Abstract
Background & Aims
5-FU-based adjuvant chemotherapy does not increase survival times of patients with colorectal tumors with microsatellite instability. We determined the response of patients with colorectal tumors with the CpG island methylator phenotype (CIMP) to 5-FU-based therapy.
Methods
We analyzed a population-based cohort of 302 patients with colorectal cancer (CRC) for a median follow-up time of 50.7 months. CIMP status was determined by analysis of the CACNAG1, SOCS1, RUNX3, NEUROG1, and MLH1 promoters; tumors were considered to be CIMP-positive (CIMP+) if at least 3 promoters were methylated.
Results
Tumors from 29.5% (89/302) of patients were CIMP+; this did not influence disease-free survival (log rank=.26). Of tumors of TNM stages II–III (n=196), 32.7% were CIMP+. Among patients with CRC stages II–III who did not receive adjuvant 5-FU chemotherapy, those with CIMP+ tumors had longest times of disease-free survival (log rank=.04); patients with CIMP+ tumors who received chemotherapy had shorter times of disease-free survival (log rank=0.02). In patients with CIMP-negative tumors, adjuvant 5-FU chemotherapy significantly increased time of disease-free survival (log-rank=.00001). However, in patients with CIMP+ tumors, adjuvant 5-FU chemotherapy did not affect time of disease-free survival (log rank=.7). Multivariate analysis showed a significant, independent interaction between 5-FU treatment and CIMP status (hazard ratio [HR]=0.6; 95% confidence interval [CI], .5–.8). Among patients with CIMP+ tumors, adjuvant chemotherapy was not an independent predictor of outcome (HR=0.8; 95% CI, 0.3–2.0). In patients who did not receive adjuvant 5-FU chemotherapy, CIMP status was the only independent predictor of survival (HR=2.0; 95% CI, 1.1–3.8)
Conclusion
Patients with CIMP+ colorectal tumors do not benefit from 5-FU–based adjuvant chemotherapy.
Keywords: Colon cancer, 5-FU adjuvant chemotherapy, DNA methylation, response to cancer therapy
INTRODUCTION
Colorectal cancer (CRC) is a common disease that accounts for a large proportion of cancer cases in Western countries. Approximately a 10–15% of CRC are caused by epigenetic silencing of the gene MLH1, provoking a characteristic molecular phenotype called microsatellite instability (MSI), displaying a form of genetic instability characterized by the accumulation of numerous mutations within repetitive sequences of DNA in non-encoding microsatellite regions 1, 2. In the last few years, there has been a growing recognition of the existence of a new pathway for CRC pathogenesis, which involves the transcriptional silencing of tumor suppressor genes by hypermethylation of CpG islands surrounding the promoter regions of various genes 3. Tumors with such features are classified as having the CpG island methylator phenotype (CIMP) 4, and it is now believed that as many as one-third to one-half of all CRCs may evolve through this pathway 5–7. CIMP tumors with methylation-induced silencing of MLH1 constitute the majority of sporadic MSI CRCs 8. However, most CIMP-positive tumors are associated with microsatellite stability (MSS) 9. These CIMP MSS tumors share certain clinical and pathological features with MSI CRCs, including a predilection for females, advanced age of disease onset, predilection for proximal colon, poor differentiation and mucinous histology 10.
Patients with MSI CRCs have a better prognosis 11 and do not obtain benefit from 5-fluorouracil (5-FU)-based adjuvant chemotherapy 12–16. Since a significant majority of CIMP tumors share features of sporadic MSI cancers, one would suspect that they would have a similar therapeutic response. However, insufficient attention has been paid to this important clinical issue, and there is only limited published data on prognosis in CRCs with CIMP 17–19, and the response to chemotherapy in this type of CRC is unclear or worse, contradictory 20–22. The present study was designed to better understand the prognosis of patients with CIMP-positive CRCs, and to determine the response to adjuvant 5-FU chemotherapy using a large population-based collection of CRCs. This information could be critical in the choice of chemotherapeutic treatment, especially considering recently developed new inhibitors of DNA methylation 23. Our results suggest that CIMP-positive CRCs, similarly to MSI tumors, do not obtain a significant benefit from 5-FU-based adjuvant chemotherapy.
MATERIALS AND METHODS
Clinical specimens
This study included 302 CRC patients that were enrolled as part of the EPICOLON project, a population-based study on CRC 24, 25. These patients were randomly selected from a previously described cohort of patients for which follow-up data were available 14, 15. The study was approved by the institutional ethics committee of each participant hospital and written informed consent was obtained from all patients.
The integrity of the mismatch repair system was evaluated by MSI testing and immunostaining for MLH1, MSH2, MSH6 and PMS2 proteins. Tumor mismatch repair (MMR) deficiency was defined by either finding MSI-high (see below) or the loss of MLH1, MSH2, MSH6 or PMS2 protein expression by immunohistochemistry.
Adjuvant chemotherapy was administered according to standard clinical criteria. Adjuvant chemotherapy with 5-fluorouracil (5-FU) was given to patients with stage II and III tumors following standard schedules and doses. Oncologists that decided to administer adjuvant treatment were blinded to the MMR or CIMP tumor status.
Tumor MSI analysis
MSI testing was performed using either the 5-marker panel proposed by the National Cancer Institute or a pentaplex of mononucleotide repeats, and tumors were classified as microsatellite stable (MSS) or unstable (MSI) as previously described 26–28.
Mismatch repair protein immunohistochemistry
One block of formalin-fixed, paraffin-embedded tumor tissue was selected per case. Sections were then incubated for 20 minutes at room temperature with mouse monoclonal antibodies against MLH1 protein (clone G168-15, dilution 1:40; PharMingen, San Diego, CA), MSH2 protein (clone FE11, dilution 1:35; Oncogene Research Products, Boston, MA), MSH6 (Calbiochem, clon FE11, dilution 1:30) and PMS2 (BD Pharmingen, clone A16-4, dilution 1:100). The Ultra-Vision streptavidin-biotin peroxidase detection kit (DAKO, Carpinteria, CA) was used as the secondary detection system. Tumor cells were judged to be negative for protein expression only if they lacked staining in a sample in which normal colonocytes and stromal cells were stained. If no immunostaining of normal tissue could be demonstrated, the results were considered ambiguous.
