Abstract
Vibrio cholerae represents both an environmental pathogen and a widely distributed microbial species comprised of closely related strains occurring in the tropical to temperate coastal ocean across the globe (Colwell RR, Science 274:2025–2031, 1996; Griffith DC, Kelly-Hope LA, Miller MA, Am. J. Trop. Med. Hyg. 75:973–977, 2006; Reidl J, Klose KE, FEMS Microbiol. Rev. 26:125–139, 2002). However, although this implies dispersal and growth across diverse environmental conditions, how locally successful populations assemble from a possibly global gene pool, relatively unhindered by geographic boundaries, remains poorly understood. Here, we show that environmental Vibrio cholerae possesses two, largely distinct gene pools: one is vertically inherited and globally well mixed, and the other is local and rapidly transferred across species boundaries to generate an endemic population structure. While phylogeographic analysis of isolates collected from Bangladesh and the U.S. east coast suggested strong panmixis for protein-coding genes, there was geographic structure in integrons, which are the only genomic islands present in all strains of V. cholerae (Chun J, et al., Proc. Natl. Acad. Sci. U. S. A. 106:15442–15447, 2009) and are capable of acquiring and expressing mobile gene cassettes. Geographic differentiation in integrons arises from high gene turnover, with acquisition from a locally cooccurring sister species being up to twice as likely as exchange with conspecific but geographically distant V. cholerae populations.
IMPORTANCE
Functional predictions of integron genes show the predominance of secondary metabolism and cell surface modification, which is consistent with a role in competition and predation defense. We suggest that the integron gene pool’s distinctness and tempo of sharing are adaptive in allowing rapid conversion of genomes to reflect local ecological constraints. Because the integron is frequently the main element differentiating clinical strains (Chun J, et al., Proc. Natl. Acad. Sci. U. S. A. 106:15442–15447, 2009) and its recombinogenic activity is directly stimulated by environmental stresses (Guerin E, et al., Science 324:1034, 2009), these observations are relevant for local emergence and subsequent dispersal.
INTRODUCTION
Vibrio cholerae is in many ways representative of globally successful bacterial species. However, its widespread occurrence is of additional concern since it harbors strains responsible for the diarrheal disease cholera, and environmental reservoirs play an important role in the evolution and transmission of pathogenic variants (1, 2). How local populations assemble from the V. cholerae “pangenome” (3, 4)—the sum of all genes in the species—is therefore a central question in linking the overall ecology of the species to understanding the emergence, spread, and persistence of local, sometimes pathogenic variants. Comparison of several V. cholerae genomes has shown that isolates typically share 98 to 100% nucleotide identity in the 1,520 genes which are common to the species (5). However, these “core” genes make up only 60 to 80% of the total within any one strain, and with every new genome that is not part of a known pandemic clonal complex, 100 to 400 new genes are discovered (5, 6). Like in many other bacterial groups, much of this variation is confined to genomic islands (the “flexible” genome) whose turnover can be astonishingly high and is largely responsible for the vast, currently immeasurable pangenome of each species. At the same time, the rapid spread of pathogenic V. cholerae strains exemplifies the potential for global population mixing on decadal time scales by both natural (e.g., currents and attachment to larger organisms) and artificial (e.g., ship ballast) avenues. Indeed, local origin followed by global spread appears to have happened repeatedly in pathogenic V. cholerae (1, 7). Thus, while fast gene turnover in genomic islands has the potential to produce local differentiation, global mixing across the ocean increases the rates of gene flow and blurs boundaries between populations by allowing access to a large reservoir of genetic material. How these opposing forces of gene turnover and gene flow interact to assemble local populations in the context of V. cholerae’s global occurrence remains an open question.
RESULTS AND DISCUSSION
To test for population structure, we compared relatedness in both the core and flexible genome among V. cholerae isolates from two sites separated by over 12,000 km, the Dhaka Delta in Bangladesh (BGD) and Oyster Pond, a brackish pond in Cape Cod, MA, on the east coast of the United States (USA). We further included a novel sister species, which we discovered to cooccur in Oyster Pond. Prior to this study, this novel species was known from a single isolate for which the genome had been sequenced and which was provisionally named Vibrio metecus (8). It serves as a crucial outgroup, since it enables us to estimate to what extent core and flexible gene pools are specific for V. cholerae or are affected by admixture from outside the species. Despite being V. cholerae’s closest relative, V. metecus is a clearly genetically distinct species and shares <95% sequence identity in housekeeping genes with its sister taxon (Fig. 1; Table 1). Interestingly, clinical isolates that were previously considered atypical V. cholerae are here identified as V. metecus. Clinical strains of V. cholerae and V. metecus are, however, excluded from a direct comparison of gene pools to avoid bias, which may arise because population dynamics of clinical strains are influenced by epidemiological factors (e.g., transmission dynamics, clonal expansions during epidemics, and travel with patients) and selective pressures from drugs. Nonetheless, previous studies have shown that the potential for gene flow among environmental and clinical strains exists, likely when the latter are released into natural waters (9). Our comparison of three environmental populations, V. cholerae (BGD), V. cholerae (USA), and V. metecus (USA), is based on a set of fifteen isolates from each group. These are considered representative, as they were selected to maximize the genetic diversity in the analyses. Here we operationally define a population as isolates that are closely related genetically (96 to 100% average nucleotide sequence identity at housekeeping protein-coding loci) and originate from locations that are geographically close (same estuary, delta, or pond).
