Abstract
Objectives
The antistaphylococcal pyrrolobenzodiazepine dimer ELB-21 forms multiple adducts with duplex DNA through covalent interactions with appropriately spaced guanine residues; it is now known to form interstrand and intrastrand adducts with oligonucleotide sequences of variable length. We determined the DNA sequence preferences of ELB-21 in relation to its capacity to exert a bactericidal effect by damaging DNA.
Methods
Formation of adducts by ELB-21 and 12- to 14-mer DNA duplexes was investigated using ion-pair reversed phase liquid chromatography and mass spectrometry. Drug-induced changes in gene expression were measured in prophage-free Staphylococcus aureus RN4220 by microarray analysis.
Results
ELB-21 preferentially formed intrastrand adducts with guanines separated by three nucleotide base pairs. Interstrand and intrastrand adducts were formed with duplexes both longer and shorter than the preferred target sequences. ELB-21 elicited rapid bactericidal effects against prophage-carrying and prophage-free S. aureus strains; cell lysis occurred following activation and release of resident prophages. Killing appeared to be due to irreparable damage to bacterial DNA and susceptibility to ELB-21 was governed by the capacity of staphylococci to repair DNA lesions through induction of the SOS DNA damage response mediated by the RecA-LexA pathway.
Conclusions
The data support the contention that ELB-21 arrests DNA replication, eliciting formation of ssDNA-RecA filaments that inactivate LexA, the SOS repressor, and phage repressors such as Cl, resulting in activation of the DNA damage response and de-repression of resident prophages. Above the MIC threshold, DNA repair is ineffective.
Keywords: antistaphylococcal activity, pyrrolobenzodiazepine dimer, DNA cross-linking, DNA adduct formation, MRSA
Introduction
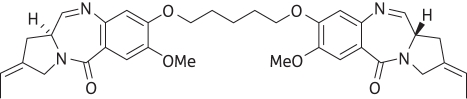
ELB-21 (Figure 1) is a pyrrolo[2,1-c][1,4]benzodiazepine (PBD) dimer that shows potent in vitro bactericidal activity against a wide range of Gram-positive clinical isolates, including methicillin-resistant strains of Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE).1 This symmetrical agent belongs to a class of molecules that binds in a sequence-selective manner the minor groove of the DNA helix and cross-links opposing DNA strands by forming covalent bonds with appropriately separated guanine residues.2–4 The five-carbon diether linking the PBD units (Figure 1) facilitates preferential interstrand binding to 5′-Pu-GATTC-Py and 5′-Pu-GAATC-Py base-pair sequences (Pu, purine; Py, pyrimidine; reactive guanines shown in bold). Recently DNA footprinting studies using substrates containing symmetrical hexanucleotide sequences enabled the identification of further high-affinity binding sites: 5′-GAATTC-3′, 5′-GTTAAC-3′ and 5′-GTATAC-3′, flanked by a Pu at the 5′ and a Py at the 3′ end.3 In addition, a number of intrastrand binding sites were identified that may also block DNA replication and contribute to the potent antistaphylococcal profile of ELB-21. These sequences, which were not predicted from initial molecular modelling studies,5 included short (5′-GATG-3′) and extended (5′-GTTAACCG-3′) flanked targets. These observations increased the number of potential DNA binding sites within the genome of the nosocomial MRSA isolate EMRSA-16 to around 40 000.3 As the footprinting substrates were designed to investigate binding to the preferred interstrand sequences,6 the genomes of target Gram-positive bacteria could contain further as-yet-unidentified interstrand and intrastrand binding sites.
Figure 1.
Structure of the pyrrolobenzodiazepine dimer ELB-21.
At supra-inhibitory concentrations, ELB-21 elicited potent bactericidal activity against MRSA and VRE isolates, with a greater than 4 log reduction in viability over a 2 h period;1 DNA extracted from ELB-21-treated MRSA was cross-linked, as evidenced by substantial increases in DNA melting temperature and increased molecular weight of DNA adducts identified by Southern blot analysis.7 Exposure of EMRSA-16 to subinhibitory concentrations of ELB-21 resulted in up-regulation of more than 160 genes in both the logarithmic and stationary phases of bacterial growth3 and, surprisingly in view of the capacity of PBD dimers to prevent DNA strand separation and transcription initiation,8 twofold or greater down-regulation of only 16 genes. Almost half of the genes affected by ELB-21 were induced prophage genes associated with the resident S. aureus prophages φSa2 and φSa3. All sequenced S. aureus strains, with the exception of COL, carry φSa3; φSa2 is integrated into the EMRSA-16 genome.9 Genes associated with the staphylococcal pathogenicity island SaPI4 were also up-regulated, as were genes indicative of a DNA damage (SOS) response; 10 genes, including those encoding the LexA repressor, recombinase A, topoisomerase and nucleotide excision repair proteins, were modulated by ELB-21. Transcriptional and proteomic analysis provided little evidence that ELB-21 selectively down-regulated genes involved in key cellular functions or bacterial pathogenicity.3
The SOS response in S. aureus has been characterized using non-selective DNA interactive agents such as mitomycin C (MMC), an anti-tumour antibiotic with substantial anti-staphylococcal activity.10,11 MMC produces intrastrand and interstrand cross-links in addition to monofunctional alkyl lesions in DNA12 and at supra-inhibitory concentrations modulates the transcriptional expression of a large number of S. aureus genes.13 Although the SOS response elicited by subinhibitory concentrations of ELB-21 appears to be more restricted than that associated with MMC, both induce prophage into the lytic cycle with concomitant release of phage particles.14 Indeed, the prophage-inducing capacity of MMC has been extensively exploited to obtain prophages from the lysogenized host.15–17 As PBD dimers bind selectively within the minor groove of duplex DNA, they may have the potential to target sequences in bacterial genes with essential housekeeping functions or to inhibit the transcription of genes necessary for bacterial pathogenesis.
Our recent finding that PBD dimers form intrastrand as well as interstrand adducts with DNA3,18 adds a further layer of complexity to the interaction of these agents with staphylococcal cells. As interstrand cross-links appear to be more stable than intrastrand covalent adducts,18 we determined the binding kinetics of ELB-21 for a range of cross-linking sequences, differing in the position of the two reactive guanine bases, using an assay that combines high-performance liquid chromatography and mass spectrometry (HPLC/MS)19 to shed light on the oligonucleotide binding preferences of the molecule. The induction of a large number of prophage genes by ELB-21 may mask more subtle changes in bacterial transcription; we therefore examined the effects of ELB-21 and MMC on gene expression in a S. aureus strain cured of prophages to better compare the core response at subinhibitory concentrations of these drugs. The data demonstrate a comparable and restricted transcriptional response to these two agents. The lytic, but not bactericidal, activity of ELB-21 appears to be due solely to activation of resident prophages within target bacteria. We conclude that ELB-21 displays a complex DNA binding profile that elicits a general DNA damage response in staphylococci comparable to MMC; the PBD dimer preferentially forms DNA intrastrand complexes by virtue of covalent interactions with appropriately spaced guanine residues.