BRAF V600E mutation analyses
V600E BRAF mutation was detected using real-time TaqMan probes in an ABI Prism 7500 sequence detection system by allelic discrimination. The PCR conditions and details have been previously described 29 including primers and probes. Probes were labeled with 6-carboxyfluorescein (FAM) as the reporter fluorophor for the mutant allele and VIC as the reporter fluorophor for the wild-type allele. Fluorescence data were analyzed with the allelic discrimination software of the ABI Prism 7500 instrument.
CIMP status determination by bisulfite pyrosequencing
The following five markers were used to assess the methylation status of the tumor tissue: CACNAG1, SOCS1, RUNX3, NEUROG1, and MLH1 9, 30. Genomic DNA was modified with sodium-bisulfite using the EZ Methylation Gold Kit (Zymo Research, Orange, CA) and markers were analyzed by pyrosequencing. The PCR reaction contained bisulfite modified DNA, HotStar Taq polymerase, forward primers, biotinylated reverse primers and water. The primers used have been described previously 30. To assess the methylation status for each marker we calculated the mean percentage of methylation for all the CpG sites included in the assay. Each marker was classified as methylated when the mean percentage was higher than 5% for the following markers: CACNAG1, SOCS1, RUNX3, and MLH1, and 10% for the NEUROG1 marker. These cut offs were chosen after determining the average methylation level of each marker in normal colonic mucosa samples plus two standard deviations. CRC was considered CIMP+ if at least 3 out of the five studied genes showed aberrant methylation 9, 30.
Statistical analysis
Continuous variables are reported as means ± standard deviation and categorical variables as frequency or percentages. Statistical differences of baseline characteristics between groups were analyzed using the chi-square test for categorical data after application of the Yates’s correction (when required), and by using the Mann-Whitney U test for quantitative data.
The primary outcome was disease-free survival (DFS). Analyses were performed in the entire series as well as in the subset of patients with stage II and III tumors in order to specifically evaluate the effect of CIMP status on the benefit from adjuvant chemotherapy. DFS was defined as the time from study entry to death from any cause or the first relapse. Data on DFS were censored at 73 months from the date of diagnosis. Differences in DFS were analyzed using the chi-square test. Probability curves were generated according to the Kaplan and Meier method and univariate survival distributions were compared with the use of the log-rank test. Confidence intervals were calculated using the standard error of survival according to the Greenwood method.
A multivariate analysis of hazard risk of death or tumor recurrence (DFS) was performed using Cox proportional-hazards regression to test the effect of CIMP, MMR status, adjuvant chemotherapy, age, TNM stage, gender, and the interaction between CIMP status and different variables such as chemotherapy, age, MMR status and BRAF V600E mutation. Hazard ratios and 95 percent confidence intervals (95% CI) for death or tumor recurrence (DFS) were computed using the Cox survival modelling.
All reported P values are two-sided, and P values of less than 0.05 were considered to indicate significance. All calculations were performed using the SPSS 16.0 software.
RESULTS
1. Influence of CIMP status on overall disease-free survival (DFS)
During the course of this study, 302 CRC patients were included. The median follow-up was 50.7 months (range 0–73 months). A total of 89 patients had CIMP+ tumors (29.5%). MMR deficiency was present in 8.3% (25/302) of tumors. Clinical and pathological differences between patients with CIMP+ or CIMP-tumors are illustrated in Table 1. Thirteen patients (4.3%) were lost to follow-up and, accordingly, they were censored at that time-point. At the end of the follow-up period, 125 patients had died (41.4%), and the median follow-up for this group was 18.2 months (range 0–68 months). Seven patients died during the immediate post-operative period (the first 30 days post-surgery), and 86 patients died due to tumor progression. The causes of death for the remaining patients were: late post-surgical complications (4 patients), complications of chemotherapy (6 patients) and other causes (22 patients). Tumor recurrence was seen in 25 cases (8.2%), at a median of 19.4 months after surgery (range 2–52 months).
Table 1.
Characteristics of patients according to CIMP status
| CIMP positive (n=89) | CIMP negative (n=213) | p | |
|---|---|---|---|
| Age, mean (SD) | 72.2 (10.9) | 69.5 (11.8) | 0.06 |
| Gender, n (%) | |||
| Male | 43 (48.3) | 142 (66.7) | 0.002 |
| Female | 46 (51.7) | 71 (33.3) | |
| TNM stage, n (%) | |||
| I | 8 (9) | 41 (18.2) | 0.1 |
| II | 37 (41.6) | 71 (33.3) | |
| III | 27 (30.3) | 61 (28.6) | |
| IV | 17 (19.1) | 40 (18.8) | |
| MMR deficient, n (%) | |||
| Yes | 20 (22.5) | 5 (2.3) | 0.0001 |
| No | 69 (77.5) | 208 (97.7) | |
| BRAF mutation, n (%) | |||
| Yes | 20 (22.5) | 1 (0.5) | 0.0001 |
| No | 69 (78.5) | 212 (95.5) | |
| Chemotherapy, n (%) | |||
| Yes | 33 (37.1) | 103 (48.4) | 0.07 |
| No | 56 (62.9) | 110 (51.6) | |
| Disease-free survival, n (%) | |||
| Yes | 43 (48.3) | 109 (51.2) | 0.6 |
| No | 46 (51.7) | 104 (48.8) | |
Chemotherapy was given to 136 patients (45.0%), which included 93 (68.4%) patients who received adjuvant chemotherapy, 8 (5.9%) neoadjuvant chemotherapy and 35 (25.7%) palliative chemotherapy. Chemotherapy was 5-FU based in all patients.
