Abstract
Morphological and functional changes during ameloblast and odontoblast differentiation suggest that enamel and dentin formation is under circadian control. Circadian rhythms are endogenous self-sustained oscillations with periods of 24 hours that control diverse physiological and metabolic processes. Mammalian clock genes play a key role in synchronizing circadian functions in many organs. However, close to nothing is known on clock genes expression during tooth development. In this work, we investigated the expression of four clock genes during tooth development. Our results showed that circadian clock genes Bmal1, clock, per1, and per2 mRNAs were detected in teeth by RT-PCR. Immunohistochemistry showed that clock protein expression was first detected in teeth at the bell stage (E17), being expressed in EOE and dental papilla cells. At post-natal day four (PN4), all four clock proteins continued to be expressed in teeth but with different intensities, being strongly expressed within the nucleus of ameloblasts and odontoblasts and down-regulated in dental pulp cells. Interestingly, at PN21 incisor, expression of clock proteins was down-regulated in odontoblasts of the crown-analogue side but expression was persisting in root-analogue side odontoblasts. In contrast, both crown and root odontoblasts were strongly stained for all four clock proteins in first molars at PN21. Within the periodontal ligament (PDL) space, epithelial rests of Malassez (ERM) showed the strongest expression among other PDL cells. Our data suggests that clock genes might be involved in the regulation of ameloblast and odontoblast functions, such as enamel and dentin protein secretion and matrix mineralization.
Keywords: Clock genes, Tooth development, Bmal1, Clock, Per1, Per2, expression pattern, immunohistochemistry
1. Introduction
Tooth development or odontogenesis is the complex process by which teeth form from embryonic cells, grow, and erupt into the mouth (Bei, 2009; Thesleff and Sharpe, 1997). Enamel, dentin, and cementum form by additive modes of growth that preserve within the hard tissues short- and long-period lines of incremental growth (Dean, 2000; Erickson, 1996). In human dental enamel for instance, there are daily cross-striations and long-period weekly daily lines or striae of Retzius (Guatelli-Steinberg et al., 2004; Li and Risnes, 2004). These regularly occurring incremental markers suggest that dental tissues formation is tightly controlled in time (Antoine et al., 2009). This hypothesis is further supported by studies that showed that collagen production in dentin follow a 12 hour-pattern (Lopez Franco et al., 2006). Circadian rhythms have been demonstrated using 3H-proline tracers that label collagen in dentin formation. Twice as much collagen is secreted during the 12 h of daylight when compared with the 12 h of night time (Ohtsuka et al., 1998). Furthermore, during the enamel maturation stage, ameloblasts oscillate between smooth-ended and ruffle-ended morphologies every 8 hours in rats (Lu et al., 2008; Smith, 1998). By periodically and rapidly altering their morphology from a tight configuration (ruffle-ended) to a more leaky configuration (smooth-ended) which re-establishes the equivalent of a new mineralization front over the same patch of maturing enamel, the ameloblast manage to fully mature the enamel matrix (Simmer et al., 2010). Taken together, these observations suggest that enamel and dentin formation and maturation are under circadian control. However, direct evidence of the expression and potential roles of clock genes (the genes that control circadian rhythms) in tooth is lacking.
The central clock is composed of about 20,000 neurons, all of which express clock genes that oscillate in synchrony (Ikeda, 2004; Welsh et al., 1995). Clock genes are defined by a set of criteria that include rhythm in activity or amount as well as molecular evidence of a feedback mechanism (Myers et al., 2003; Williams and Sehgal, 2001). Clock genes transmit output signals that drive rhythms of gene expression in central and peripheral tissues (Cassone and Stephan, 2002). The most direct mechanism by which clock genes drive circadian gene expression is through regulation of promoter activity of clock-controlled genes (Kondratov et al., 2006).
The expression of clock genes has never been investigated in dental tissues. We analyzed the RNA expression of the main clock genes, aryl hydrocarbon receptor nuclear translocator-like (Bmal1), circadian locomoter output cycles kaput (Clock), period homolog 1 (Per1) and period homolog 2 (Per2) in extracts of whole teeth and localized the main clock proteins on mouse teeth serial sections from embryonic day (E) 13 to day-21 post-natal (PN21) using immunohistochemistry. This paper is a first step towards the understanding of the role of clock genes in enamel and dentin formation.
