340-bp sequence approximately 3740-bp upstream of the Tpbpa transcription start site required for trophoblast-specific expression [21]. This promoter region has been used to drive expression of Cre-recombinase and green fluorescent protein (GFP) exclusively in junctional zone trophoblast cells [22], demonstrating its efficacy for directing transgene expression in this region of the placenta. The promoter regions of other genes that have been characterized to drive in vivo transgene expression in the placenta include renin (expressed in trophoblast giant cells) [23], growth hormone-releasing hormone (expressed in junctional zone trophoblast cells) [24], and eomesodermin (expressed in early trophoblast progenitors) [25]. However, transgene expression in tissues other than trophoblast was also noted for each gene, so the promoter regions for these genes may not yet be ideal for directing placenta-specific transgenesis.
The promoter regions of the genes discussed above have been used to drive transgene expression within the junctional zone. Modulation of promoters active in rodent labyrinth zone syncytiotrophoblast has been less successful, although characterization of the promoter regions of the recently identified Syncytin genes [26], may be a possible means to drive transgenes exclusively in rodent labyrinth. Regardless, the most effective means thus far for genetic manipulation of rodent syncytiotrophoblast has been to generate animals expressing transgenes driven by promoter regions active in human syncytiotrophoblast. For instance, the upstream locus control region (LCR) of human chorionic somatomammotropin (hCS), a member of the human growth hormone (hGH) gene cluster activated exclusively in human syncytiotrophoblast, is effective in driving transgene expression specifically in rodent syncytiotrophoblast despite the lack of a rodent ortholog for this gene [27–29]. Therefore, the necessary transcriptional machinery required to activate the LCR and upstream promoter sequences for hCS is present in mouse syncytiotrophoblast. Likewise, the CYP19 (aromatase) gene, which facilitates the conversion of androgens to estrogens, is expressed in human syncytiotrophoblast but not in rodent placenta. Using reporter constructs driven by the CYP19 promoter, Kamat et al. have observed that 501-bp upstream of CYP19 exon I.1, an exon encoding the 5′-untranslated region of CYP19 transcripts exclusively transcribed in human placenta, is sufficient to induce reporter expression in mouse syncytiotrophoblast [30,31]. Another group has used this 501-bp hCYP19 promoter to drive expression of Cre recombinase in mouse placenta. However, in this case the transgene was detected in all trophoblast derivatives rather than just syncytiotrophoblast [32]. Regardless, the promoter regions that drive expression of hCS and CYP19 in human syncytiotrophoblast are effective means of driving transgene expression specifically in the rodent placenta.
3. Transplantation of cells to modulate placental gene expression and function
Transplantation of cells is an effective means to introduce genetically modified cells into a placenta. This methodology involves combining two or more populations of cells from different genetic backgrounds to produce a conceptus comprised of several populations of genetically-distinct cells. Two approaches have been used to accomplish this feat: intraplacental engraftment and chimerism.
3.1. Intraplacental engraftment
Engraftment of cells to the placenta was a technique established by Senut et al. [33]. In their study, cultured rat or human cells genetically modified to express either LacZ or human growth hormone (hGH) were injected into rat placentas. Following injection, cells successfully engrafted to the placenta as evidenced by beta-galactosidase activity and the detection of hGH in placental and fetal circulations [33]. Therefore, this methodology may be an effective means to continuously deliver biologically active molecules in utero. The primary limitation of this procedure was the injurious nature of injection, which dramatically reduced fetal viability when performed earlier than embryonic day (E)15 in rats. In addition, engraftment of foreign cells was associated with instigation of an inflammatory response, which could affect the delicate immune balance required for successful pregnancy. Regardless, refinements may allow for this technique to be an effective means for experimentally manipulating the placenta.