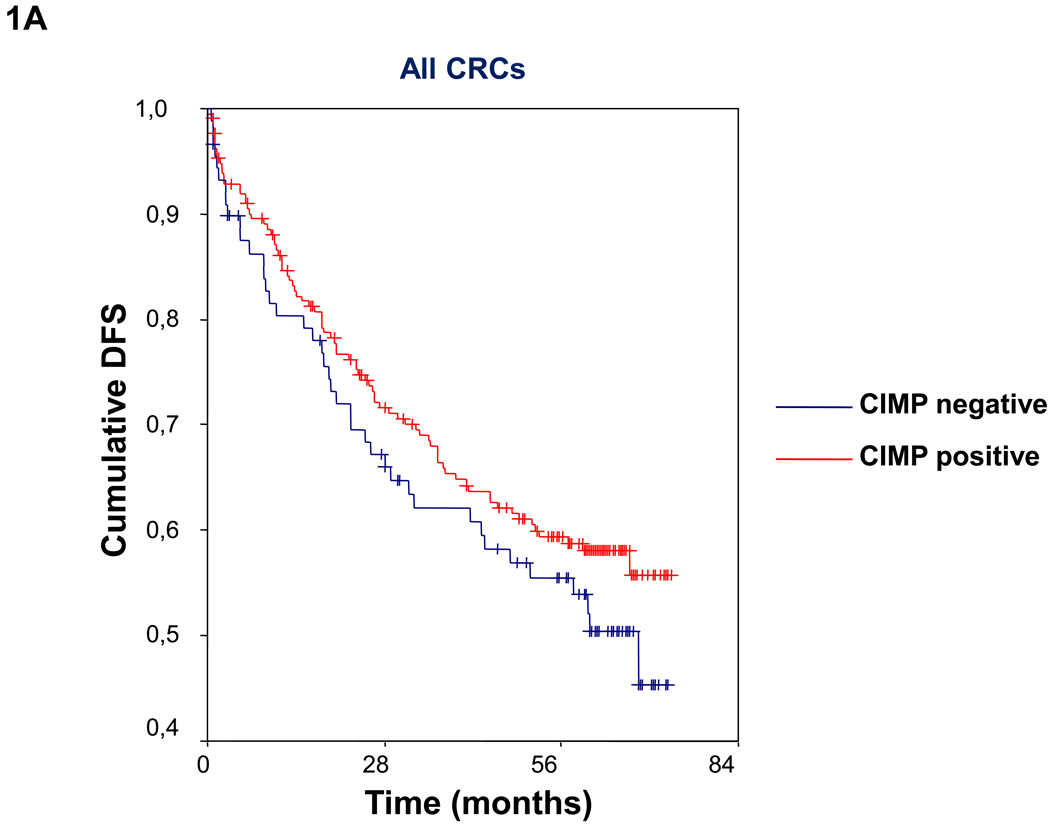
Patients with CIMP- tumors were significantly more likely to be male, MMR proficient, and BRAF mutation negative (Table 1). Overall, in the whole cohort, there were no significant differences in the percentage of disease-free survivors at the end of follow-up between patients with CIMP+ and CIMP− tumors (CIMP+: 48.3%; CIMP−: 51.2%; chi square p=0.6; log rank=0.3) (Table 1. Figure 1A). This lack of difference in DFS remained unchanged when we excluded patients who died in the postoperative period. Moreover, we did not find changes in DFS when we excluded patients with MMR deficient tumors (data not shown).
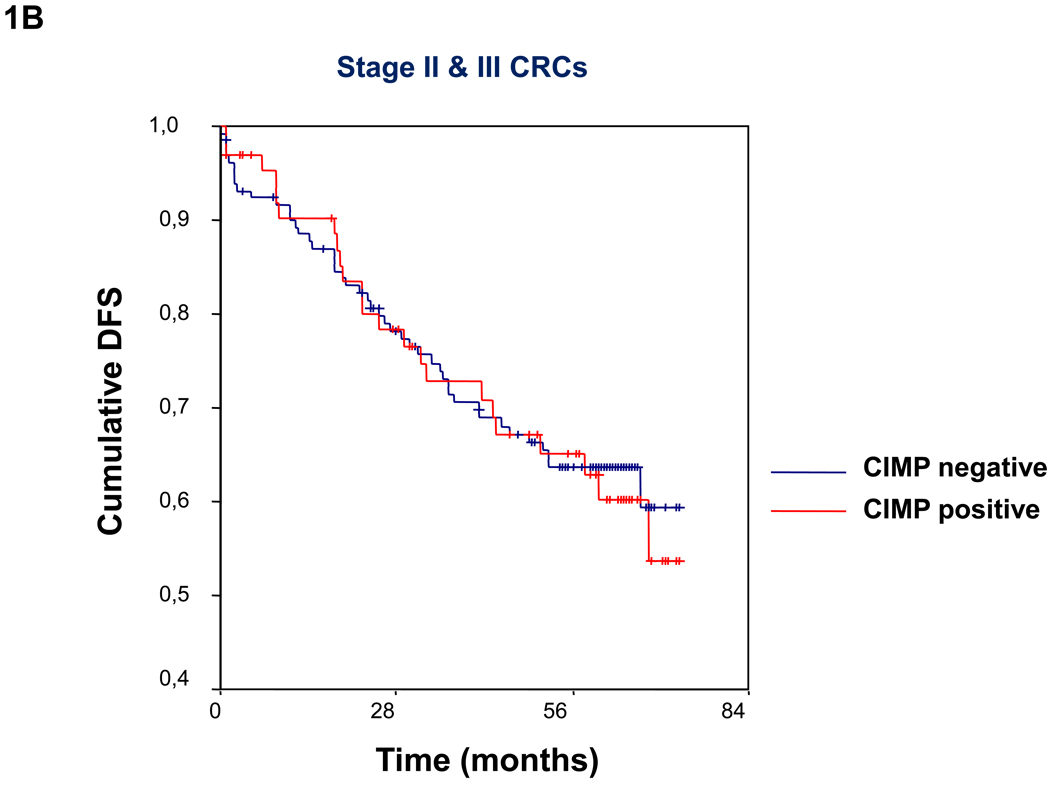
Figure 1.