FIG 1 .
Phylogenetic relationships among clinical and environmental V. cholerae, V. metecus, and other Vibrionaceae strains. Maximum likelihood phylogeny based on the concatenated DNA sequences of six protein-coding genes (mdh, adk, pgi, gyrB, recA, and rpoB). Bootstrap support values are displayed on the nodes. Isolates are color coded according to species and origins. Unless otherwise noted, all V. cholerae strains are non-O1/non-O139.
TABLE 1 .
DNA sequence identity across six housekeeping loci for all V. cholerae and V. metecus isolates from Bangladesh (BGD) and the United States (USA)
| % DNA sequence identity |
|||||
|---|---|---|---|---|---|
| Isolates compared | Avg | SD | Minimum | Maximum | |
| V. cholerae (BGD) | V. cholerae (BGD) | 98.9 | 0.4 | 98.3 | 100.0 |
| V. cholerae (USA) | V. cholerae (USA) | 99.0 | 0.3 | 98.3 | 100.0 |
| V. cholerae (USA) | V. cholerae (BGD) | 98.8 | 0.3 | 98.2 | 99.3 |
| V. metecus (USA) | V. metecus (USA) | 97.9 | 0.6 | 96.5 | 99.6 |
| V. metecus (USA) | V. cholerae (BGD) | 93.0 | 0.4 | 91.9 | 94.3 |
| V. metecus (USA) | V. cholerae (USA) | 93.0 | 0.4 | 92.2 | 94.0 |
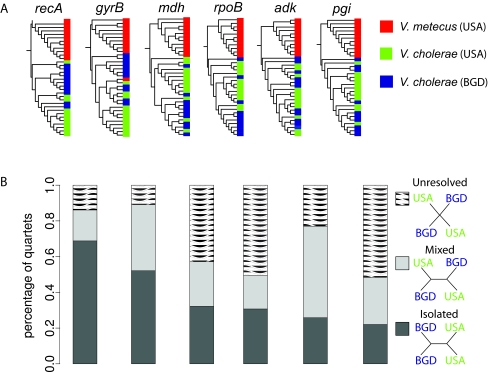
Comparison of representative core genes shows that there is little biogeographic structure in V. cholerae and that it is nearly completely genetically isolated from its sister species. Both V. cholerae populations (USA and BGD) displayed no variation in 16S rRNA gene sequences, and average DNA sequence identity in protein-coding genes was the same for within- and between-locality comparisons (~99%). The phylogenetic trees of six core protein-coding genes show only moderate, statistically nonsignificant clustering of V. cholerae isolates from the same geographical location (Fig. 2A). In four out of the six genes, >40% of all resolved tree quartets, built by joining pairs of cooccurring isolates from each location, show clustering of the BGD and USA isolates (Fig. 2B). This value is surprisingly close to a perfect admixture scenario, in which this proportion is expected to be ~50%, assuming equal probability of exchanging genes with members of the different populations, and thus contradicts biogeographic differentiation. Only recA, which is highly recombinogenic, supports strong biogeographic differentiation, suggesting that the extent of genetic isolation differs according to the frequency of homologous recombination. The same analysis between the two cooccurring sister species shows that for all six alleles, >97% of the quartets support the species partition. This apparently strong global mixing of V. cholerae’s core genome is in good agreement with weak population structure in the cosmopolitan marine bacterium Alteromonas macleodii (10) but differs from hot spring Archaea, which are geographically isolated (11, 12). Thus, while recombination between cooccurring bacteria may temporarily increase similarity in part of their core genomes, population isolation in the ocean does not appear to persist long enough to create a significant phylogeographic signal across the genome. This is especially surprising for V. cholerae, as populations of this primarily estuarine bacterial species are discontinuously distributed.
FIG 2 .
Phylogenetic relationship of representative “core” genes from Vibrio cholerae and V. metecus strains from the Dhaka Delta in Bangladesh (BGD) and from the U.S. east coast (USA). Phylogenetic trees derived from individual protein-coding housekeeping genes (A) and tree quartets (B) were built in TREE-PUZZLE (49) by joining pairs of V. cholerae isolates from BGD and the USA. The bars indicate the proportion of all quartets having topologies consistent (structured) and inconsistent (mixed) with geography, as well as the proportion of unstructured tree quartets, which have a multifurcating maximum likelihood topology. Isolates are color coded according to species and origins.
Determination of geographic structure in the flexible genome is complicated by the vast diversity of genes circulating in and out of genomes (3), so relative rates of diversification can currently not be determined from the available genomic and metagenomic data. Moreover, most genomic islands are found only in a small proportion of natural isolates or occur as singletons (6), making direct comparisons of strains intractable. Quantitative analysis of their variation would also be extremely difficult, as there is no consistent unit to measure change. The integron, on the other hand, has several characteristics making it a well-suited model system to study the flexible genome. It universally occurs in V. cholerae and also represents the largest and most rapidly changing genomic island in this species (6, 13). Its role is to facilitate the acquisition and expression of gene cassettes in a contiguous array, which can contain from 100 to 200 genes, representing up to 3% of the genome (6, 13). Gene cassettes are flanked by conserved attC repeats (14), which can be targeted for their amplification by PCR, regardless of the genes they contain. Using this approach, we obtained an average of 65 nonidentical cassettes per isolate (>2,900 cassettes from 45 isolates).