2. Results and Discussion
2.1. Detection clock RNAs using RT-PCR
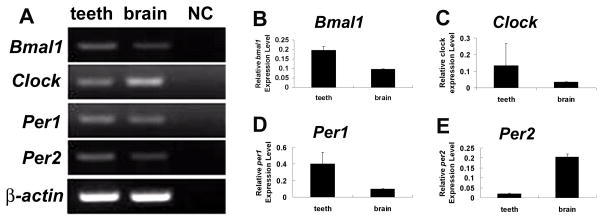
Bmal1, Clock, Per1 and Per2 mRNAs were detected in PN1 whole tooth extracts by conventional RT-PCR (Fig. 1), as well as in brain (positive control). All the PCR products were sequenced to confirm clock RNA expression. In addition, quantitative real-time PCR (qRT-PCR) was used to evaluate relative expression levels between tooth and brain (Fig. 1).
Figure 1.
Expression of clock genes RNAs in teeth and brain at post-natal (PN) day 1. Conventional RT-PCR showed that Bmal1, Clock, Per1, and Per2 RNAs are expressed in mouse teeth as well as in the brain. No amplification bands are detected in negative control samples (omission of cDNA) (A). β-actin was also amplified using the same cDNA and served as a positive RT-PCR control (A). Real time PCR was performed to quantify the relative expression levels of Bmal1, Clock, Per1 and Per2 in teeth versus the brain (B–E). Of interest, Bmal1, Clock and Per1 RNA expression levels were relative higher in teeth than in brain (B–D). In contrast, Per2 expression was higher in brain (E). All data were normalized to β-actin. Data were expressed as means ± SD at four determinations. NC, negative control.
2.2. Clock proteins expression in teeth at E13, E14 and E17
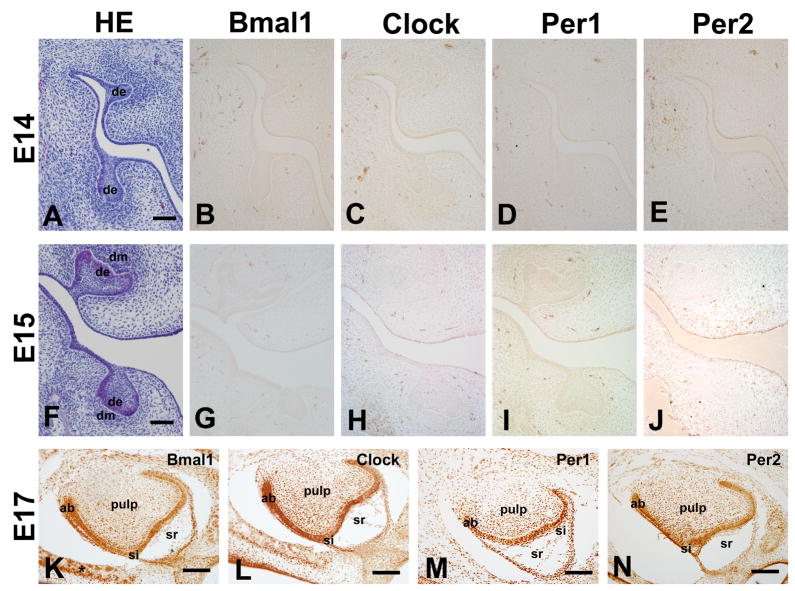
We have then characterized the protein expression patterns. At the bud stage (E13), the dental epithelium invades into presumptive dental mesenchyme. In this stage, clock proteins were undetectable in dental epithelium or condensed dental mesenchyme (Fig. 2). Similarly, no expression of clock proteins was detected in tooth germs of E14-E16 old mice (Fig. 2).
Figure 2.
Expression of clock proteins in mouse teeth at E14, E15 and E17. None of the four clock proteins can be detected in teeth bud and cap stages as shown here for E14 (A) and E 15 (F) (B–E and G–J). Clock protein expression in teeth was first detected at E17 (K–N). Bmal1 protein was expressed in ameloblasts, and odontoblast (K) as well as in alveolar bone osteoblasts (K). Clock is detected in ameloblasts and odontoblasts but also in dental pulp cells (L). Per1 protein expression was the strongest among the four proteins characterized, being strongly expressed in ameloblasts, odontoblasts, dental pulp, and stratum intermedium cells (M). Per2 showed similar localization with Clock but relatively stronger expression levels (N). Ameloblasts, odontoblasts and dental pulp cells were positive for Per2. Star indicated positive expression of Bmal1 in osteoblast (K). ab, ameloblast; od, odontoblast, de, dental epithelium; dm, dental mesenchyme;. HE, Hematoxylin-Eosin; Scale bars = 50 μm.
At the bell stage (E17), the neural crest-derived mesenchyme cells differentiate into dentin-secreting odontoblasts, while the jaw epithelium differentiates into the enamel-secreting ameloblasts. At this stage all four clock proteins showed similar expression patterns but with different intensities within the nucleus of ameloblasts and odontoblasts (Fig. 2). Bmal1 protein expression was relatively weak but clearly detected in ameloblasts and odontoblasts; Clock and Per2 showed stronger expression levels than Bmal1; and Per1 protein expression was the strongest among the four proteins characterized. Dental pulp cells, stratum intermedium (mainly for Per1) and osteoblasts (mainly for Bmal1) showed also variable levels of sporadic protein expression (Fig. 2).