(A)Kaplan-Meier survival curves depicting the effect of CIMP status on disease-free survival (DFS) in the entire series of CRCs. Vertical tick marks indicate censored events. (B) Kaplan-Meier survival curves illustrating the effect of CIMP status on disease-free survival (DFS) in patients with TNM stage II and III CRCs. Vertical tick marks indicate censored events.
2. Influence of CIMP status on prognosis in patients with locally advanced, non-metastatic colorectal cancer
Our cohort included follow-up on 196 patients with stage II or III CRCs. The median follow-up time was 55.0 months (range 0–73). Characteristics of this subset of patients according to CIMP status are summarized in Table 2. The patients with CIMP- tumors were significantly more likely to be younger, male, have tumors that were MMR proficient and BRAF mutation negative. Eleven patients (5.6%) were lost to follow-up. At the end of follow-up, 69 (35.2%) patients had died and 21 (10.7%) had experienced tumor recurrence. One-hundred and one patients received adjuvant 5-FU-based chemotherapy; 47 (46.5%) were stage II and 54 (53.5%) were stage III CRC patients.
Table 2.
Characteristics of stage II–III patients
| CIMP positive (n=64) | CIMP negative (n=132) | p | |
|---|---|---|---|
| Age, mean (SD) | 73.7 (9.6) | 69.7 (11.8) | 0.02 |
| Gender, n (%) | |||
| Male | 27 (42.9) | 142 (65.4) | 0.003 |
| Female | 36 (57.1) | 71 (34.6) | |
| TNM stage, n (%) | |||
| II | 37 (57.8) | 71 (53.8) | 0.6 |
| III | 27 (42.2) | 61 (46.2) | |
| MMR deficient, n (%) | |||
| Yes | 15 (23.4) | 3 (2.3) | 0.0001 |
| No | 49 (76.6) | 129 (97.7) | |
| BRAF mutation, n (%) | |||
| Yes | 13 (20.3) | 1 (0.8) | 0.0001 |
| No | 51 (79.7) | 131 (99.2) | |
| Chemotherapy, n (%) | |||
| Yes | 26 (40.6) | 75 (56.8) | 0.03 |
| No | 38 (69.4) | 57 (43.2) | |
| Disease-free survival, n (%) | |||
| Yes | 36 (56.3) | 70 (53.0) | 0.7 |
| No | 28 (43.8) | 62 (47.0) | |
Disease free survival is the proportion of survivors at the end of follow-up.
Patients who received chemotherapy were younger (64.7±7.5 vs. 77.6±10.6; p=0.0001) and there were no differences in gender, duration of follow-up or percentage of lost to follow-up compared to patients who received adjuvant chemotherapy. Adjuvant chemotherapy was more frequently administered to CIMP negative stage II and stage III cancer patients (Table 2).
Overall, in patients with stage II–III CRC, there were no differences in the percentage of disease-free survivors at the end of follow-up according to CIMP status (CIMP+: 56.3%; CIMP−: 53.0%; chi square p=0.7; log rank=0.8) (Table 2, Figure 1B). In the univariate analysis, adjuvant chemotherapy, TNM stage and age less than 60 were the only variables with a significant influence on DFS (Table 3). In the multivariate analysis (Table 4), adjuvant chemotherapy and TNM stage were the only independent predictors of survival; however, a significant interaction between adjuvant chemotherapy and CIMP status on DFS was also observed (hazard ratio 0.6, 95% CI 0.5–0.8). No significant interaction was found between CIMP status and age (hazard ratio 1.2, 95% CI 0.9–1.5) or age and adjuvant chemotherapy (hazard ratio 1.1, 95% CI 0.9–1.3) in terms of DFS.
Table 3.
Univariate survival analysis in stage II and III patients
| Univariate survival analysis | p-value | |
|---|---|---|
| Probability of DFS (95%CI) | ||
| Adjuvant Chemotherapy | ||
| Yes | 61.4% (51.7–71.1%) | |
| No | 46.3% (36.1–56.5%) | 0.001 |
| CIMP status | ||
| Positive | 56.3% (43.9–68.7%) | |
| Negative | 53.0% (44.3–61.7%) | 0.8 |
| TNM stage | ||
| II | 60.2% (50.8–69.6%) | |
| III | 46.6% (36.0–57.2%) | 0.02 |
| Age | ||
| ≤60 | 74.2% (58.5–89.9%) | |
| >60 | 50.0% (42.1–57.9%) | 0.02 |
| Microsatellite instability | ||
| Stable | 52.6% (45.1–60.1%) | |
| Unstable | 65.2% (42.7–87.7%) | 0.2 |
| BRAF mutation | ||
| Mutated | 42.8% (13.8–66.2%) | |
| Not mutated | 55.6% (52.5–67.5%) | 0.8 |
| Gender | ||
| Male | 51.8% (44.1–59.5%) | |
| Female | 56.8% (47.2–66.4%) | 0.7 |
| Univariate survival analysis according to treatment received | ||
| Probability of DFS (95%CI) | p-value | |
| CIMP + patients (n=64) | ||
| Adjuvant chemotherapy | 57.7% (38.3–77.1%) | 0.7 |
| No adjuvant chemotherapy | 68.4% (53.3–83.5%) | |
| CIMP – patients (n=132) | ||
| Adjuvant chemotherapy | 80.0% (70.8–89.2%) | 0.00001 |
| No adjuvant chemotherapy | 45.6% (32.4–58.8%) | |
| Univariate survival analysis according to CIMP status | ||
| Probability of DFS (95%CI) | p-value | |
| Adjuvant chemotherapy (n=101) | ||
| CIMP + | 57.7% (38.3–77.1%) | 0.02 |
| CIMP − | 80.0% (70.8–89.2%) | |
| No adjuvant chemotherapy (n=95) | ||
| CIMP + | 68.4% (53.3–83.5%) | 0.04 |
| CIMP − | 45.6% (32.4–58.8%) | |
p-value: log-rank test
Table 4.