Materials and methods
Bacterial strains, culture and antibacterial agents
S. aureus EMRSA-16 is a MRSA isolate from the Royal Free Hospital (London, UK). S. aureus RN4220, a derivative of NCTC 8325 cured of all prophages,20,21 was the gift of Friedrich Götz (University of Tübingen, Germany). S. aureus RN981 and RN1030 are recA− mutants derived from, respectively, RN450 and RN451; RN450 (8325-4) is a prophage-free derivative of NCTC 8325 and RN451 is RN450 but lysogenic for ϕ11.22,23 RN450, RN451, RN981 and RN1030 were kindly provided by José Penadés (Instituto Valenciano de Investigaciones Agrarias, Segorbe, Spain). Bacteria were grown in Mueller-Hinton (MH) broth (Oxoid) or on MH agar plates at 37°C. The PBD dimer ELB-21 was obtained from Spirogen Ltd. (London, UK) and was synthesized as described.4 MMC was obtained from Sigma (Poole, UK). MICs were determined by the CLSI (formerly NCCLS) broth microplate assay as previously described.7 For MIC determination, agents were dissolved in DMSO prior to dilution in broth; at the concentrations used, the solvent had no effect on bacterial growth. For cross-linking studies, ELB-21 was maintained as a 3 mM stock solution in 50/50 v/v methanol/water at −20°C for no longer than 4 months. Working solutions of 200 μM were prepared by diluting stock solution with nuclease-free water; these were stored at −20°C for no more than 1 week. Lytic phage particles in bacterial cultures were enumerated using the soft agar overlay method21 and RN450 as indicator strain.
Transmission electron microscopy
Phages were fixed in 1.5% glutaraldehyde for at least 2 h at room temperature, treated with osmium tetroxide and embedded in epoxy resin. Sectioning and staining with uranyl acetate was followed by Reynolds’ lead citrate. Ultrathin sections were viewed and photographed using a Philips 201 transmission electron microscope.
Microarray analysis
Total bacterial RNA was purified from each sample, labelled and hybridized to the BμG@S SAv1.1.0 microarray as described previously.3 This array has been described elsewhere24 and contains PCR products representing all predicted open reading frames from the initial seven S. aureus genome sequencing projects. The array design is available in BμG@Sbase (accession number: A-BUGS-17; http://bugs.sgul.ac.uk/A-BUGS-17) and also ArrayExpress (accession number: A-BUGS-17). Hybridization data were analysed using an Affymetrix 428 scanner then quantified using Bluefuse for Microarrays 3.5 software (BlueGnome). Data analysis was performed in GeneSpring GX 7.3 (Agilent Technologies) using median-normalized Cy5/Cy3 ratio intensities for three biological replicates. Only genes whose expression ratio showed at least a twofold difference with a Benjamini and Hochberg false discovery rate ≤0.05% in the presence of ELB-21 were regarded as being significantly different from the control. Fully annotated microarray data have been deposited in BμG@Sbase (accession number E-BUGS-115 http://bugs.sgul.ac.uk/E-BUGS-115) and also ArrayExpress (accession number E-BUGS-115).
Quantitative RT–PCR (qRT–PCR)
qRT–PCR was performed as described previously.3 Gene-specific primer pairs were designed for S. aureus RN4220 genes of interest to yield amplicons of 100–150 bp and the oligonucleotides used are listed in Table S1 (available as Supplementary data at JAC Online). Data shown are the median of three biological and two technical replicates; each replicate was performed in duplicate.
Kinetics of covalent binding of drug to dsDNA
The interaction of ELB-21 and short (12- to 14-mer) oligonucleotides incorporating putative binding sites was investigated using a combination of ion-pair reversed phase liquid chromatography (RPLC) and mass spectrometry (MS) as described earlier.18,19 Sequences of the single-stranded (ss) oligonucleotides used are given in Table 1 and were obtained in lyophilized form from AtdBio Ltd. (Southampton, UK). They were annealed to form dsDNA: each oligonucleotide was dissolved in 100 mM CH3COONH4 to give a 2 mM stock solution that was diluted in annealing buffer (10 mM Tris, 50 mM NaCl, 1 mM EDTA pH 8.5) immediately prior to annealing to give a 1 mM DNA solution. The complementary ssDNA sequences were held at 70°C for 10 min, allowed to cool slowly and then kept at −20°C overnight to ensure completion of the annealing process; dsDNA duplex solutions were adjusted to a DNA concentration of 50 μM with 20 mM CH3COONH4 prior to use. Ligand-DNA complexes were prepared by incubating ELB-21 with ss or duplex oligonucleotides at a molar ratio of 4:1 and the reactions followed at room temperature. RPLC was performed on a Thermo Electron HPLC system incorporating a 4.6 × 50 mm Xterra MS C18 column packed with 2.5 μM particles (Waters Ltd., Elstree, UK). A gradient system of 100 mM triethyl ammonium bicarbonate as buffer A and 40% acetonitrile in water (HPLC grade, Fisher Scientific UK) as buffer B were used. The gradient was ramped from 90% buffer A at 0 min to 50% buffer A at 20 min and finally to 10% buffer A at 35 min. UV absorbance was monitored at 254 nm and fractions containing separated components were collected manually, combined as appropriate, lyophilized and analysed by matrix assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF MS): covalently bound DNA-ELB-21 adduct samples were prepared by diluting with matrix (37 mg 2′,4′,6′-trihydroxyacetophenone in 1 mL acetonitrile, 45 mg ammonium citrate in 1 mL water – mix 1:1 for matrix) either 2:1, 1:1 or 1:5 (sample:matrix), then 1 μL of sample was spotted onto the MALDI target plate and allowed to dry. Samples were analysed with an Applied Systems Voyager-DE Biospectrometry Workstation MALDI-TOF mass spectrometer with a nitrogen laser in positive linear mode using delayed extraction (500 ns) and an accelerating voltage of 25 000 V. Acquisition was between 4000 and 15 000 Da with 100 shots/spectrum.
Table 1.