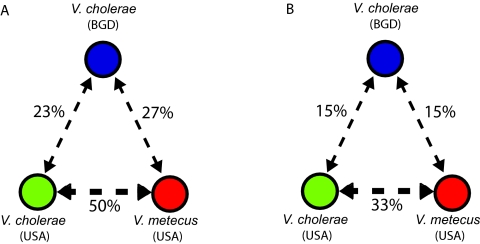
In contrast to the core genome, the integron displays strong geographical differentiation, apparently caused by recent gene transfer among cooccurring bacteria. Phylogenetic analysis of gene cassettes found in all three biogeographic groups revealed that cassettes from the V. cholerae (USA) group cluster with those from V. metecus (USA) up to 2 times as often as they do with cassettes from geographically distinct V. cholerae (BGD) (Fig. 3A). This bias is statistically significant, as demonstrated by the analysis of relative transition rates between the different biogeographic groups along the branches of gene cassette trees (see Fig. S1 in the supplemental material). Consequently, cooccurring V. cholerae and V. metecus from the United States have more overlap in the composition pan-genome of their gene cassette pools than either of them has with geographically separate V. cholerae from Bangladesh. This can be expressed by the Chao-Sørensen similarity index (15). When looking at a recent evolutionary time scale (i.e., using 95% DNA sequence identity to define cassette families), this index is 2-fold higher among cooccurring strains than geographically separate strains, even if from the same species (Fig. 3B). It decreases if longer time scales are used and becomes similar among all groups when a DNA sequence identity criterion of 75% or lower is applied (see Table S1 in the supplemental material). This strongly suggests that cassettes are locally exchanged more rapidly than they can be dispersed geographically, thus resulting in endemic (sub)populations sharing a location-specific gene pool.
FIG 3 .
Similarity in integron cassette families among three biogeographic groups. Phylogenetic comparison (A) and Chao-Sørensen index (B) showing that geographically cooccurring V. cholerae and V. metecus are more similar to each other than to geographically distinct V. cholerae. For the phylogenetic comparison, phylogenetic trees for all 54 gene cassette families occurring in all three biogeographic groups were created. The proportions of clades grouping any two of these three biogeographic groups were counted if their bootstrap support was >80%. When multiple nested clades could have been counted, only the most inclusive clade was. The Chao-Sørensen index calculates the similarity of two samples corrected for bias due to incomplete sampling (15). When calculating the index, cassette families were defined as having >95% sequence similarity to account for recent gene transfers (see Table S1 in the supplemental material for additional sequence similarity cutoffs). All index values in this analysis had a 3% confidence interval.
Strikingly, the integron comprises a nearly completely separate gene pool with regards to the rest of the genome. When using >30% amino acid identity to define protein families, only 2.5% of families found in gene cassettes are also found in the rest of the genome, which decreases to <1% if selfish elements, such as transposons and toxin-antitoxin systems, are excluded (see Table S2 in the supplemental material). V. cholerae integron gene cassettes can, in some cases, have noncassette homologs in other bacteria, highlighting their diverse evolutionary origins. However, within the species, there seems to be almost no movement between the cassette and noncassette gene pools. This is further supported by the G+C content of gene cassettes, which is markedly lower than that of other genes from the same genome (<1% of gene cassettes have a G+C content of >45%, while >88% of noncassette genes do). Therefore, in addition to showing different dispersal properties, the integron and core genome have been genetically segregated over long periods of time. The source of segregation is not known, but if the integron genes confer environment-specific fitness, their transfer into the core genome might be selected against since the genes may be disadvantageous under different environmental contexts (16).
How quickly does the integron have to change to allow evolution of endemicity relative to that of the core genome? Comparison of integron gene cassette compositions scaled against core genome divergence shows that the integron is completely reshuffled by the time 1% nucleotide divergence has accumulated in core genes (i.e., the similarity between two integrons is equivalent to the similarity between two randomly selected sets of gene cassettes) (Fig. 4). However, the true rate of change is probably much higher, as indicated by the few available V. cholerae genomes identical at six or more housekeeping loci, which can still differ significantly in their integrons (Fig. 4). In fact, rapid differentiation can occur by the gain or loss of dozens of cassettes in a single recombination event (17). Such rapid change may be triggered by environmental stressors, such as UV light, oxidative damage, and antibiotic exposure, since these have recently been shown to increase the activity of the V. cholerae integron integrase and thus the rate of cassette acquisition or loss (18). This sensitivity might predispose integrons to adaptation to rapidly changing environmental factors, since cassettes are immediately expressed after integration and do not disrupt the host genome.
FIG 4 .