Cox regression multivariate analysis of disease-free survival in stage II and III patients
| Variable | Hazard Ratio | 95% CI | p | |
|---|---|---|---|---|
| All patients | Age (<60) | 1.2 | 0.8–1.9 | 0.3 |
| Adjuvant CT | 0.5 | 0.3–0.8 | 0.002 | |
| TNM stage | 1.8 | 1.2–2.8 | 0.005 | |
| CIMP status | 1.2 | 0.8–2.0 | 0.4 | |
| All patients | Interaction CIMP-Adjuvant CT | 0.6 | 0.5–0.8 | 0.0001 |
| Interaction CIMP-Age | 1.2 | 0.9–1.5 | 0.17 | |
| Interaction Age-Adjuvant CT | 1.1 | 0.9–1.3 | 0.4 | |
| CIMP-positive | Age | 0.8 | 0.3–2.0 | 0.7 |
| Adjuvant CT | 0.8 | 0.3–2.0 | 0.6 | |
| TNM stage | 3.4 | 1.5–7.7 | 0.004 | |
| CIMP-negative | Age | 1.5 | 0.9–2.6 | 0.13 |
| Adjuvant CT | 0.4 | 0.2–0.6 | 0.0001 | |
| TNM stage | 1.3 | 0.8–2.2 | 0.3 | |
| Adjuvant CT | Age | 1.4 | 0.7–2.7 | 0.3 |
| CIMP status | 0.6 | 0.3–1.2 | 0.1 | |
| TNM stage | 2.1 | 1.1–4.0 | 0.03 | |
| No adjuvant CT | Age | 1.2 | 0.6–2.2 | 0.6 |
| CIMP status | 2.0 | 1.1–3.8 | 0.03 | |
| TNM stage | 1.6 | 0.9–2.8 | 0.1 | |
CT. Chemotherapy; CI: confidence interval.
2.1. Effect of CIMP status on survival according to chemotherapy treatment
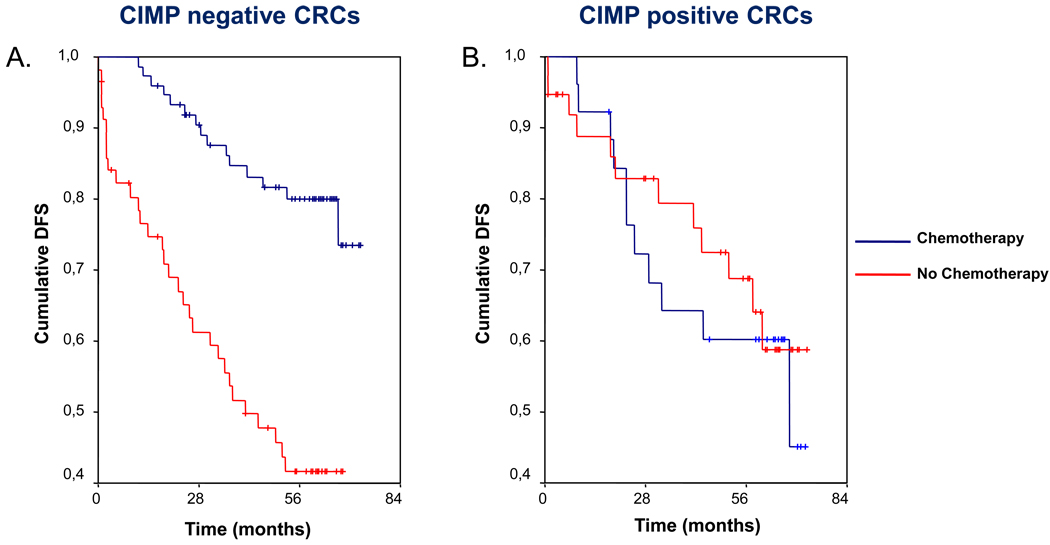
Patients with CIMP-negative tumors who received adjuvant chemotherapy had a higher probability of DFS (Table 3, Figure 2A). However, in patients with CIMP-positive tumors, adjuvant chemotherapy did not improve the probability of DFS (Table 3, Figure 2B). In order to evaluate the combined influence of different factors on DFS, a multivariate Cox regression analysis was performed. In patients with CIMP-negative tumors, adjuvant chemotherapy was the only factor to influence DFS (Table 4). On the other hand, only the TNM stage independently predicted DFS in patients with CIMP-positive tumors, without any effect of adjuvant chemotherapy (Table 4). In this subset of patients neither MMR status (hazard ratio 0.8, 95% CI: 0.5–1.2), nor BRAF V600E mutation (hazard ratio 0.9, 95% CI: 0.5–2.7) was significantly related to DFS.
Figure 2.
Kaplan-Meier survival curves demonstrating the effect of CIMP status on survival according to 5-FU based chemotherapy treatment. In patients with CIMP-negative tumors (panel A), adjuvant chemotherapy clearly provides a survival benefit with a higher probability of disease-free survival (DFS). In patients with CIMP-positive tumors (panel B), the use of 5-FU based adjuvant chemotherapy does not improve DFS. Vertical tick marks indicate censored events.