3.2. Chimerism
Chimerism is a technique whereby embryos from different genetic backgrounds are aggregated together to form a single embryo that has two or more populations of genetically distinct cells. Initially this methodology was restricted to the aggregation of early mouse embryos, resulting in chimerism in both embryonic and extraembryonic (derivatives of trophectoderm and primitive endoderm) lineages. Advances in the stability and culture of ES cells have facilitated an alternative source of embryonic cells in chimera generation. Despite the technically arduous nature of these procedures, chimeras have been an invaluable tool in the production of mutant animals. In keeping with the placenta-centric theme of this review, we will focus only on the use of chimeras for deciphering the effects of genetic modifications on extraembryonic lineage development. For a detailed description of chimera generation and applications, see Tam and Rossant [34].
Since ES cells differentiate into embryonic structures and are incapable of contributing to primitive endoderm or trophectoderm derivatives, ES cell-embryo chimeras are a useful tool to delineate lineage-specific consequences of genetic mutations [35]. Determining the effect of a mutation on embryonic versus extraembryonic tissues then depends on the source of the genetic modification i.e. whether the mutation is carried by ES cells or the embryo. However, regardless of the source the genetic constitution of the embryo is a mixture of mutant and wild-type cells, making results obtained by this technique somewhat inconclusive.
To more specifically determine the impact of genetic modifications on embryonic versus extraembryonic tissues, an amendment to conventional chimera generation can be performed to exclusively direct cells towards extraembryonic development. This adjustment entails electrofusion of a two-cell stage embryo, resulting in a single cell with twice the normal number of chromosomes, i.e. tetraploid. Generation of a chimera using a pre-implantation tetraploid embryo aggregated with diploid ES cells results in lineage exclusivity, with tetraploid cells contributing wholly to the formation of extraembryonic structures and ES cells restricted to producing embryonic entities [36].
Tetraploid complementation has been used to decipher whether embryonic lethality as a result of gene targeting is due to deficiencies in embryonic or extraembryonic structures. For example, rescue of embryonic lethality in SOCS3-deficient embryos, which normally die in utero, was facilitated by aggregation of SOCS3-deficient embryos with tetraploid wild-type embryos [37]. Unfortunately, tetraploid complementation does not allow for discerning whether failure of extraembryonic tissue development is due to defects in trophoblastic or primitive endodermal derivatives. To address this issue, Takahashi et al. performed a novel chimeric analysis by microinjecting wild-type TS cells into the blastocoel of SOCS3-defective embryos [38]. These injected TS cells successfully contributed to the formation of the placenta, resulting in partial rescue of embryonic lethality. Although these authors found this methodology to be less effective than tetraploid complementation, future refinements to this technique could provide an effective means to rescue lethalities due to placental insufficiency and also to assess the competencies of TS cells possessing specific mutations.
4. Viral delivery strategies for genetic manipulation of the placenta
An alternative approach to produce genetic modification of the placenta is to directly introduce novel genetic material to the organ itself. One of the most effective means to accomplish this is to use viral gene delivery. Recombinant viruses have successfully been used in vitro as gene delivery vehicles to isolated cells or tissues, but in vivo studies have been somewhat slower to adopt this technology because of the potential for carcinogenicity, immunogenicity, and inconsistency of transgene expression [39]. However, due to the transitory, extraembryonic nature of the placenta, this organ could be an ideal future candidate for viral-mediated gene delivery in animal models and in patients who exhibit a high risk for some pregnancy complications. For research purposes, the most commonly used vectors for mediation of gene delivery have been adenoviruses and retroviruses.
4.1. Adenoviruses
Adenoviruses are non-enveloped icosahedral viruses composed of a nucleocapsid and a double-stranded linear DNA genome. They are capable of transducing both dividing and non-dividing cells and confer high levels of transgene expression. Once inside the host cell, adenoviral DNA enters the nucleus but is not integrated into the genetic material of the host; hence it is not transmitted to daughter cells. Moreover, adenoviral particles are recognized by the innate immune system, resulting in efficient clearance of virus. Therefore, because adenoviruses are not transmitted to daughter cells and are efficiently cleared by the immune system, recombinant adenoviral vectors are useful options for gene delivery to a slow-replicating population of cells for a defined period of time.