Rapid decay of integron similarity as V. cholerae strains diverge in “core” genes. Integron and core gene similarities are measured as the proportion of all gene cassette families (defined by >90% nucleotide identity) shared by pairs of isolates and as nucleotide sequence identity at six protein-coding housekeeping loci among the same pairs, respectively. Because integron data combine whole-genome sequences and PCR-based gene cassette sampling, a control for the expected shared portion of gene cassettes between identical integrons was included. This is based on random subsamplings of 65 gene cassettes, corresponding to the average number of cassettes available for isolates in this study.
If rapid change in integron gene cassette content is adaptive, what type of environmental challenges may the gene cassettes meet? Studies of gene cassettes recovered from a variety of organisms have shown that they can encode a broad range of functions (19). However, the functional profile of the gene cassette “pangenome” from a single species has never been determined, as this requires an extensive and phylogenetically specific data set. From the large data set of V. cholerae gene cassettes that we have assembled, only 25% of genes can be assigned to broad functional categories, and a few functions clearly dominate among those that can be annotated (Table 2). The five most abundant protein superfamilies, and most of the less abundant ones, are related to one of three functions: (i) cell envelope modification, (ii) secondary metabolite production/modification, or (iii) stress response. Importantly, all of these categories can have a significant and rapid impact on ecological fitness in both environmental and clinical settings, as their function suggests a role in organismal interactions, e.g., a role in adapting strains to cohabitation with different populations of organisms. For example, cassette-encoded plasmid Achromobacter secretion (PAS) factors (thought to facilitate toxin secretion) and capsular polysaccharide biosynthesis proteins have been directly implicated in virulence (20) but could also make cells more resistant towards increased protozoan grazing pressure (21). Similarly, antibiotic resistance carried by acetyltransferases, vicinal oxygen chelate superfamily proteins, and ld-carboxypeptidases may ensure survival in both clinical and environmental settings. The overrepresentation of lipid-binding lipocalins and ld-carboxypeptidases in the gene cassettes of clinical V. cholerae compared to that in the gene cassettes of environmental isolates suggests that these genes could facilitate colonization/infection (see Table S3 in the supplemental material). Consistent with the suggestion that an important role of the flexible genome lies in changing cellular properties as protection against virus attack (22, 23), the functional predictions of cassette-carried genes indicate broad adaptation towards predatory and competitive interactions.
TABLE 2 .
Comparison of the functional classifications of cassette- and noncassette-encoded proteins from V. cholerae isolates
| Functional classification | Value for: |
|
|---|---|---|
| Genome | Cassette | |
| Total no. of proteinsa | 3,441 | 3,441 |
| No. of nonclassified proteins | 871 | 2,587 |
| No. of classified proteins: | 2,570 | 854 |
| GCN5-related N-acetyltransferases | 15 | 404 |
| Toxin-antitoxin systems | 1 | 111 |
| Vicinal oxygen chelate proteins | 4 | 105 |
| Nudix hydrolases | 3 | 81 |
| Isochorismatase-like amidohydrolases | 2 | 26 |
| ATP-independent intracellular proteases | 2 | 11 |
| OsmC organic hydroperoxide reductases | 0 | 10 |
| NAD(P)+-binding redox proteins | 13 | 9 |
| Retron-type reverse transcriptases | 2 | 8 |
| Sulfate-binding proteins (Sbp) | 0 | 7 |
| Alpha/beta hydrolase folds | 0 | 6 |
| Those in other classifications | 2,527 | 76 |
All V. cholerae cassettes from this study and from public databases were included. Noncassette-encoded proteins have been randomly sampled from all nonclonal V. cholerae genomes (minus their integrons) to match the number of available cassette-encoded proteins.
Overall, these observations add an important dimension to how microbial species may succeed globally by adapting locally. The core genome, which overall appears to be well mixed geographically, is responsible primarily for exploitation of metabolic resources, which can be distributed on global scales. For example, V. cholerae associates with copepods (24) and possesses the ability to digest chitin (25, 26). Additional metabolic capabilities can be acquired via horizontally transferred genes (27) and can be enriched in local populations (28, 29). In contrast, the integron represents a gene pool, which appears to be designed for rapid differentiation, never reaching fixation within populations. In fact, it is the main (and sometimes only) source of genetic difference between pandemic strains of V. cholerae (6) and thus may diversify on ecological time scales, i.e., as strains are dispersed into new locations. Because its genes are efficiently shared among cooccurring species, the integron displays a high level of endemicity.
Several factors could be contributing to this surprisingly efficient gene sharing across species boundaries. Although gene cassette transfer has been empirically observed to be limited by the genetic distance between the donor and the host (30), the capacity of some integron integrases, such as that from V. cholerae, to recognize a broad range of attC recombination sites could explain the apparent lack of a genetic barrier between V. cholerae and V. metecus integrons (14). Furthermore, two vectors facilitating the penetration of novel DNA into V. cholerae cells are likely to contribute to such efficient gene sharing. First, most V. cholerae cells display chitin-induced natural competence, making environmental DNA available to them for recombination (31). Second, conjugation between V. cholerae cells, besides providing novel DNA to the recipient, has been shown to induce the SOS response, which in turn is known to increase the rate of gene cassette insertion in integrons (32). As gene acquisition by integrons is directly stimulated by various environmental stresses which trigger the SOS response (18), the types of genes being shared by these genetic elements may be adaptive towards local environmental challenges, whether they stem from interactions within microbial communities in coastal New England or an antibiotic regime in a clinic.