2.2. Effect of chemotherapy on survival according to CIMP status
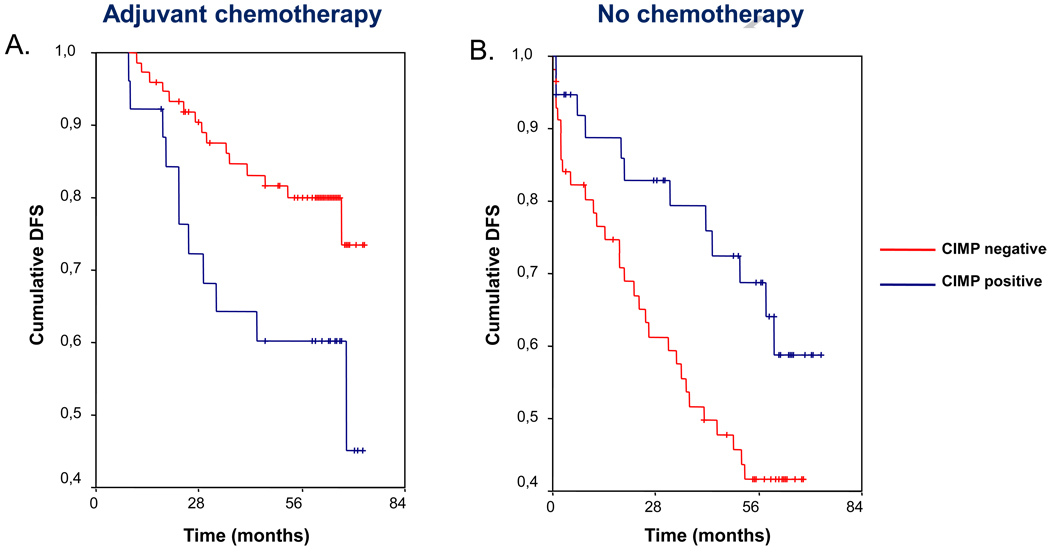
In patients treated with 5-FU adjuvant chemotherapy, DFS was significantly different depending on CIMP status (Table 3, Figure 3A). A multivariate analysis performed in this subset of patients reveals that CIMP status does not independently predict DFS, and the only independent factor significantly associated with survival was TNM stage (Table 4). Contrarily, in patients who did not receive adjuvant chemotherapy, the probability of DFS was positively affected by CIMP status (Table 3, Figure 3B). In this setting, CIMP status was the only independently significant factor related to DFS in the Cox regression multivariate analysis (Table 4). In this subset of patients, DFS was not affected by either MMR status (hazard ratio 0.6, 95% CI: 0.4–1.1) or BRAF V600E mutation (hazard ratio 1.5, 95% CI: 0.5–4.9).
Figure 3.
Kaplan-Meier survival curves indicating the effect of chemotherapy on survival according to CIMP status. In patients who received 5-FU based adjuvant chemotherapy (panel A), the existence of a CIMP-positive tumor confers a worse prognosis, with lower disease-free survival (DFS). In contrast, in patients who did not receive adjuvant chemotherapy (panel B) the situation reverses, and those with CIMP-positive tumors had a better prognosis. Vertical tick marks indicate censored events.
DISCUSSION
The present study was designed to better understand the contribution of CIMP in the prognosis of patients with CRC, and to determine its effect in response to 5-FU based adjuvant chemotherapy in a population-based collection of patients. Our results indicate that, in this cohort of CRC patients, CIMP status does not independently affect DFS. However, when we select only patients with TNM stage II or III, CIMP status directly influences the patient response to 5-FU based adjuvant chemotherapy. Our data suggest that adjuvant 5-FU chemotherapy improves DFS in this study group, but the effect depends on CIMP status. The key observation is that the benefit from adjuvant 5-FU chemotherapy is limited to patients with CIMP-negative CRC. When 5-FU based adjuvant chemotherapy is administered to patients with CIMP-positive tumors, this does not improve DFS. Moreover, survival in stage II or III patients with CIMP-positive CRC who did not receive adjuvant chemotherapy was clearly better.
Our findings corroborate previous evidence for the effects of 5-FU based chemotherapy in MMR deficient CRCs 16. It is important to point out that, in our cohort, only a small proportion of the tumors were MMR-deficient, and the differential responses are lost when analyzing the whole group. This highlights that the correlation between the response to chemotherapy and CIMP status is independent of the MMR status in these CRCs. Our cohort comes from an observational study and decisions about adjuvant chemotherapy were clinically based, rather than randomized. Since factors like patient age can influence the decision to treat stage II or III patients, we performed a multivariate analysis between several clinical-pathological and molecular variables. Although the median age of patients receiving adjuvant chemotherapy was almost 13 years younger than those who did not, no benefit was observed in the patients with CIMP-positive tumors. Our data revealed a significant independent interaction for survival between CIMP status and chemotherapy, whereas we did not find any interaction between age and CIMP status or age and chemotherapy in terms of survival. These results indicate that CIMP status, rather than age, influenced the response to chemotherapy in this study.