Single-strand (ss) and double-strand (ds) oligonucleotides designed as substrates for RPLC-MS investigations of DNA:drug adduct formation
| Label | ssDNA Sequence | Average mass (Da) | Label | dsDNA Sequence | Average mass (Da) | ELB-21:dsDNA adduct mass (Da) |
|---|---|---|---|---|---|---|
| Seq-1 | 5′-TATAGAATCTATA-3′ | 3956.5 | Duplex-1 | 5′-TATAGAATCTATA-3′ | 7904.1 | 8516.8 |
| Seq-2 | 5′-TATAGATTCTATA-3′ | 3946.6 | 3′-ATATCTTAGATAT-5′ | |||
| Seq-3 | 5′-TATAGAATGTATA-3′ | 3996.6 | Duplex-2 | 5′-TATAGAATGTATA-3′ | 7904.1 | 8516.8 |
| Seq-4 | 5′-TATACATTCTATA-3′ | 3906.5 | 3′-ATATCTTACATAT-5′ | |||
| Seq-5 | 5′-TATAGATCTATA-3′ | 3643.7 | Duplex-3 | 5′-TATAGATCTATA-3′ | 7287.4 | 7900.1 |
| 3′-ATATCTAGATAT-5′ | ||||||
| Seq-6 | 5′-TATAGATGTATA-3′ | 3683.4 | Duplex-4 | 5′-TATAGATGTATA-3′ | 7286.7 | 7899.5 |
| Seq-7 | 5′-TATACATCTATA-3′ | 3603.4 | 3′-ATATCTACATAT-5′ | |||
| Seq-8 | 5′-TATAGAAATCTATA-3′ | 4269.9 | Duplex-5 | 5′-TATAGAAATCTATA-3′ | 8522.05 | 9106.66 |
| Seq-9 | 5′-TATAGATTTCTATA-3′ | 4251.9 | 3′-ATATCTTTAGATAT-5′ | |||
| Seq-10 | 5′-TATAGAAATGTATA-3′ | 4309.9 | Duplex-6 | 5′-TATAGAAATGTATA-3′ | 8522.05 | 9106.66 |
| Seq-11 | 5′-TATACATTTCTATA-3′ | 4211.8 | 3′-ATATCTTTACATAT-5′ |
Results
Formation of covalent adducts with ds oligonucleotides
Predictive modelling and DNA cross-linking and footprinting studies2,5 have indicated that the preferred interstrand binding sites for ELB-21 are 5′-Pu-GATTC-Py and 5′-Pu-GAATC-Py, but more recent work3 has identified further interstrand targets as well as at least 10 potential intrastrand sites comprising 2 to 6 bases separating same-strand guanines. To establish the preferences for ELB-21 covalent adduct formation with DNA we investigated real-time covalent binding of the drug to the DNA duplexes shown in Table 1; these duplexes incorporate the major interstrand and intrastrand sites identified by our previous work.3
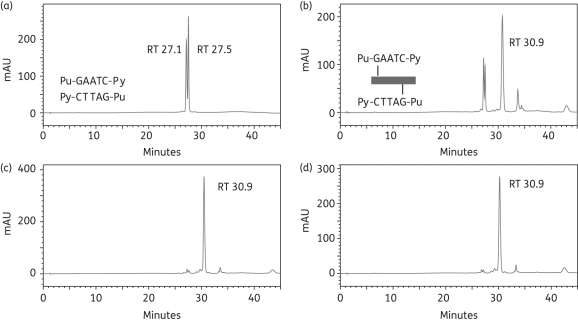
The formation of interstrand cross-links within the 5′-Pu-GAATC-Py sequence was investigated using the 13-mer oligonucleotide duplex-1 (Table 1). RPLC analysis of the annealed duplex formed from the ss sequences Seq-1 and Seq-2 (Table 1) resulted in two peaks with retention times (RTs) of 27.1 min and 27.5 min (Figure 2a), equivalent to denatured Seq-1 and Seq-2 as determined by MALDI-TOF MS and consistent with our previous finding that short dsDNA duplexes denature under the HPLC conditions used. Immediately following mixing of duplex-1 and ELB-21, a major adduct peak (RT 30.9 min) was apparent (Figure 2b), confirmed by MALDI-TOF MS as the 1:1 ELB-21/Seq-1/Seq2 duplex interstrand cross-linked adduct and representing approximately 70% of the DNA in the sample. A minor (<10%) peak (RT 33.7 min) was also observed and was identified as the ELB-21/Seq-1 ssDNA adduct; its abundance declined over time. Further incubation determined that interstrand 5′-Pu-GAATC-Py adduct formation was complete (∼95%) within 3 h (Figure 2c and d).
Figure 2.
Formation of ELB-21 interstrand adducts with dsDNA 13-mer duplex-1. Ion-pair RPLC chromatograms of (a) duplex-1, showing denaturation to Seq-1 (RT 27.1 min) and Seq-2 (RT 27.5 min) under RPLC conditions; (b) immediately (0 h) after mixing of ELB21 with duplex-1 showing the appearance of interstrand 5′-Pu-GAATC-Py adduct (RT 30.9 min); (c) 3 h after mixing of ELB-21 with duplex-1 showing completion of reaction; (d) 12 h after mixing of ELB-21 with duplex-1. Peaks were characterized using MALDI-TOF MS. Binding sites and cross-linking profiles are indicated within the appropriate chromatograms in this and subsequent figures.
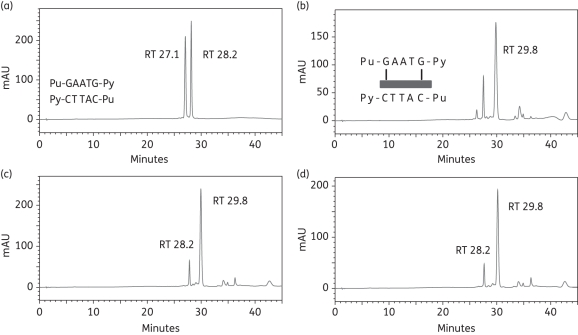
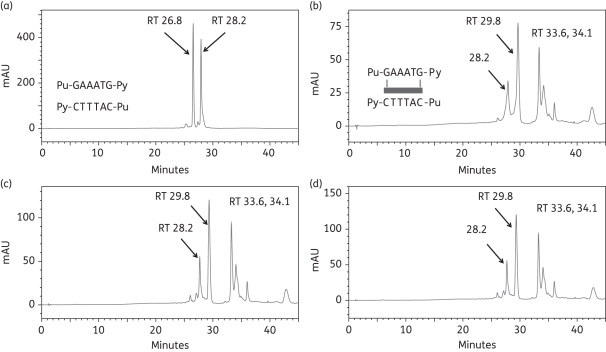
As we have obtained evidence that ELB-21 may form intrastrand cross-links,3 we investigated the potential for intrastrand adduct formation using duplex-2 (Table 1); this duplex differs from duplex-1 with respect to replacement of 9-cytosine with a guanine residue on the 5′ strand, yielding 5′-Pu-GAATG-Py. Analysis of the annealed duplex under RPLC conditions revealed two peaks (RT 27.1 min and RT 28.2 min), confirmed by MALDI-TOF MS to be Seq-3 and Seq-4, respectively (Figure 3a). A prominent peak was apparent at RT 29.8 min immediately (0 h) after mixing of ELB-21 and duplex-2 (Figure 3b), confirmed by MALDI-TOF MS as the 1:1 ELB-21/Seq-3/Seq-4 duplex adduct. During the time course of these experiments, two further peaks emerged at RT 34.2 min and RT 36.3 min (Figure 3b–d), although the RT 29.8 min peak remained prominent throughout and accounted for 90% of the adducts formed at completion. Adduct formation with 5′-Pu-GAATG-Py progressed very rapidly and was complete within 1 h (Figure 3c). Even at 0 h, it was noticeable that that the ratio of the unreacted oligonucleotides Seq-3 and Seq-4 changed in favour of Seq-4, indicating preferential ELB-21 reactivity with the 5′-Pu-GAATG-Py-containing strand Seq-3 (Figure 3b). The chromatograms indicated that ELB-21 initially formed the duplex intrastrand cross-linked RT 29.8 min adduct with the Seq3/Seq-4 duplex, consuming both ssDNA species at an equal rate. However, in contrast to the duplex-1 interstrand cross-linked adduct, which is stable under RPLC conditions due to the covalent bonding of both oligonucleotide strands,19 a fraction of the intrastrand ELB-21/Seq-3/Seq-4 adduct dissociated as the ELB-21/Seq-3 intrastrand adduct (RT 34.2 min; m/z 4609) and the free Seq-4 strand (RT 28.2 min), causing a relative increase in the RT 28.2 min Seq-4 peak in comparison to the RT 27.1 min Seq-3 peak. MALDI-TOF MS data (not shown) indicated that the RT 36.3 min peak corresponded to the 2:1 ELB-21/Seq-3 adduct, with one ELB-21 unit attached to each of the guanine residues of the 5′-Pu-GAATG-Py binding site (Figure 3b).
Figure 3.
Formation of ELB-21 intrastrand adducts with dsDNA 13-mer duplex-2. Ion-pair RPLC chromatograms of (a) duplex-2, showing denaturation to Seq-3 (RT 27.1 min) and Seq-4 (RT 28.2 min) under RPLC conditions; (b) immediately (0 h) after mixing of ELB-21 with duplex-2, showing the rapid appearance of intrastrand 5′-Pu-GAATG-Py adduct (RT 29.8 min). Seq-3, which contains the 5′-Pu-GAATG-Py intrastrand binding site, has almost disappeared; (c) 1 h after mixing of ELB-21 with duplex-2, showing emergence of two minor peaks (RT 32.4 min and RT 36.3 min) in addition to a major peak (RT 29.8 min). Seq-3 is completely absent, indicating completion of the reaction; (d) 12 h after mixing of ELB-21 with duplex-2. Peaks were characterized using MALDI-TOF MS.