MATERIALS AND METHODS
Strain isolation and typing.
Three 1-liter water samples were collected from Oyster Pond (Falmouth, MA, U.S. east coast) on three different days in October 2006. The water temperature was 20°C, and the salt concentration was 5 ppt. Five-milliliter aliquots of the water samples were filtered on hydrophilic 0.22-µm-pore-size membranes (Pall Scientific). The filters were placed on thiosulfate citrate bile salts sucrose (TCBS) plates (Difco). After 2 days of growth at 30°C, sucrose-positive (yellow) colonies were counted, and several were restreaked a total of three times alternatingly on tryptic soy broth (TSB) (Difco) and on TCBS media. The ability to utilize sucrose is found only in a few species of vibrios, including Vibrio cholerae, and produces yellow colonies on TCBS media. Environmental V. cholerae strains from the Dhaka Delta, Bangladesh, were a gift from Shah Faruque (ICDDR, Dhaka, Bangladesh) (33).
For typing of strains by sequencing, isolates were grown in TSB overnight. DNA was extracted using either a tissue DNA kit (Qiagen) or Lyse-and-Go (Pierce). The 16S rRNA gene was PCR amplified using primers 27F-1492R and sequenced using the 27F primer (34). The 16S rRNA gene sequence was used to identify the organism using the RDP classifier (35) and BLAST (36). Strains identified as V. cholerae were selected for further analysis. Following the rationale of multilocus sequence analysis (MLSA), housekeeping genes were used for strain characterization since these are unlikely to be under environmental selection (37). Fragments of the six following genes were partially amplified using primers matching most Vibrio species: malate dehydrogenase (mdh), 452 bp (Mdh.for, 5′-GAT CTG AGY CAT ATC CCW AC-3′, and Mdh.rev, 5′-GCT TCW ACM ACY TCR GTA CCC G-3′); adenylate kinase (adk), 463 bp (Adk.for, 5′-GTA TTC CAC AAA TYT CTA CTG G-3′, and Adk.rev, 5′-GCT TCT TTA CCG TAG TA-3′); DNA gyrase subunit B (gyrB), 713 bp (GyrB_VFmod.for, 5′-CGT TTY TGG CCR AGT G-3′, and GyrB.rev, 5′-TCM CCY TCC ACW ATG TA-3′); recombinase A (recA), 618 bp (recA.for, 5′-TGG ACG AGA ATA AAC AGA AGG C-3′, and recA.rev, 5′-CCG TTA TAG CTG TAC CAA GCG CCC-3′); glucose phosphate isomerase (pgi), 437 bp (Pgi_primo.rev, 5′-CMG CRC CRT GGA AGT TGT TRT-3′, and Pgi_primo.for, 5′-GAC CTW GGY CCW TAC ATG GT-3′); and RNA polymerase subunit B (rpoB), 910 bp (CM32b, 5′-GGA ACT GCC TGA CGT TGC AT-3′, and 1110F, 5′-GTA GAA ATC TAC CGC ATG ATG-3′). The primers used for the amplification of mdh, recA, rpoB, and adk have been tested for the typing of Vibrio isolates in previous studies (38–41). The primers targeting pgi and gyrB have been developed in this study and can reliably amplify their target genes not only from V. cholerae and V. metecus but also from most Vibrio species. All genes in our MLSA scheme were amplified using the following PCR conditions: 2 min at 94°C, followed by 32 cycles of 1 min each at 94°C (adk, 46°C; pgi, 40°C; mdh, 42°C; gyrB, 52°C; recA, 52°C; and rpoB, 50°C) and 72°C, with a final step of 6 min at 72°C. Genes were sequenced at least twice using the forward and reverse primers. All sequencing was performed at the Bay Paul Center at the Marine Biological Laboratory, Woods Hole, MA.
Amplification and sequencing of gene cassettes.
Gene cassettes were obtained from fifteen strains of each biogeographical group (V. metecus from the U.S. east coast, V. cholerae from the U.S. east coast, and V. cholerae from the Dhaka Delta, Bangladesh). Cassettes were amplified by targeting the conserved attC sites flanking them, an approach termed gene cassette PCR (13). The primers used were developed to specifically target V. cholerae attC sites, as follows: HS721, 5′-AGC CCC TTA RSC GGG CGT TA-3′, and HS722, 5′-TCC CTC TTG ARS CGT TTG TTA-3′ (17). The PCR conditions used were 1 cycle of 80°C for 2 min; 30 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 90 s; and 1 cycle of 72°C for 10 min. PCR products amplified from each isolate were separately cloned in the TOPO TA vector (Invitrogen), yielding 45 gene cassette clone libraries. A total of 96 clones from each library were sequenced twice. Identical sequences obtained from the same isolate were eliminated so that all cassettes recovered from the same strain had unique sequences. This yielded an average of 65 unique gene cassette sequences for each of the 45 isolates (total of 2,908 sequences).
Phylogenetic and phylogeographic analysis of core genes.