Several different studies have investigated the relationship between CIMP status and survival in CRC, however, the results have been inconsistent 18, 19, 22, 31. A few studies have suggested an adverse effect of CIMP on survival, specifically in the subgroup of CIMP-positive/MSI-negative patients 18, 22, 31, 32. In addition, some reports suggest that there is resistance to chemotherapy in CIMP-positive tumors 20, 21, while others have indicated the opposite 22. Although these discrepancies have not been systematically analyzed, it is likely that some of the contradictory results are influenced by the relative proportion of MSI-positive tumors, BRAF V600E mutant CRCs, and technical issues related to evaluating the methylation status. Prior studies that have reported a lack of chemotherapeutic response often included only patients with metastatic disease when comparing different chemotherapeutic schedules 20, 21. The biggest caveat for such studies is that they lack a control arm, which would have permitted appropriate comparisons between treatment and no treatment groups. In support of this, we did not observe a poorer prognosis for CIMP-positive patients treated with 5-FU, but experienced only a lack of benefit in stage II–III patients. On the other hand, only one study has suggested a beneficial response to chemotherapy for CIMP positive CRC in stage III CRCs 22. Possible explanations for differences between that study and ours are likely methodological, because methylation analysis in the previous study used only three methylation loci, which may have resulted in an erroneous categorization of CIMP tumors, as a minimum of 5 markers are typically analyzed to define CIMP status in CRC. In addition, the prior study used non-quantitative methylation-specific PCR. Lastly, this study included patients who were treated between 1985 and 1999, and perhaps does not account for the important differences in clinical management of CRC over time, as well as the improvement in overall prognosis for CRC 33. Moreover, the patient population used in this study provided results that are conspicuous outliers for response to chemotherapy in CRCs with MSI as well 22. Contrary to what has been suggested by a previous study 19, we did not find an influence of BRAF mutations on survival or the response to chemotherapy,. However, in our cohort, the number of patients with BRAF mutated tumors was too small to identify small differences, particularly in the subgroup of CIMP-negative CRC.
Another possible factor that could influence heterogeneity between studies is the methylation markers employed as well as the methodology for determination of gene methylation. A variety of methylation markers have been used, and recently a robust new marker panel has been proposed that outperforms prior panels and recognizes a distinct, heavily methylated subset of CRCs 9. We have used this new panel of CIMP markers, and the methylation status of CIMP markers was defined using pyrosequencing, a quantitative technique with several advantages over qualitative techniques. Given the strengths of our unbiased population of CRC patients, our experience with methylation analysis using pyrosequencing 30, and the fact that our results are consistent with most of the prior studies, we believe that this interpretation is a more reliable reflection of the role of CIMP in CRC.
CIMP-positive CRCs are genetically different from CIMP-negative tumors, and microarray analysis has identified a number of genes that are differentially expressed between these groups of tumors 34. The metabolic activity responsible for converting 5-FU into its active metabolites may be different between CIMP-positive and CIMP-negative tumors, resulting in differences in chemosensitivity between these groups of patients. The lack of benefit of 5-FU in MSI CRC has been repeatedly reported, and the majority of MSI tumors are also CIMP positive. Our results suggest that this apparent lack of response to adjuvant 5-FU seems to be related to hypermethylation and not to the MMR status of the tumors; a clinically important topic that should be the focus of research in the immediate future. In addition, identification of CIMP-positive tumors is not easy and is not commonly a part of routine laboratory workup. Molecular or immunohistochemical markers of CIMP-positive tumours should be developed for the practical identification of this important subset of CRC.
In summary, our results suggest that patients with CIMP-positive CRC do not benefit from adjuvant 5-FU based chemotherapy. This behavior has been previously described for MMR deficient CRC, suggesting a common mechanism related to hypermethylation. These results should be confirmed in larger series of patients coming from clinical trials in future. Moreover identification of CIMP tumours
Acknowledgments
Grant Support: The present work was supported by grants R01 CA72851 and R01 CA129286 to AG and CRB from the National Cancer Institute, National Institutes of Health, funds from Fundación de la CV para la Investigación en el Hospital General Universitario de Alicante to RJ, LPC, AP and CA, grant SAF 07-64873 from the Ministerio de Educación y Ciencia to AC, grants from the Asociación Española contra el Cáncer (Fundación Científica and Junta de Barcelona) to AC, funds from the AGAUR, (2009 SGR 849) to AC, and funds from the Baylor Research Institute. Lucia Pérez-Carbonell is a recipient of a predoctoral grant from Instituto de Salud Carlos III (FI07/00303). CIBERehd is funded by the Instituto de Salud Carlos III.
Abbreviations
- 5-FU
5-Fluorouracil
- CIMP
CpG Island Methylator Phenotype
- CRC
Colorectal Cancer
- DFS
Disease-Free Survival
- MMR
Mismatch Repair
- MSI
Microsatellite Instability
- MSS
Microsatellite Stability
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: None of the authors have any potential conflicts to disclose.