Administration of recombinant adenoviral vectors to pregnant mice, rats, rabbits, and sheep has been used to confer transgene expression in both the fetus and placenta. Although systemic administration of these vectors is effective in transducing fetal and placental cells, in utero inoculation is more effective in its specificity and infectivity for the fetal-placental unit [40]. In particular, direct inoculation of the placenta with recombinant adenovirus has been successfully used to alter gene expression specifically in this organ. For example, Xing et al. used an intra-placental injection of recombinant adenovirus to pregnant rats on E14 to induce expression of the human glucose transporter GLUT3 [40]. A later study in pregnant mice used intra-placental injection of adenovirus expressing various growth factor transgenes to determine the effect of these mitogens on late gestation placental and fetal growth [41].
Despite the promising foundations provided by these studies, adenoviral-mediated gene delivery to the placenta exhibits several limitations. First, adenovirus infection is capable of eliciting an immune response, and this could have repercussions for the delicate immune balance required for pregnancy. Second, efficient adenoviral infection and transgene expression is restricted to a narrow time-window. In mice and rats, researchers have been limited to injecting mid- to late-gestation placentas, partly because of the lack of adenoviral transmittance to daughter cells and because intra-placental injection is an injurious technique that may reduce fetal viability if performed earlier than E13.5. Others also noted that if injection of adenovirus was delayed until later than E16.5 in rat pregnancy, transgene expression in the placenta was diminished, indicating resistance to infection [40]. Despite these limitations, the transient nature of adenovirus infection, failure to stably integrate into the host genome, and high levels of transgene expression make adenovirus-mediated gene delivery a good candidate for some types of placental research.
4.2. Retroviruses
Retroviruses are enveloped viruses containing an RNA genome and proteins including reverse transcriptase. Once inside a host cell, reverse transcriptase catalyzes the conversion of the viral RNA genome to complementary DNA, which then stably integrates into the genetic material of the host cell and is transmitted to cell progeny. Although most retroviruses are only capable of low-level transgene expression and infection is restricted to dividing cells, a subset of retroviruses - lentiviruses - can transduce dividing or non-dividing cells and is less prone to transgene silencing than conventional retroviruses [42]. Thus, lentivirus-based vectors are currently the preferred choice for retroviral-mediated transgene delivery.
Due to the stable transmittance of lentiviral infection to cell progeny, researchers have achieved extensive transgene expression at later stages of development in embryonic and extraembryonic tissues after exposure of early embryos to lentiviral vectors. Further research using these vectors was able to accomplish more specific extraembryonic transgene expression by exploiting the physical properties of blastocysts. In a landmark study using rhesus monkey blastocysts, Wolfgang et al. showed that transgene delivery could be restricted to trophectoderm and primitive endoderm derivatives by microinjecting lentiviral vectors into the blastocoel [43]. More recent studies in mice have achieved even greater specificity for trophectoderm and its derivatives by simply immersing zona pellucida-free blastocysts in medium containing lentivirus. Due to the peripheral positioning of the trophectoderm, these cells were readily transduced by lentivirus, but inner cells were shielded from infection by the epithelial tight junctions of the trophectoderm [44,45]. Specificity for the trophoblast lineage and efficiency of infection was confirmed by transferring blastocysts infected with lentivirus carrying reporter constructs to pseudopregnant dams, and observing reporter expression later in pregnancy exclusively in the placenta. Since these initial studies were performed, transgene expression via blastocyst transduction with lentiviral vectors has been effectively performed in mice [45], rats [46], and sheep [47], indicating that this is a feasible methodology for altering gene expression in the placenta in many species.