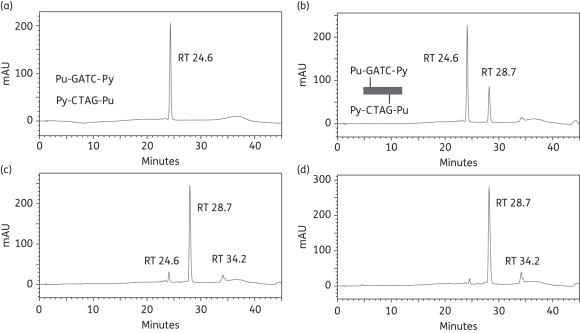
Similar experiments were conducted with the self-complementary 5′-Pu-GATC-Py duplex-3 (Seq5/Seq5) and 5′-Pu-GATG-Py duplex-4 formed from Seq-6 and Seq-7. The latter contains an intrastrand binding site as a consequence of the replacement of cytosine-8 in Seq-5 with a guanine residue. Figure 4 shows that ELB-21 immediately formed a covalent adduct (RT 28.7 min) with duplex-3 and the reaction was essentially complete within 3 h. The formation of covalent adducts between ELB-21 and duplex-4 was evident immediately after mixing, but the reaction failed to go to completion, even after 24 h (Figure S1, Supplementary data). Thus, in contrast to experiments with the predicted3 pentanucleotide binding sequences 5′-Pu-GAATC-Py (Figure 2) and 5′-Pu-GAATG-Py (Figure 3), ELB-21 preferentially formed interstrand, rather than intrastrand, cross-links with these tetranucleotide sequences. The experiments demonstrate that ELB-21 can form cross-linked adducts with binding sites that are smaller than the ideal binding sequences.
Figure 4.
Formation of ELB-21 interstrand adducts with dsDNA 12-mer duplex-3. Ion-pair RPLC chromatograms of (a) duplex-3, showing denaturation to Seq-5 (RT 24.6 min); (b) immediately (0 h) after mixing of ELB-21 with duplex-3, showing the rapid appearance of interstrand 5′-Pu-GATC-Py duplex adduct (RT 28.7 min); (c) 3 h after mixing of ELB-21 with duplex-3, showing the emergence of a minor peak at RT 34.2 min in addition to the major peak at RT 28.7 min; (d) 12 h after mixing of ELB-21 with duplex-3. Peaks were characterized using MALDI-TOF MS.
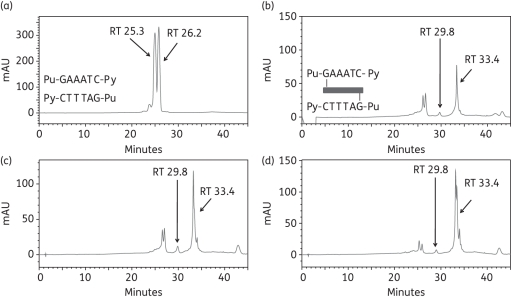
Finally, to investigate binding to an extended sequence, two oligonucleotides were designed with one additional A/T base pair between the two guanines of duplexes 1 and 2. Thus duplex-5 (Seq-8/Seq-9) offered the extended interstrand Pu-GAAATC-Py cross-linking site and duplex-6 (Seq-10/Seq-11) offered the extended intrastrand Pu-GAAATG-Py site (Table 1). After incubation of ELB-21 with duplex-5, a minor peak at RT 29.8 min and a major peak at RT 33.4 min were apparent (Figure 5). The identities of the peaks were confirmed by MALDI-TOF MS; the minor RT 29.8 min peak was found to be the interstrand ELB-21/Seq-8/Seq-9 adduct and the RT 33.4 min peak contained the co-eluting ELB-21/Seq-8 and ELB-21/Seq-9 mono-adducts. Adduct formation was not complete even after 24 h incubation and the RT 29.8 min peak (representing the interstrand cross-linked adduct) remained as a minor peak throughout the experiment. ELB-21 was then incubated with the extended intrastrand sequence. Annealed duplex-6 (Seq-10/Seq-11) containing the sequence Pu-GAAATG-Py resolved as peaks at RT 26.8 min and RT 28.2 min (Figure 6a), identified by MALDI-TOF MS as Seq-10 and Seq-11, respectively. Incubation of ELB-21 with duplex-6 resulted in the rapid emergence of a major peak at RT 29.8 min and two minor peaks at RT 33.6 min and RT 34.1 min (Figure 6b). As anticipated, the RT 29.8 min peak was identified as the intrastrand ELB-21/Seq-10/Seq-11 duplex adduct. Both the RT 33.6 min and RT 34.1 min peaks gave mass values corresponding to ELB-21/Seq-10 adducts. Based on previous studies with non-nucleophilic inosine-containing oligonucleotides, which are unable to form covalent linkages with PBD,18 the RT 33.6 min peak was identified as the intrastrand cross-linked ELB-21/Seq-10 adduct and the RT 34.1 min peak was identified as the ELB-21/Seq-10 mono-alkylated adduct. The intrastrand cross-linked duplex adduct was the most abundant in this experiment (Figure 6).
Figure 5.
Formation of ELB-21 interstrand adducts with dsDNA 14-mer duplex-5. Ion-pair RPLC chromatograms of (a) duplex-5, showing denaturation to Seq-8 (RT 00.0 min) and Seq-9 (RT 00.0 min); (b) immediately (0 h) after mixing of ELB-21 with duplex-5, showing the rapid appearance of the minor interstrand Pu-GAAATC-Py duplex adduct at RT 29.8 min; (c) 4 h after mixing of ELB-21 with duplex-5; (d) 24 h after mixing of ELB-21 with duplex-5, showing that the interstrand ELB-21/Seq-8/Seq-9 adduct at RT 29.8 min remains as a minor peak. Peaks were characterized using MALDI-TOF MS.
Figure 6.
Formation of ELB-21 intrastrand adducts with dsDNA 14-mer duplex-6. Ion-pair RPLC chromatograms of (a) duplex-6, showing denaturation to Seq-10 (RT 26.8 min) and Seq-11 (RT 28.2 min); (b) immediately (0 h) after mixing of ELB-21 with duplex-6, showing the rapid appearance of the major intrastrand Pu-GAAATG-Py duplex adduct (RT 29.8 min); (c) 3 h after mixing of ELB-21 with duplex-6; (d) 24 h after mixing of ELB-21 with duplex-6, showing that the duplex intrastrand ELB-21/Seq-14/Seq-15 adduct at (RT 29.8 min) remains the major product of the incubation.
Thus the rank order of ELB-21 binding sequences was determined in this study to be 5′-Pu-GAATG-Py ≥ 5′-Pu-GAATC-Py > 5′-Pu-GATC-Py >>> 5′-Pu-GATG-Py ∼ 5′-Pu-GAAATG-Py ∼ 5′-Pu-GAAATC-Py.