The 16S rRNA gene and all six housekeeping loci sequenced from V. cholerae and V. metecus isolates in this study, as well those available from other closely related strains in GenBank, were individually aligned using MUSCLE (42). All housekeeping loci also were concatenated to obtain an alignment of 3,593 bp. The average, minimum, and maximum percent identities between different biogeographical groups of strains were calculated from the concatenated alignment using the pairwise distance option in PAUP* 4.0 (Sinauer Associates) (Table 1). Maximum likelihood (ML) nucleotide phylogeny was performed for each gene alignment (Fig. 2) as well as for the concatenated alignment using RAxML (43) (Fig. 1). All free model parameters were estimated by RAxML using the GAMMA+P-Invar model of rate heterogeneity, with an ML estimate of the alpha parameter. Bootstrap support values were calculated with the same parameters (100 replicates).
A subset of the strains shown in Fig. 1 were selected to compare the geographical structures of the core genes and integron cassettes. This subset was selected to maximize genetic diversity within each biogeographical group and to ensure that each group was equally represented in the data set. This was achieved by including only environmental V. cholerae and V. metecus strains from Bangladesh (BGD) or the U.S. east coast (USA), displaying less than three identical alleles out of the six housekeeping loci sequenced. The total number of cassettes from each biogeographical group was roughly equal (V. cholerae [USA], 742 cassettes; V. metecus [USA], 746; V. cholerae [BGD], 750).
Phylogeographic analysis of core genes (Fig. 2) was performed for each of the six housekeeping genes by building all possible tree quartets made by joining pairs of cooccurring isolates, as described in Results and Discussion. The percentage of structured quartets supporting the global mixing or local structure hypothesis between the two populations was calculated for each gene. Structured quartets have a bifurcating maximum likelihood topology, as opposed to unstructured quartets, which are multifurcating. This was done for V. cholerae from both of the geographic locations and for cooccurring V. cholerae and V. metecus.
Analysis of gene cassette phylogenetic trees.
To analyze the geographical structures of the V. cholerae and V. metecus gene cassette pools, we chose the same set of strains used for phylogeographic analysis (i.e., environmental isolates from the Dhaka Delta, Bangladesh, and the U.S. east coast having less than three identical alleles out of the six housekeeping loci sequenced). All gene cassettes present in this data set were grouped into families, in which each member shares at least 80% nucleotide identity with all other members of the family. Fifty-four gene cassette families harbored at least one cassette from each of the three biogeographical groups under study (V. metecus from the United States, V. cholerae from the United States, and V. cholerae from Bangladesh). For these cassette families, a sequence alignment and phylogenetic tree (using MUSCLE and RAxML, as described above) were created and subsequently inspected for clades containing only cassettes from (i) V. cholerae (USA) and V. metecus (USA), (ii) V. cholerae (USA) and V. cholerae (BGD), and (iii) V. metecus (USA) and V. cholerae (BGD). Nested clades of the same type were collapsed in order to account for only the largest bipartitions that joined the different populations. The number of clades with a bootstrap value of >80% was counted across all 54 trees, and the proportion of the three different types of clades was compiled (Fig. 3A).
To investigate the frequency of gene cassette movement among three biogeographical populations (V. cholerae from the United States, V. cholerae from Bangladesh, and V. metecus from the United States), we analyzed the rates of gene cassette transition among these populations along the branches of the trees. This was done using an approach much like that used for the estimation of nucleotide substitution rates in phylogenetic analysis (44), but using biogeographical groups as character states rather than the four nucleotide types. We computed the matrix of transition rates that best describes the distribution of the three populations at the tips of the 54 gene trees, using a maximum likelihood approach (see Fig. S1 in the supplemental material). Since the trees are unrooted, we used a symmetric model for the rate matrix. With rates being relative, we arbitrarily fixed the V. cholerae (USA)-V. metecus (USA) rate to 1.0 and optimized the remaining two parameters, V. cholerae (USA)-V. cholerae (BGD) and V. metecus (USA)-V. cholerae (BGD). Optimization was performed through the phangorn phylogenetic package for R (K. Schliep, personal communication), using a partition model in which one partition consists in exactly one gene tree. Confidence intervals were estimated by optimizing the rate matrix for 100 replicate data sets of 54 trees. Replicate data sets were built by randomly picking 1 tree among 100 bootstrapped trees for each of the 54 gene families. For numerical reasons and to avoid instabilities, zero-length branches were set to a length of 1 × 10E−5.
Determination of gene cassette pool similarity.
The Chao-Sørensen index (15) was used to determine the similarity of the gene cassette pools of the three biogeographical groups investigated (Fig. 3B). This index is a modification of the Sørensen index, corrected for bias due to incomplete sampling. It represents the probability that two randomly chosen cassette families, one from each of the two samples considered, are cassette families present in both samples. Cassette families were defined at different DNA sequence identity thresholds (75% to 95%, with 5% intervals), in which each member shares DNA sequence identity equal to or higher than the threshold with all other members of the family. The Chao-Sørensen index was calculated using SONS 1.0 (45) for each of these cassette family definitions among V. cholerae (USA)-V. metecus (USA), V. cholerae (USA)-V. cholerae (BGD), and V. metecus (USA)-V. cholerae (BGD) (see Table S1 in the supplemental material).
Pairwise comparison of isolates for their integron and core gene similarity.