Authors’ contributions
R.J. and A.G. developed the study concept and design and drafted the manuscript
R.J., T.P.N., A.G., F.B., L.P.C, P.Z., A.P., C.A., E.S., J.C., J.D.M., J.C., L.B., J.M.R., X.B., R.M.X., D.N.P., acquired and analyzed the data
A.C., M.A. and X.L. provided the study materials
REFERENCES
- 1.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wheeler JM, Bodmer WF, Mortensen NJ. DNA mismatch repair genes and colorectal cancer. Gut. 2000;47:148–153. doi: 10.1136/gut.47.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 4.Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, O'Connor T, Ward R. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–1387. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- 5.Goel A, Arnold CN, Niedzwiecki D, Chang DK, Ricciardiello L, Carethers JM, Dowell JM, Wasserman L, Compton C, Mayer RJ, Bertagnolli MM, Boland CR. Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res. 2003;63:1608–1614. [PubMed] [Google Scholar]
- 6.Nagasaka T, Koi M, Kloor M, Gebert J, Vilkin A, Nishida N, Shin SK, Sasamoto H, Tanaka N, Matsubara N, Boland CR, Goel A. Mutations in both KRAS and BRAF may contribute to the methylator phenotype in colon cancer. Gastroenterology. 2008;134:1950–1960. doi: 10.1053/j.gastro.2008.02.094. 1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, Compton C, Mayer RJ, Goldberg R, Bertagnolli MM, Boland CR. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 8.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 9.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 10.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 11.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 12.Carethers JM, Smith EJ, Behling CA, Nguyen L, Tajima A, Doctolero RT, Cabrera BL, Goel A, Arnold CA, Miyai K, Boland CR. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology. 2004;126:394–401. doi: 10.1053/j.gastro.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 13.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, Tu D, Redston M, Gallinger S. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jover R, Zapater P, Castells A, Llor X, Andreu M, Cubiella J, Pinol V, Xicola RM, Bujanda L, Rene JM, Clofent J, Bessa X, Morillas JD, Nicolas-Perez D, Paya A, Alenda C. Mismatch repair status in the prediction of benefit from adjuvant fluorouracil chemotherapy in colorectal cancer. Gut. 2006;55:848–855. doi: 10.1136/gut.2005.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jover R, Zapater P, Castells A, Llor X, Andreu M, Cubiella J, Balaguer F, Sempere L, Xicola RM, Bujanda L, Rene JM, Clofent J, Bessa X, Morillas JD, Nicolas-Perez D, Pons E, Paya A, Alenda C. The efficacy of adjuvant chemotherapy with 5-fluorouracil in colorectal cancer depends on the mismatch repair status. Eur J Cancer. 2009;45:365–373. doi: 10.1016/j.ejca.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 16.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, Ribic C, Grothey A, Moore M, Zaniboni A, Seitz JF, Sinicrope F, Gallinger S. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahlin AM, Palmqvist R, Henriksson ML, Jacobsson M, Eklof V, Rutegard J, Oberg A, Van Guelpen BR. The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status. Clin Cancer Res. 2010;16:1845–1855. doi: 10.1158/1078-0432.CCR-09-2594. [DOI] [PubMed] [Google Scholar]
- 18.Kim JH, Shin SH, Kwon HJ, Cho NY, Kang GH. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch. 2009;455:485–494. doi: 10.1007/s00428-009-0857-0. [DOI] [PubMed] [Google Scholar]
- 19.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogino S, Meyerhardt JA, Kawasaki T, Ryan DP, Kulke MH, Enzinger PC, Wolpin BM, Loda M, Fuchs CS. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch. 2007;450:529–537. doi: 10.1007/s00428-007-0398-3. [DOI] [PubMed] [Google Scholar]
- 21.Shen L, Catalano PJ, Benson AB, III, O'Dwyer P, Hamilton SR, Issa JP. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res. 2007;13:6093–6098. doi: 10.1158/1078-0432.CCR-07-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van RM, Elsaleh H, Joseph D, McCaul K, Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res. 2003;9:2898–2903. [PubMed] [Google Scholar]
- 23.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 24.Pinol V, Andreu M, Castells A, Paya A, Bessa X, Rodrigo J. Frequency of hereditary non-polyposis colorectal cancer and other colorectal cancer familial forms in Spain: a multicentre, prospective, nationwide study. Eur J Gastroenterol Hepatol. 2004;16:39–45. doi: 10.1097/00042737-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 25.Pinol V, Castells A, Andreu M, Castellvi-Bel S, Alenda C, Llor X, Xicola RM, Rodriguez-Moranta F, Paya A, Jover R, Bessa X. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293:1986–1994. doi: 10.1001/jama.293.16.1986. [DOI] [PubMed] [Google Scholar]
- 26.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 27.Xicola RM, Llor X, Pons E, Castells A, Alenda C, Pinol V, Andreu M, Castellvi-Bel S, Paya A, Jover R, Bessa X, Giros A, Duque JM, Nicolas-Perez D, Garcia AM, Rigau J, Gassull MA. Performance of different microsatellite marker panels for detection of mismatch repair-deficient colorectal tumors. J Natl Cancer Inst. 2007;99:244–252. doi: 10.1093/jnci/djk033. [DOI] [PubMed] [Google Scholar]
- 28.Goel A, Nagasaka T, Hamelin R, Boland CR. An optimized pentaplex PCR for detecting DNA mismatch repair-deficient colorectal cancers. PLoS One. 2010;5:e9393. doi: 10.1371/journal.pone.0009393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benlloch S, Paya A, Alenda C, Bessa X, Andreu M, Jover R, Castells A, Llor X, Aranda FI, Massuti B. Detection of BRAF V600E mutation in colorectal cancer: comparison of automatic sequencing and real-time chemistry methodology. J Mol Diagn. 2006;8:540–543. doi: 10.2353/jmoldx.2006.060070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goel A, Xicola RM, Nguyen TP, Doyle BJ, Sohn VR, Bandipalliam P, Rozek LS, Reyes J, Cordero C, Balaguer F, Castells A, Jover R, Andreu M, Syngal S, Boland CR, Llor X. Aberrant DNA methylation in hereditary nonpolyposis colorectal cancer without mismatch repair deficiency. Gastroenterology. 2010;138:1854–1862. doi: 10.1053/j.gastro.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward RL, Cheong K, Ku SL, Meagher A, O'Connor T, Hawkins NJ. Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. J Clin Oncol. 2003;21:3729–3736. doi: 10.1200/JCO.2003.03.123. [DOI] [PubMed] [Google Scholar]
- 32.Barault L, Charon-Barra C, Jooste V, de la Vega MF, Martin L, Roignot P, Rat P, Bouvier AM, Laurent-Puig P, Faivre J, Chapusot C, Piard F. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–8546. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- 33.Bujanda L, Sarasqueta C, Hijona E, Hijona L, Cosme A, Gil I, Elorza JL, Asensio JI, Larburu S, Enriquez-Navascues JM, Jover R, Balaguer F, Llor X, Bessa X, Andreu M, Paya A, Castells A. Colorectal cancer prognosis twenty years later. World J Gastroenterol. 2010;16:862–867. doi: 10.3748/wjg.v16.i7.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferracin M, Gafa R, Miotto E, Veronese A, Pultrone C, Sabbioni S, Lanza G, Negrini M. The methylator phenotype in microsatellite stable colorectal cancers is characterized by a distinct gene expression profile. J Pathol. 2008;214:594–602. doi: 10.1002/path.2318. [DOI] [PubMed] [Google Scholar]