Antistaphylococcal effects of ELB-21
ELB-21 displayed potent antibacterial activity against 38 MRSA isolates, with an MIC90 value of 0.03 mg/L;1 the MIC for EMRSA-16 was confirmed as 0.015 mg/L (Table 2). The agent exhibited potent bactericidal activity against EMRSA-16 and other bacteria, with an immediate reduction in viable count following exposure to the drug,1 and it induced genes associated with prophages φSa2 and φSa3,3 common residents within S. aureus clinical isolates.25 These observations raised the possibility that the bactericidal effect associated with ELB-21 is due to ‘lysis from within’ determined by activation of resident prophages. We therefore examined the effect of ELB-21 on the viability, optical density and prophage induction of prophage-carrying and prophage-cured strains of S. aureus in comparison to MMC, a well-established inducer of prophage.
Table 2.
Susceptibility of S. aureus strains to ELB-21 and mitomycin C (MMC)
| MIC (mg/L) |
|||
|---|---|---|---|
| Bacterial strain | Description | ELB-21 | MMC |
| EMRSA-16 | Hospital-acquired MRSA | 0.015 | 0.03 |
| NCTC 8325 | Isolated in 1960 from sepsis patient | 0.015 | 0.125 |
| RN4220 | Prophage-cured derivative of NCTC 8325 | 0.03 | 0.25 |
| RN450 | Prophage-cured derivative of NCTC 8325 | 0.03 | 0.25 |
| RN451 | RN450 lysogenic for φ11 | 0.03 | 0.25 |
| RN981 | ΔrecA derivative of RN450 | 0.00015 | 0.0075 |
| RN1030 | ΔrecA derivative of RN451 | 0.000075 | 0.0075 |
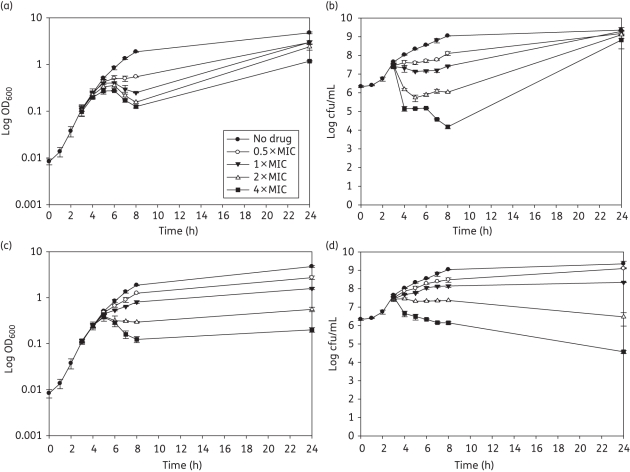
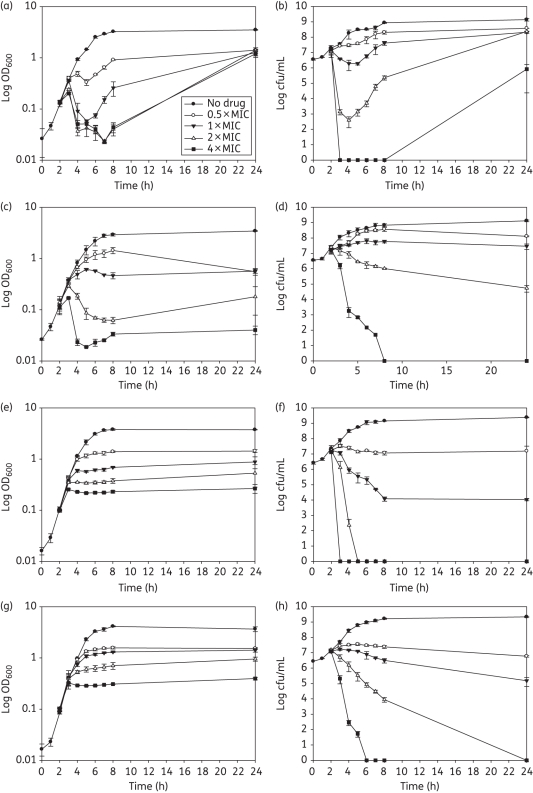
The addition of ELB-21 at concentrations of 2–4 × MIC to 3 h mid-logarithmic EMRSA-16 cells produced an immediate and large reduction in viability; at 1 × MIC the bactericidal effect was much less pronounced (Figure 7b). Cell viability began to decline approximately 1 h after addition of the drug; with supra-inhibitory concentrations, a threefold reduction in cfu/mL was evident after 8 h (Figure 7a). Phage release occurred 3 h after drug exposure and the pfu/mL increased up to 5 h, timepoints which are coincident with maximal reduction in OD600; phage numbers declined rapidly following this delayed burst (Figure S2, Supplementary data). Interestingly, more infectious phage particles were induced by 1 × MIC compared with 4 × MIC; only one morphological phage type, typical of the Siphoviridae,26 was evident after extensive transmission electron microscopy (TEM) investigation of ELB-21-induced supernatants. Growth of the surviving fraction of EMRSA-16 cells was evident after 24 h incubation. The response was comparable to that induced by MMC (Figure 7c and d), except that recovery of MMC-exposed bacteria was not evident after 24 h (Figure 7d). MMC was a far more potent inducer of prophage than ELB-21 (Figure S2, Supplementary data). To further investigate if induction of prophage contributed wholly or in part to the bactericidal or lytic profile of ELB-21, we compared the response to ELB-21 exposure of NCTC 8325, a strain carrying φSa2 and φSa3, with that of its prophage-free derivative RN4220. As shown in Figure 8, there was a clear difference in the responses of these two strains to ELB-21 and MMC at concentrations at or above the MIC (0.015–0.03 and 0.125–0.25 mg/L, respectively). The immediate reduction in OD600 at 2 h following addition of supra-inhibitory ELB-21 concentrations (Figure 8a) was far greater than that found with the more recent clinical isolate EMRSA-16 (Figure 7a) and was accompanied by a corresponding immediate reduction in viability (Figure 8b). Similar profiles were obtained with MMC (Figure 8c and d) and, again, in contrast to ELB-21, there was no recovery of viability after 24 h incubation (Figure 8d). Although loss of viability of RN4220 was comparable to the NCTC 8325 parent strain with both ELB-21 (Figure 8f) and MMC (Figure 8h), there was no reduction in optical density for either drug (Figure 8e and g). Thus prophage induction does not account for killing of the prophage-replete parent NCTC 8325, but bacterial lysis, observed with a proportion of the bacteria, appears to be due solely to activation of resident prophages. Exposure to both ELB-21 and MMC resulted in levels of induction of resident NCTC 8325 prophages comparable to those found with EMRSA-16 (data not shown).
Figure 7.
Response of S. aureus EMRSA-16 to subinhibitory and supra-inhibitory concentrations of ELB-21 and mitomycin C (MMC). ELB-21 (a, b) and MMC (c, d) were added to mid-logarithmic phase cultures (OD600 ≈ 0.1; 3 h), OD600 measured (a, c) and viable numbers determined (b, d): Error bars represent ±1 SD; n = 3.
Figure 8.
Response of S. aureus NCTC 8325 (a–d) and the prophage-cured derivative RN4220 (e–h) to subinhibitory and supra-inhibitory concentrations of ELB-21 and mitomycin C (MMC). ELB-21 (a, b, e, f) and MMC (c, d, g, h) were added to mid-logarithmic phase cultures (OD600 ≈ 0.1; 2 h), OD600 measured (a, c, e, g) and viable numbers determined (b, d, f, h). Error bars represent ±1 SD; n = 3.