Lentivirus infection of blastocysts has been used as a means to deliver transgenes to the placenta resulting in gain-of-function or loss-of-function phenotypes. As an example of the former, in Ets2−/− or Mapk14−/− mice that both die in utero as a result of deficient placentation, reversal of embryonic lethality was achieved following blastocyst transduction by lentivirus carrying functional transgenes for Ets2 or Mapk14, respectively [44]. Loss-of-function phenotypes have been observed following lentiviral delivery of short hairpin-RNA (shRNA) specific for target genes. Although to-date transgenic shRNA-mediated gene silencing in rodents has targeted reporter genes [45,46], Purcell et al. have used this methodology to decrease Proline-Rich 15 gene expression in sheep trophectoderm in order to determine the role of this protein in conceptus elongation [47]. Thus, lentiviral delivery should be an effective strategy for genetically manipulating the trophoblast lineage from any mammalian species.
An alternative and more robust means for loss-of-function phenotypes is through lentiviral-mediated delivery of transgenes encoding Cre recombinase in animals expressing a targeted floxed gene. This methodology was first established by Georgiades et al., who infected trophectoderm with a viral vector encoding Cre recombinase. Following genomic integration of Cre recombinase, floxed reporter genes were excised as indicated by alterations in the expression of the appropriate reporters [45]. Further studies by Morioka et al. used lentiviral vectors for delivery of Cre recombinase to trophectoderm in order to excise floxed Ets2 in the host genome [48]. The obvious advantage of this technique is that a placenta-specific promoter is not required to drive expression of Cre recombinase, thereby increasing the feasibility of generating placenta-specific knockouts using animals containing a desired floxed gene. The limitations of this latter research approach is the need to manipulate endogenous genes via homologous recombination in a pluripotent cell population such as ES cells, which to date is only a routine procedure in mice.
5. Overview
In this review, we summarized currently established techniques for specific genetic modification of the rodent placenta. Rodent transgenesis has greatly facilitated scientific advancement in many fields, but has been of limited use in understanding placental biology due to embryonic lethality. However, the incorporation of new research strategies has enabled researchers to decipher the effect of a genetic modification on placental development and function. Given that placental irregularities are associated with various pregnancy complications, a further understanding of the processes governing placental development and function is clinically important. Moreover, refinement of these techniques may eventually allow for genetic modification of the human placenta to facilitate pregnancy success in high-risk cases of pregnancy complications due to placental irregularities.
Acknowledgments
We would like to thank Stanton Fenton for illustrative assistance, and all past and current members of our laboratory. SJR is supported by a post-doctoral fellowship awarded by the Lalor Foundation. This work was supported by grant HD020676 from the National Institutes of Health.
Footnotes
Conflict of Interest
The authors wish to declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Benirschke KKP, Baergen RN. Pathology of the Human Placenta. 5. Springer-Verlag; New York: 2006. [Google Scholar]
- 2.Simmons DG, Cross JC. Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev Biol. 2005;284:12–24. doi: 10.1016/j.ydbio.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Rossant J. Stem cells and lineage development in the mammalian blastocyst. Reprod Fertil Dev. 2007;19:111–8. doi: 10.1071/rd06125. [DOI] [PubMed] [Google Scholar]