All V. cholerae isolates from this study or from public databases (for which housekeeping loci and integron gene cassette sequences were available) were selected for pairwise comparison of integron and core gene similarity (Fig. 4). Both comparisons were done using the PAUP* 4.0 software (Sinauer Associates). Integron similarity is defined as 100 × (the number of gene cassette families found in both isolates being compared/the number of gene cassette families found in either of the two isolates being compared). Core gene similarity is defined as the percent DNA sequence identity among six protein-coding housekeeping loci of the isolates being compared (mdh, adk, pgi, gyrB, recA, and rpoB). Because integron data combine whole-genome sequences and PCR-based gene cassette sampling, a control for the expected shared portion of gene cassettes in identical integrons was included. This is based on random subsamplings of 65 gene cassettes, corresponding to the average number of cassettes available for each isolate.
Comparison of the gene complements of the genomes and integrons of V. cholerae.
All publicly available closed and draft quality V. cholerae genomes were obtained from GenBank, and their integron gene cassette arrays were located. All genes located in gene cassettes were removed from the genomes, yielding a total of 35,150 noncassette-encoded proteins. All publicly available V. cholerae gene cassettes were downloaded from the ACID database (46) and combined with those obtained from this study, yielding a data set containing 3,441 cassette-encoded proteins. The overlap between the two data sets was determined using MG-DOTUR, with an operational protein family definition of 0.30 (OPF0.30) (47) (see Table S2 in the supplemental material). This definition means that the BLAST score ratio between two proteins has to be 0.30 or higher for them to be considered part of the same family (30% amino acid identity, corrected for length).
To determine the functional overlap between cassette and noncassette genes (as opposed to the simple identification of protein families found in both data sets described above), 3,441 proteins were randomly sampled from V. cholerae genomes that had their integron removed and had less than three out of six housekeeping loci (adk, gyrB, recA, rpoB, pgi, and mdh) identical in sequence. The size of this sample of noncassette-encoded proteins is equivalent to the size of the V. cholerae cassette-encoded protein data set. The functional profiles of these two data sets were compared using MG-RAST (48) (Table 2).
Functional analysis of gene cassettes from V. cholerae and V. metecus.
All V. cholerae and V. metecus gene cassettes obtained from this study and those publicly available were functionally annotated using the MG-RAST server (48). For each protein superfamily detected, the total number of member proteins and protein families (OPF0.30) was determined. The origins of the proteins from the most abundant superfamilies were inspected to determine if they were from clinical or environmental isolates. The distribution of proteins from a given superfamily was considered biased when a category (clinical/environmental) was significantly overrepresented in a protein superfamily in relation to the representation of that category in the whole data set (see Table S3 in the supplemental material).
SUPPLEMENTAL MATERIAL
ACKNOWLEDGMENTS
This work was supported by grants from the National Science Foundation and the National Institutes of Health-cosponsored Woods Hole Center for Oceans and Human Health, the Moore Foundation, and the Department of Energy to M.F.P., as well as postdoctoral fellowships from the MIT-Merck alliance to Y.B. and from the Dutch Government to O.X.C.
We thank Frédérique Leroux, Colleen Cavanaugh, Ford Doolittle, and Eric Alm for critical comments on the manuscript.
Footnotes
Citation Boucher Y, et al. 2011. Local mobile gene pools rapidly cross species boundaries to create endemicity within global Vibrio cholerae populations. mBio 2(2):e00335-10. doi:10.1128/mBio.00335-10.
REFERENCES
- 1. Colwell RR. 1996. Global climate and infectious disease: the cholera paradigm. Science 274:2025–2031 [DOI] [PubMed] [Google Scholar]
- 2. Nelson EJ, Harris JB, Morris JG, Calderwood SB, Camilli A. 2009. Cholera transmission: the host, pathogen and bacteriophage dynamic. Nat. Rev. Microbiol. 7:693–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lapierre P, Gogarten JP. 2009. Estimating the size of the bacterial pan-genome. Trends Genet. 25:107–110 [DOI] [PubMed] [Google Scholar]
- 4. Tettelin H, Riley D, Cattuto C, Medini D. 2008. Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 12:472–477 [DOI] [PubMed] [Google Scholar]
- 5. Vesth T, et al. 2010. On the origins of a Vibrio species. Microb. Ecol. 59:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chun J, et al. 2009. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 106:15442–15447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Safa A, Nair GB, Kong YC. 2009. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 18:46–54 [DOI] [PubMed] [Google Scholar]
- 8. Haley BJ, et al. 2010. Comparative genomic analysis reveals evidence of two novel Vibrio species closely related to V. cholerae. BMC Microbiol. 10:154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Faruque SM, Albert MJ, Mekalanos JJ. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ivars-Martinez E, et al. 2008. Biogeography of the ubiquitous marine bacterium Alteromonas macleodii determined by multilocus sequence analysis. Mol. Ecol. 17:4092–4106 [DOI] [PubMed] [Google Scholar]