We have previously shown that the addition of ELB-21 to logarithmic phase EMRSA-16 cells led to a twofold or greater change in expression of 184 genes, the large majority of which were upregulated;3 almost half were induced prophage genes as well as genes associated with the SaPI4 pathogenicity island. To determine the transcriptional response of ELB-21 in a prophage-free background, we exposed mid-logarithmic MH broth-grown RN4220 to the agent at a concentration of 0.5 MIC. One hour after addition of ELB-21, 27 genes were found to be up-regulated (Table S2, Supplementary data) and 32 genes down-regulated (Table S3, Supplementary data) at least twofold compared with drug-free RN4220 cultures. Ten of the genes activated by exposure to the drug are involved in DNA damage repair and the pattern of ELB-21-mediated induction of SOS-associated proteins was comparable to that induced by MMC. Other ELB-21-activated genes included small numbers encoding membrane proteins, transporter components and metabolic enzymes. No clear pattern of down-regulation by ELB-21 emerged, with a variety of regulatory, transport, membrane and metabolic proteins affected (Table S3), and may reflect specific binding to primary sequences at subinhibitory concentrations within these genes. Interestingly, the number of RN4220 genes down-regulated by the PBD dimer was twice that found with EMRSA-16, an isolate carrying prophages φSa2 and φSa3. The pattern of genes with increased expression following exposure to ELB-21 was markedly similar to that associated with induction by a subinhibitory concentration of MMC (Table S2), even though the latter compound is considered to be a less specific DNA binding agent than PBD dimers such as ELB-21.13 Thus exposure to MMC at 0.5 MIC resulted in the twofold or greater up-regulation of 25 genes and down-regulation of 16 (Tables S2 and S3), with a higher degree of complementarity between ELB-21 and MMC for up-regulated compared with down-regulated genes. Changes in the expression of eight genes by ELB-21 and MMC, as determined by microarray analysis, were validated by qRT–PCR (Table S4, Supplementary data); there was a high degree of correlation between the two methods.
That the relative susceptibility of DNA repair-competent staphylococci is critically dependent on induction of the SOS response was emphasized by large decreases in MIC values for both ΔrecA mutant RN1030 and prophage-cured ΔrecA mutant RN981; both ΔrecA mutants were more than 100-fold more susceptible to ELB-21 than the parent strains (Table 2).
Discussion
PBD dimers were designed to arrest DNA replication, inhibit binding of enzymes and transcription factors to DNA and prevent transcription by stably cross-linking opposing DNA strands.2,4,27,28 The drugs are shaped to fit the minor groove of duplex DNA without distortion of the helix and to form covalent bonds with appropriately spaced guanine residues. The five-carbon diether linking moiety of ELB-21 facilitates interstrand tethering of guanines separated by 3 bp, and hydrogen bonds between the N10 protons of the PBD units and neighbouring adenine residues provide further sequence selectivity.29 ELB-21 is the most potent antibacterial in this group of compounds examined to date, displaying bactericidal killing kinetics against susceptible Gram-positive bacteria and cross-linking DNA in situ.1,7 However, recent investigation of ELB-21 binding sites using DNase I footprinting methods identified a range of further interstrand DNA binding sites both longer and shorter than the predicted target sequences and indicated that the drug forms intrastrand as well as interstrand DNA adducts.3 These observations complement those made with SJG-136, a PBD dimer with a three-carbon diether bridge that recognizes guanines on opposing DNA strands separated by 2 bp.4 It has successfully completed phase I clinical trials in patients with advanced solid tumours;30 this promising, clinically effective agent has recently been found to form interstrand and intrastrand adducts with a wider variety of DNA targets than previously recognized.18 As the nature of the adducts formed between ELB-21 and dsDNA sequences has implications for its mode of antibacterial action, the capacity of recombination-competent bacteria to repair damaged sites on DNA and the potential for resistance to this class of agent, we investigated the preferred binding patterns of ELB-21 to short oligonucleotide target sequences using a new method19 that provides precise, quantitative kinetic data and mass spectrometric identification of the adducts formed.
Our study has provided direct evidence that ELB-21 forms interstrand cross-links at the preferred binding site 5′-Pu-GAATC-Py;2,5 cross-links were evident immediately after mixing of the drug with duplex DNA and were complete within 3 h (Figure 2), confirming the high affinity of ELB-21 for one of its designed target sequences. As anticipated from previous footprinting studies,3 the drug also avidly cross-links shorter sequences within duplex DNA, including 5′-Pu-GATC-Py (Figure 4), one of the main targets for SJG-136.8 The capacity of ELB-21 to cross-link sequences longer than the preferred target, such as 5′-Pu-GAAATC-Py (Figure 5), emphasizes the considerable number of potential binding sites that almost certainly contribute towards the potent antistaphylococcal activity of PBD dimers such as ELB-21.1,7 The preference of ELB-21 for formation of the intrastrand adduct Pu-GAATG-Py (Figure 6) over the interstrand cross-linking of Pu-GAATC-Py was unexpected and represents a key finding of this study, along with the capacity of the drug to slowly form intrastrand adducts with the shorter sequence 5′-Pu-GATG-Py. The observation that ELB-21 formed intrastrand adducts with duplex sequences of ideal length for interstrand cross-linking contrasts with data obtained using SJG-136;18 this PBD dimer also forms intrastrand adducts, but preferences are for sequences longer and shorter than the ideal length interstrand targets. Insights into adduct formation with SJG-136 were obtained using energy-minimized molecular models; the agent could be readily accommodated in the minor groove by participating guanines for both interstrand and intrastrand adducts but could be accommodated through binding to extended sequences only at the expense of some distortion of the DNA helix at the points of attachment.18 We anticipate similar effects with ELB-21 that may explain induction of the RecA-LexA pathway in EMRSA-16 that we have observed in this and prior3 studies.
We previously determined that exposure of EMRSA-16 to ELB-21 resulted in a pattern of gene up-regulation indicative of a DNA damage response with concomitant induction of prophage.3 The rapid bactericidal action of the drug was accompanied by delayed cell lysis that appeared to be due to release of induced phages, as strains cured of all resident prophages were rapidly killed, but not lysed, by ELB-21. In accord with other studies,31,32 a relatively low proportion of the EMRSA-16 population released viable phage particles following drug treatment, as evidenced by lysis of around 5% of bacteria and low plaque counts of culture supernatants. It is unclear in our study whether phages are released in delayed fashion from the non-viable or the viable portion of bacteria in the culture. Although ELB-21 was designed as a sequence-selective agent with the capacity to bind to a restricted set of motifs within target DNA, the response of EMRSA-16 to this agent was similar to that obtained with the non-selective DNA interactive agent MMC, suggesting that the antibacterial effects observed were due to an inadequate response to DNA damage.
Changes in bacterial gene expression following addition of ELB-21 and MMC to mid-logarithmic S. aureus cultures further emphasized these common features of the bacterial response. We have shown3 that exposure of EMRSA-16 to subinhibitory concentrations of ELB-21 for 1 h elicited the up-regulation of more than 160 genes, almost half of which were induced prophage genes; also prominent among ELB-21-activated genes were those involved in DNA damage repair and genes encoding toxins within the SaPI4 pathogenicity island. As the bacterial transcription profile is likely to have been influenced in an unpredictable way by the activation of a large number of prophage genes and the subsequent release of viable phage particles, we determined the transcriptional response to ELB-21 and MMC of S. aureus RN4220. In this prophage-free background, only a limited number of genes were induced more than twofold, and the profiles obtained with each agent were remarkably similar. With ELB-21, the majority encoded components of the DNA damage repair system and included the positive (RecA; recombinase A) and negative (LexA) regulators of the SOS response and products of genes in the LexA regulon involved in error-prone DNA repair.