- 4.Cross JC. How to make a placenta: mechanisms of trophoblast cell differentiation in mice--a review. Placenta. 2005;26 (Suppl A):S3–9. doi: 10.1016/j.placenta.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Ain R, Canham LN, Soares MJ. Gestation stage-dependent intrauterine trophoblast cell invasion in the rat and mouse: novel endocrine phenotype and regulation. Dev Biol. 2003;260:176–90. doi: 10.1016/s0012-1606(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 6.Pijnenborg R, Robertson WB, Brosens I, Dixon G. Review article: trophoblast invasion and the establishment of haemochorial placentation in man and laboratory animals. Placenta. 1981;2:71–91. doi: 10.1016/s0143-4004(81)80042-2. [DOI] [PubMed] [Google Scholar]
- 7.Georgiades P, Ferguson-Smith AC, Burton GJ. Comparative developmental anatomy of the murine and human definitive placentae. Placenta. 2002;23:3–19. doi: 10.1053/plac.2001.0738. [DOI] [PubMed] [Google Scholar]
- 8.Cox B, Kotlyar M, Evangelou AI, Ignatchenko V, Ignatchenko A, Whiteley K, Jurisica I, Adamson SL, Rossant J, Kislinger T. Comparative systems biology of human and mouse as a tool to guide the modeling of human placental pathology. Mol Syst Biol. 2009;5:279. doi: 10.1038/msb.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winston JH, Hanten GR, Overbeek PA, Kellems RE. 5′ flanking sequences of the murine adenosine deaminase gene direct expression of a reporter gene to specific prenatal and postnatal tissues in transgenic mice. J Biol Chem. 1992;267:13472–9. [PubMed] [Google Scholar]
- 10.Shi D, Winston JH, Blackburn MR, Datta SK, Hanten G, Kellems RE. Diverse genetic regulatory motifs required for murine adenosine deaminase gene expression in the placenta. J Biol Chem. 1997;272:2334–41. [PubMed] [Google Scholar]
- 11.Soares MJ, Konno T, Alam SM. The prolactin family: effectors of pregnancy-dependent adaptations. Trends Endocrinol Metab. 2007;18:114–21. doi: 10.1016/j.tem.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Shah P, Sun Y, Szpirer C, Duckworth ML. Rat placental lactogen II gene: characterization of gene structure and placental-specific expression. Endocrinology. 1998;139:967–73. doi: 10.1210/endo.139.3.5838. [DOI] [PubMed] [Google Scholar]
- 13.Shida MM, Jackson-Grusby LL, Ross SR, Linzer DI. Placental-specific expression from the mouse placental lactogen II gene promoter. Proc Natl Acad Sci U S A. 1992;89:3864–8. doi: 10.1073/pnas.89.9.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J, Linzer DI. Identification of trophoblast-specific regulatory elements in the mouse placental lactogen II gene. Mol Endocrinol. 1998;12:418–27. doi: 10.1210/mend.12.3.0078. [DOI] [PubMed] [Google Scholar]
- 15.Cho JH, Kimura H, Minami T, Ohgane J, Hattori N, Tanaka S, Shiota K. DNA methylation regulates placental lactogen I gene expression. Endocrinology. 2001;142:3389–96. doi: 10.1210/endo.142.8.8347. [DOI] [PubMed] [Google Scholar]
- 16.Ng YK, George KM, Engel JD, Linzer DI. GATA factor activity is required for the trophoblast-specific transcriptional regulation of the mouse placental lactogen I gene. Development. 1994;120:3257–66. doi: 10.1242/dev.120.11.3257. [DOI] [PubMed] [Google Scholar]
- 17.Shida MM, Ng YK, Soares MJ, Linzer DI. Trophoblast-specific transcription from the mouse placental lactogen-I gene promoter. Mol Endocrinol. 1993;7:181–8. doi: 10.1210/mend.7.2.8469232. [DOI] [PubMed] [Google Scholar]
- 18.Vuille JC, Cattini PA, Bock ME, Verstuyf A, Schroedter IC, Duckworth ML, Friesen HG. Rat prolactin-like protein A partial gene and promoter structure: promoter activity in placental and pituitary cells. Mol Cell Endocrinol. 1993;96:91–8. doi: 10.1016/0303-7207(93)90099-6. [DOI] [PubMed] [Google Scholar]