- 11. Reno ML, Held NL, Fields CJ, Burke PV, Whitaker RJ. 2009. Biogeography of the Sulfolobus islandicus pan-genome. Proc. Natl. Acad. Sci. U. S. A. 106:8605–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Whitaker RJ, Grogan DW, Taylor JW. 2003. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976–978 [DOI] [PubMed] [Google Scholar]
- 13. Mazel D, Dychinco B, Webb VA, Davies J. 1998. A distinctive class of integron in the Vibrio cholerae genome. Science 280:605–608 [DOI] [PubMed] [Google Scholar]
- 14. MacDonald D, Demarre G, Bouvier M, Mazel D, Gopaul DN. 2006. Structural basis for broad DNA-specificity in integron recombination. Nature 440:1157–1162 [DOI] [PubMed] [Google Scholar]
- 15. Chao A, Chazdon RL, Colwell RK, Shen TJ. 2005. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett. 8:148–159 [Google Scholar]
- 16. Martin W. 1999. Mosaic bacterial chromosomes: a challenge en route to a tree of genomes. Bioessays 21:99–104 [DOI] [PubMed] [Google Scholar]
- 17. Labbate M, et al. 2007. Use of chromosomal integron arrays as a phylogenetic typing system for Vibrio cholerae pandemic strains. Microbiology 153:1488–1498 [DOI] [PubMed] [Google Scholar]
- 18. Guerin E, et al. 2009. The SOS response controls integron recombination. Science 324:1034 [DOI] [PubMed] [Google Scholar]
- 19. Gillings MR, Holley MP, Stokes HW, Holmes AJ. 2005. Integrons in Xanthomonas: a source of species genome diversity. Proc. Natl. Acad. Sci. U. S. A. 102:4419–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim YR, et al. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71:5461–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matz C, Kjelleberg S. 2005. Off the hook—how bacteria survive protozoan grazing. Trends Microbiol. 13:302–307 [DOI] [PubMed] [Google Scholar]
- 22. Kunin V, et al. 2008. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res. 18:293–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rodriguez-Valera F, et al. 2009. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7:828–836 [DOI] [PubMed] [Google Scholar]
- 24. Huq A, et al. 1983. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 45:275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hunt DE, Gevers D, Vahora NM, Polz MF. 2008. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl. Environ. Microbiol. 74:44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pruzzo C, Vezzulli L, Colwell RR. 2008. Global impact of Vibrio cholerae interactions with chitin. Environ. Microbiol. 10:1400–1410 [DOI] [PubMed] [Google Scholar]
- 27. Pál C, Papp B, Lercher MJ. 2005. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat. Genet. 37:1372. [DOI] [PubMed] [Google Scholar]
- 28. Hehemann JH, et al. 2010. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464:908–912 [DOI] [PubMed] [Google Scholar]
- 29. Martiny AC, Kathuria S, Berube PM. 2009. Widespread metabolic potential for nitrite and nitrate assimilation among Prochlorococcus ecotypes. Proc. Natl. Acad. Sci. U. S. A. 106:10787–10792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boucher Y, et al. 2006. Recovery and evolutionary analysis of complete integron gene cassette arrays from Vibrio. BMC Evol. Biol. 6:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824–1827 [DOI] [PubMed] [Google Scholar]
- 32. Baharoglu Z, Bikard D, Maze D. 2010. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet. 6:e1001165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dziejman M, et al. 2005. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc. Natl. Acad. Sci. U. S. A. 102:3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lane DJ. 1991. 16S/23S rRNA sequencing, p. 115–175 In Stackebrandt E, Goodfellow M, Nucleic acid techniques in bacterial systematics. Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 35. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 37. Maiden MC. 2008. Population genomics: diversity and virulence in the Neisseria. Curr. Opin. Microbiol. 11:467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O’Shea YA, Reen FJ, Quirke AM, Boyd EF. 2004. Evolutionary genetic analysis of the emergence of epidemic Vibrio cholerae isolates on the basis of comparative nucleotide sequence analysis and multilocus virulence gene profiles. J. Clin. Microbiol. 42:4657–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Preheim SP, et al. 2011. Metapopulation structure of Vibrionaceae among coastal marine invertebrates. Environ. Microbiol. 13:265–275 [DOI] [PubMed] [Google Scholar]
- 40. Tarr CL, et al. 2007. Identification of Vibrio isolates by a multiplex PCR assay and rpoB sequence determination. J. Clin. Microbiol. 45:134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thompson CC, et al. 2004. Use of recA as an alternative phylogenetic marker in the family Vibrionaceae. Int. J. Syst. Evol. Microbiol. 54:919–924 [DOI] [PubMed] [Google Scholar]
- 42. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 44. Rodriguez F, Oliver JL, Marin A, Medina JR. 1990. The general stochastic model of nucleotide substitution. J. Theor. Biol. 142:485–501 [DOI] [PubMed] [Google Scholar]
- 45. Schloss PD, Handelsman J. 2006. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 72:6773–6779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Joss MJ, et al. 2009. ACID: annotation of cassette and integron data. BMC Bioinformatics 10:118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schloss PD, Handelsman J. 2008. A statistical toolbox for metagenomics: assessing functional diversity in microbial communities. BMC Bioinformatics 9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Meyer F, et al. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schmidt HA, von Haeseler A. 2007. Maximum-likelihood analysis using TREE-PUZZLE. Curr. Protoc. Bioinformatics Chapter 6 :Unit 6.6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.