Induction of RecA by both ELB-21 and MMC in all likelihood accounts for the central features of the response of S. aureus to these agents. The extremely high susceptibility of ΔrecA strains to ELB-21 and MMC suggests that the bactericidal action of these agents occurs above the threshold of repair of damaged DNA, with growth negated by permanently stalled DNA polymerase and bacterial death due to irreparable strand breaks. During normal bacterial growth, the repressor LexA is bound to a specific DNA sequence, the SOS box, located in the promoter region of genes associated with the SOS system such as recA, lexA and uvrAB and it therefore down-regulates its own expression. The consensus sequence for SOS boxes in S. aureus is CGAACAAATGTTCG (conserved regions shown in bold)33 which, interestingly, contains potential target sequences for ELB-21 and other PBD dimers. LexA has high affinity for A-T-rich regions but, as SOS boxes generally include G-C pairs, the affinity of LexA for each targeted promoter is variable and this facilitates some expression of SOS genes in the repressed state. The SOS response can be induced by stalled replication forks or unrepaired defects resulting in the formation of ssDNA;34,35 the ATP-dependent assembly of ssDNA-RecA nucleoprotein filaments engenders LexA autocleavage and inactivation of the repressor with consequent induction of SOS genes. During an SOS response, some genes may be rapidly induced, partly induced or remain repressed until high or persistent DNA damage has occurred;36 such incremental induction patterns may explain why some SOS genes remain silent after ELB-21-induced damage. In similar fashion, ssDNA-RecA filaments mediate autocleavage of phage repressors such as Cl, inducing phage release by up-regulation of prophage genes.31,35,37
Some S. aureus virulence proteins, such as β-toxin (Hlb), fibronectin binding protein (Fnb) and staphylokinase (Sak), are encoded by prophage genes and phage induction is likely to activate the expression of these virulence effectors.31,34 Indeed, ELB-21 up-regulates virulence-associated, SaPI4-encoded genes hlb, tst (toxic shock syndrome toxin-1) and ssp (V8 serine protease) in EMRSA-16. Antibiotics that de-repress resident staphylococcal prophages have been shown to induce mobilization and high-frequency horizontal transmission of co-resident pathogenicity islands.23,38 Such phage-related chromosomal islands play a major role in the dissemination of genes that impact human health and bacterial adaptation to animal hosts39 and may affect the resolution of infections during the course of therapy with SOS-inducing chemotherapeutic agents. Further understanding of these complex interactions will aid the selection of chemotherapeutic agents for the treatment of severe infections in the human host. Although ELB-21 has the capacity to damage host cell DNA, it exerts potent antibacterial effects at concentrations below its cytotoxicity threshold; we will describe the selective toxicity profile of the compound in a future publication. Although ELB-21 has a toxicity profile that does not bear comparison with established antibiotics, the need for agents with the potential to supplement the current arsenal of antibiotics is so great that the systemic use of colistin is now commonplace.40 The growing body of clinical data for SJG-136 suggests that PBD dimers elicit therapeutic effects below the toxicity threshold, and we will continue to investigate ELB-21 as a potential ‘agent of last resort’ for severe systemic Gram-positive infections.
Funding
This study was supported by Wellcome Trust project grant 078669/Z/05/Z and by BSAC project grant GA840.
Transparency declarations
David Thurston is chief scientific officer at Spirogen Ltd. All other authors: none to declare.
Supplementary data
Acknowledgements
We acknowledge BμG@S (the Bacterial Microarray Group at St George's, University of London) for supply of the microarray and the Wellcome Trust for funding the multi-collaborative microbial pathogen microarray facility under its Functional Genomics Resources Initiative.
References
- 1.Hadjivassileva T, Thurston DE, Taylor PW. Pyrrolobenzodiazepine dimers: novel sequence-selective, DNA-interactive, cross-linking agents with activity against Gram-positive bacteria. J Antimicrob Chemother. 2005;56:513–8. doi: 10.1093/jac/dki256. doi:10.1093/jac/dki256. [DOI] [PubMed] [Google Scholar]
- 2.Smellie M, Bose DS, Thompson AS, et al. Sequence-selective recognition of duplex DNA through covalent interstrand cross-linking: kinetic and molecular modeling studies with pyrrolobenzodiazepine dimers. Biochemistry. 2003;42:8232–9. doi: 10.1021/bi034313t. doi:10.1021/bi034313t. [DOI] [PubMed] [Google Scholar]
- 3.Doyle M, Feuerbaum E-A, Fox KR, et al. Response of Staphylococcus aureus to subinhibitory concentrations of a sequence-selective, DNA minor groove cross-linking pyrrolobenzodiazepine dimer. J Antimicrob Chemother. 2009;64:949–59. doi: 10.1093/jac/dkp325. doi:10.1093/jac/dkp325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregson SJ, Howard PW, Gullick DR, et al. Linker length modulates DNA cross-linking reactivity and cytotoxic potency of C8/C8′ ether-linked C2-exo-unsaturated pyrrolo[2,1-c][1,4]benzodiazepine (PBD) dimers. J Med Chem. 2004;47:1161–74. doi: 10.1021/jm030897l. doi:10.1021/jm030897l. [DOI] [PubMed] [Google Scholar]
- 5.Jenkins TC, Hurley LH, Neidle S, et al. Structure of a covalent DNA-minor groove adduct with a pyrrolobenzodiazepine dimer—evidence for sequence-specific interstrand cross-linking. J Med Chem. 1994;37:4529–37. doi: 10.1021/jm00052a012. doi:10.1021/jm00052a012. [DOI] [PubMed] [Google Scholar]
- 6.Hampshire AJ, Fox KR. Preferred binding sites for the bifunctional intercalator TANDEM determined using DNA fragments that contain every symmetrical hexanucleotide sequence. Anal Biochem. 2008;374:298–303. doi: 10.1016/j.ab.2007.10.008. doi:10.1016/j.ab.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Hadjivassileva T, Stapleton PD, Thurston DE, et al. Interactions of pyrrolobenzodiazepine dimers and duplex DNA from methicillin-resistant Staphylococcus aureus. Int J Antimicrob Agents. 2007;290:672–8. doi: 10.1016/j.ijantimicag.2007.01.005. doi:10.1016/j.ijantimicag.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Martin C, Ellis T, McGurk CJ, et al. Sequence-selective interaction of the minor-groove interstrand cross-linking agent SJG-136 with naked and cellular DNA: footprinting and enzyme inhibition studies. Biochemistry. 2005;44:4135–47. doi: 10.1021/bi0479813. doi:10.1021/bi0479813. [DOI] [PubMed] [Google Scholar]
- 9.Baba T, Takeuchi F, Kuroda M, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–27. doi: 10.1016/s0140-6736(02)08713-5. doi:10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 10.Tani K, Yamaguchi T. Studies on a new antibiotic M-92 produced by Micromonospora. IV. Bactericidal action of the component VA-2. J Antibiotics. 1983;36:289–95. doi: 10.7164/antibiotics.36.289. [DOI] [PubMed] [Google Scholar]
- 11.Kumazawa J, Yagisawa M. The history of antibiotics: the Japanese story. J Infect Chemother. 2002;8:125–33. doi: 10.1007/s101560200022. doi:10.1007/s101560200022. [DOI] [PubMed] [Google Scholar]