- 19.Orwig KE, Soares MJ. Transcriptional activation of the decidual/trophoblast prolactin-related protein gene. Endocrinology. 1999;140:4032–9. doi: 10.1210/endo.140.9.6954. [DOI] [PubMed] [Google Scholar]
- 20.Dai G, Wolfe MW, Soares MJ. Distinct regulatory regions from the prolactin-like protein C variant promoter direct trophoblast giant cell versus spongiotrophoblast cell-specific expression. Endocrinology. 1999;140:4691–8. doi: 10.1210/endo.140.10.7078. [DOI] [PubMed] [Google Scholar]
- 21.Calzonetti T, Stevenson L, Rossant J. A novel regulatory region is required for trophoblast-specific transcription in transgenic mice. Dev Biol. 1995;171:615–26. doi: 10.1006/dbio.1995.1309. [DOI] [PubMed] [Google Scholar]
- 22.Simmons DG, Fortier AL, Cross JC. Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev Biol. 2007;304:567–78. doi: 10.1016/j.ydbio.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Jones CA, Hurley MI, Black TA, Kane CM, Pan L, Pruitt SC, Gross KW. Expression of a renin/GFP transgene in mouse embryonic, extra-embryonic, and adult tissues. Physiol Genomics. 2000;4:75–81. doi: 10.1152/physiolgenomics.2000.4.1.75. [DOI] [PubMed] [Google Scholar]
- 24.Nogues N, Del Rio JA, Perez-Riba M, Soriano E, Flavell RA, Boronat A. Placenta-specific expression of the rat growth hormone-releasing hormone gene promoter in transgenic mice. Endocrinology. 1997;138:3222–7. doi: 10.1210/endo.138.8.5295. [DOI] [PubMed] [Google Scholar]
- 25.Kwon GS, Hadjantonakis AK. Eomes::GFP-a tool for live imaging cells of the trophoblast, primitive streak, and telencephalon in the mouse embryo. Genesis. 2007;45:208–17. doi: 10.1002/dvg.20293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dupressoir A, Marceau G, Vernochet C, Benit L, Kanellopoulos C, Sapin V, Heidmann T. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci U S A. 2005;102:725–30. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elefant F, Su Y, Liebhaber SA, Cooke NE. Patterns of histone acetylation suggest dual pathways for gene activation by a bifunctional locus control region. EMBO J. 2000;19:6814–22. doi: 10.1093/emboj/19.24.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin Y, Lu SY, Fresnoza A, Detillieux KA, Duckworth ML, Cattini PA. Differential placental hormone gene expression during pregnancy in a transgenic mouse containing the human growth hormone/chorionic somatomammotropin locus. Placenta. 2009;30:226–35. doi: 10.1016/j.placenta.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su Y, Liebhaber SA, Cooke NE. The human growth hormone gene cluster locus control region supports position-independent pituitary- and placenta-specific expression in the transgenic mouse. J Biol Chem. 2000;275:7902–9. doi: 10.1074/jbc.275.11.7902. [DOI] [PubMed] [Google Scholar]
- 30.Kamat A, Graves KH, Smith ME, Richardson JA, Mendelson CR. A 500-bp region, approximately 40 kb upstream of the human CYP19 (aromatase) gene, mediates placenta-specific expression in transgenic mice. Proc Natl Acad Sci U S A. 1999;96:4575–80. doi: 10.1073/pnas.96.8.4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamat A, Smith ME, Shelton JM, Richardson JA, Mendelson CR. Genomic regions that mediate placental cell-specific and developmental regulation of human Cyp19 (aromatase) gene expression in transgenic mice. Endocrinology. 2005;146:2481–8. doi: 10.1210/en.2004-1606. [DOI] [PubMed] [Google Scholar]
- 32.Wenzel PL, Leone G. Expression of Cre recombinase in early diploid trophoblast cells of the mouse placenta. Genesis. 2007;45:129–34. doi: 10.1002/dvg.20276. [DOI] [PubMed] [Google Scholar]
- 33.Senut MC, Suhr ST, Gage FH. Gene transfer to the rodent placenta in situ. A new strategy for delivering gene products to the fetus. J Clin Invest. 1998;101:1565–71. doi: 10.1172/JCI1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tam PP, Rossant J. Mouse embryonic chimeras: tools for studying mammalian development. Development. 2003;130:6155–63. doi: 10.1242/dev.00893. [DOI] [PubMed] [Google Scholar]
- 35.Robertson E, Bradley A, Kuehn M, Evans M. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature. 1986;323:445–8. doi: 10.1038/323445a0. [DOI] [PubMed] [Google Scholar]
- 36.Tarkowski AK, Witkowska A, Opas J. Development of cytochalasin in B-induced tetraploid and diploid/tetraploid mosaic mouse embryos. J Embryol Exp Morphol. 1977;41:47–64. [PubMed] [Google Scholar]
- 37.Takahashi Y, Carpino N, Cross JC, Torres M, Parganas E, Ihle JN. SOCS3: an essential regulator of LIF receptor signaling in trophoblast giant cell differentiation. EMBO J. 2003;22:372–84. doi: 10.1093/emboj/cdg057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi Y, Dominici M, Swift J, Nagy C, Ihle JN. Trophoblast stem cells rescue placental defect in SOCS3-deficient mice. J Biol Chem. 2006;281:11444–5. doi: 10.1074/jbc.C600015200. [DOI] [PubMed] [Google Scholar]
- 39.Wilson JM. Adenoviruses as gene-delivery vehicles. N Engl J Med. 1996;334:1185–7. doi: 10.1056/NEJM199605023341809. [DOI] [PubMed] [Google Scholar]
- 40.Xing A, Boileau P, Cauzac M, Challier JC, Girard J, Hauguel-de Mouzon S. Comparative in vivo approaches for selective adenovirus-mediated gene delivery to the placenta. Hum Gene Ther. 2000;11:167–77. doi: 10.1089/10430340050016247. [DOI] [PubMed] [Google Scholar]
- 41.Katz AB, Keswani SG, Habli M, Lim FY, Zoltick PW, Midrio P, Kozin ED, Herlyn M, Crombleholme TM. Placental gene transfer: transgene screening in mice for trophic effects on the placenta. Am J Obstet Gynecol. 2009;201:499, e1–8. doi: 10.1016/j.ajog.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfeifer A, Ikawa M, Dayn Y, Verma IM. Transgenesis by lentiviral vectors: lack of gene silencing in mammalian embryonic stem cells and preimplantation embryos. Proc Natl Acad Sci U S A. 2002;99:2140–5. doi: 10.1073/pnas.251682798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolfgang MJ, Eisele SG, Browne MA, Schotzko ML, Garthwaite MA, Durning M, Ramezani A, Hawley RG, Thomson JA, Golos TG. Rhesus monkey placental transgene expression after lentiviral gene transfer into preimplantation embryos. Proc Natl Acad Sci U S A. 2001;98:10728–32. doi: 10.1073/pnas.181336098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okada Y, Ueshin Y, Isotani A, Saito-Fujita T, Nakashima H, Kimura K, Mizoguchi A, Oh-Hora M, Mori Y, Ogata M, Oshima RG, Okabe M, Ikawa M. Complementation of placental defects and embryonic lethality by trophoblast-specific lentiviral gene transfer. Nat Biotechnol. 2007;25:233–7. doi: 10.1038/nbt1280. [DOI] [PubMed] [Google Scholar]
- 45.Georgiades P, Cox B, Gertsenstein M, Chawengsaksophak K, Rossant J. Trophoblast-specific gene manipulation using lentivirus-based vectors. Biotechniques. 2007;42:317–8. 320, 322–5. doi: 10.2144/000112341. [DOI] [PubMed] [Google Scholar]
- 46.Lee DS, Rumi MA, Konno T, Soares MJ. In vivo genetic manipulation of the rat trophoblast cell lineage using lentiviral vector delivery. Genesis. 2009;47:433–9. doi: 10.1002/dvg.20518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purcell SH, Cantlon JD, Wright CD, Henkes LE, Seidel GE, Jr, Anthony RV. The involvement of proline-rich 15 in early conceptus development in sheep. Biol Reprod. 2009;81:1112–21. doi: 10.1095/biolreprod.109.076190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morioka Y, Isotani A, Oshima RG, Okabe M, Ikawa M. Placenta-specific gene activation and inactivation using integrase-defective lentiviral vectors with the Cre/LoxP system. Genesis. 2009;47:793–8. doi: 10.1002/dvg.20563. [DOI] [PubMed] [Google Scholar]