- 12.Palom Y, Kumar GS, Tang L-Q, et al. Relative toxicities of DNA cross-links and monoadducts: new insights from studies of decarbomoyl mitomycin C and mitomycin C. Chem Res Toxicol. 2002;15:1398–406. doi: 10.1021/tx020044g. doi:10.1021/tx020044g. [DOI] [PubMed] [Google Scholar]
- 13.Anderson KL, Roberts C, Disz T, et al. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J Bacteriol. 2006;188:6739–56. doi: 10.1128/JB.00609-06. doi:10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell A. The future of bacteriophage biology. Nat Rev Genet. 2003;4:471–7. doi: 10.1038/nrg1089. doi:10.1038/nrg1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giovanetti E, Brenciani A, Vecchi M, et al. Prophage association of mef(A) elements encoding efflux-mediated erythromycin resistance in Streptococcus pyogenes. J Antimicrob Chemother. 2005;55:445–51. doi: 10.1093/jac/dki049. doi:10.1093/jac/dki049. [DOI] [PubMed] [Google Scholar]
- 16.Rockney A, Kobiler O, Amir A, et al. Host responses influence on the induction of lambda prophage. Mol Microbiol. 2008;68:29–36. doi: 10.1111/j.1365-2958.2008.06119.x. doi:10.1111/j.1365-2958.2008.06119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domelier A-S, van der Mee-Marquet N, Sizaret P-Y, et al. Molecular characterization and lytic activities of Streptococcus agalactiae bacteriophages and determination of lysogenic-strain features. J Bacteriol. 2009;191:4776–85. doi: 10.1128/JB.00426-09. doi:10.1128/JB.00426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahman KM, Thompson AS, James CH, et al. The pyrrolobenzodiazepine dimer SJG-136 forms sequence-dependent intrastrand DNA cross-links and monoalkylated adducts in addition to interstrand cross-links. J Am Chem Soc. 2009;131:13756–66. doi: 10.1021/ja902986x. doi:10.1021/ja902986x. [DOI] [PubMed] [Google Scholar]
- 19.Narayanaswarmy M, Griffiths WJ, Howard PW, et al. An assay combining high-performance liquid chromatography and mass spectrometry to measure DNA interstrand cross-linking efficiency in oligonucleotides of varying sequences. Anal Biochem. 2008;374:173–81. doi: 10.1016/j.ab.2007.11.018. doi:10.1016/j.ab.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 20.Novick RP. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–66. doi: 10.1016/0042-6822(67)90105-5. doi:10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 21.Resch A, Fehrenbacher B, Eisele K, et al. Phage release from biofilm and planktonic Staphylococcus aureus cells. FEMS Microbiol Lett. 2005;252:89–96. doi: 10.1016/j.femsle.2005.08.048. doi:10.1016/j.femsle.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 22.Wyman L, Goering RV, Novick RP. Genetic control of chromosomal and plasmid recombination in Staphylococcus aureus. Genetics. 1974;76:681–702. doi: 10.1093/genetics/76.4.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Úbeda C, Maiques E, Knecht E, et al. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol Microbiol. 2005;56:836–44. doi: 10.1111/j.1365-2958.2005.04584.x. doi:10.1111/j.1365-2958.2005.04584.x. [DOI] [PubMed] [Google Scholar]
- 24.Witney AA, Marsden GL, Holden MT, et al. Design, validation, and application of a seven-strain Staphylococcus aureus PCR product microarray for comparative genomics. Appl Environ Microbiol. 2005;71:7504–14. doi: 10.1128/AEM.71.11.7504-7514.2005. doi:10.1128/AEM.71.11.7504-7514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindsay JA, Holden MTG. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 2004;12:378–85. doi: 10.1016/j.tim.2004.06.004. doi:10.1016/j.tim.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Ackermann HW. Phage classification and characterization. Methods Mol Biol. 2009;501:127–40. doi: 10.1007/978-1-60327-164-6_13. doi:10.1007/978-1-60327-164-6_13. [DOI] [PubMed] [Google Scholar]
- 27.Puvvada MS, Forrow SA, Hartley JA, et al. Inhibition of bacteriophage T7 RNA polymerase in vitro transcription by DNA-binding pyrrolo[2,1-c][1,4]-benzodiazepines. Biochemistry. 1997;36:2478–84. doi: 10.1021/bi952490r. doi:10.1021/bi952490r. [DOI] [PubMed] [Google Scholar]
- 28.Wells G, Martin CR, Howard PW, et al. Design, synthesis, and biophysical and biological evaluation of a series of pyrrolobenzodiazepine-poly(N-methylpyrrole) conjugates. J Med Chem. 2006;49:5442–61. doi: 10.1021/jm051199z. doi:10.1021/jm051199z. [DOI] [PubMed] [Google Scholar]
- 29.Gregson SJ, Howard PW, Hartley JA, et al. Design, synthesis, and evaluation of a novel pyrrolobenzodiazepine DNA-interactive agent with highly efficient cross-linking ability and potent cytotoxicity. J Med Chem. 2001;44:737–48. doi: 10.1021/jm001064n. doi:10.1021/jm001064n. [DOI] [PubMed] [Google Scholar]
- 30.Hochhauser D, Meyer T, Spanswick VJ, et al. Phase I study of sequence-selective minor groove DNA binding agent SJG-136 in patients with advanced solid tumors. Clin Cancer Res. 2009;15:2140–7. doi: 10.1158/1078-0432.CCR-08-1315. doi:10.1158/1078-0432.CCR-08-1315. [DOI] [PubMed] [Google Scholar]
- 31.Goerke C, Köller J, Wolz C. Ciprofloxacin and trimethoprim cause phage induction and virulence modulation in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:171–7. doi: 10.1128/AAC.50.1.171-177.2006. doi:10.1128/AAC.50.1.171-177.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Comeau AM, Tétart F, Trojet SN, et al. Phage-antibiotic synergy (PAS): β-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS One. 2007;2:e799. doi: 10.1371/journal.pone.0000799. doi:10.1371/journal.pone.0000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cirz RT, Jones MB, Gingles NA, et al. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J Bacteriol. 2007;189:531–9. doi: 10.1128/JB.01464-06. doi:10.1128/JB.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley WL. Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon. Mol Microbiol. 2006;62:1228–38. doi: 10.1111/j.1365-2958.2006.05444.x. doi:10.1111/j.1365-2958.2006.05444.x. [DOI] [PubMed] [Google Scholar]
- 35.Butala M, Žgur-Bertok D, Busby SJW. The bacterial LexA transcriptional repressor. Cell Mol Life Sci. 2009;66:82–93. doi: 10.1007/s00018-008-8378-6. doi:10.1007/s00018-008-8378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang APP, Pigli YZ, Rice PA. Structure of the LexA-DNA complex and implications for SOS box measurement. Nature. 2010;466:883–6. doi: 10.1038/nature09200. doi:10.1038/nature09200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das M, Ganguly T, Chattoraj P, et al. Purification and characterization of repressor of temperate S. aureus phage Φ11. J Biochem Mol Biol. 2007;40:740–8. doi: 10.5483/bmbrep.2007.40.5.740. [DOI] [PubMed] [Google Scholar]
- 38.Maiques E, Úbeda C, Campoy S, et al. β-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J Bacteriol. 2006;188:2726–9. doi: 10.1128/JB.188.7.2726-2729.2006. doi:10.1128/JB.188.7.2726-2729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novick RP, Christie GE, Penadés JR. The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol. 2010;8:541–51. doi: 10.1038/nrmicro2393. doi:10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Nation RL, Turnidge JD, et al. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. 2006;6:589–601. doi: 10.1016/S1473-3099(06)70580-1. doi:10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.