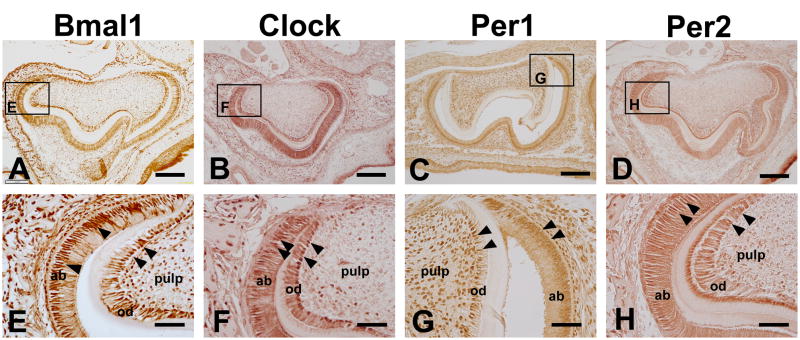
2.3. Clock proteins localization in teeth at PN4
At PN4 stage, the ameloblast and odontoblast are well-differentiated. They are induced to synthesize enamel- and dentin-specific proteins and form enamel, pre-dentin and dentin. At this stage, Bmal1 was up-regulated in the nucleus of ameloblasts, odontoblasts (Fig. 3). Clock, Per1 and Per2 protein were continued to be strongly expressed in the nucleus of ameloblasts and odontoblasts (Fig. 3). At this stage, the staining of the dental pulp cells was down-regulated for Clock, Bmal1, and Per2 (Fig. 3). In contrast, Per1 was still strongly expressed in dental pulp cells (Fig. 3). All four clock proteins were also strongly expressed by the alveolar bone osteoblasts at this stage (Fig. 3).
Figure 3.
Bmal1, Clock, Per1 and Per2 expression in mouse teeth at P4. Bmal1 was expressed in the nucleus of ameloblasts and odontoblasts (A and E). Bmal1 protein was not uniformly localized in ameloblasts and odontoblasts and some cells were devoid of staining (A, E). Clock, Per1 and Per2 proteins were strongly detected in the nucleus of ameloblasts and odontoblasts (B–D and F–H). Clock protein was also detected in some of the osteoblasts in the alveolar bone (F), and Per1 was also strongly expressed in dental pulp cells at this stage (G). ab, ameloblast; od, odontoblast. Scale bars = 100 μm in A–D, 20 μm in E–H.
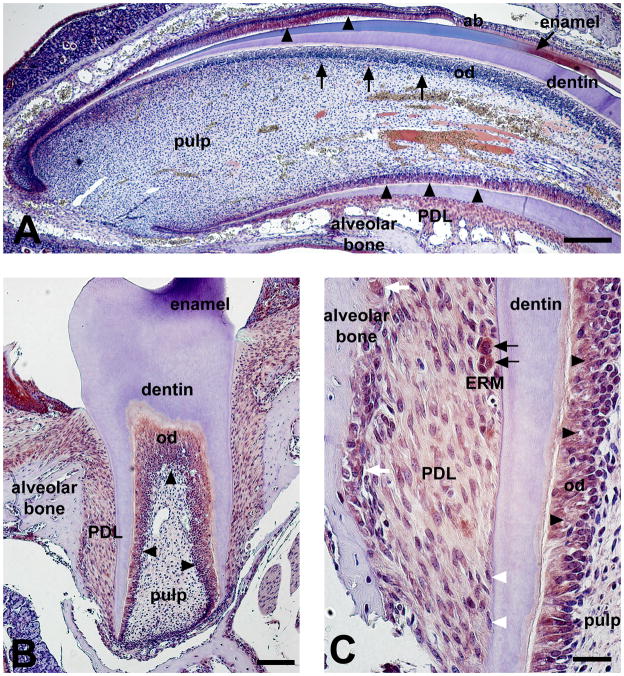
2.4. Per2protein expression in teeth and bone at PN21
At PN21 stage, the first molars are being erupted. Interestingly, all clock genes showed distinct expression pattern at the crown-analogue side versus the root-analogue side of the incisor. Bmal1, Clock, Per1 and Per2 are expressed in incisor root odontoblasts strongly. In contrast, clock proteins expression was low/undetectable at the crown-analogue side odontoblasts (Fig. 4). Odontoblasts in first molars were also strongly stained for all four clock proteins but dental pulp cells were devoid of staining (Fig. 4). Periodontal dental ligament (PDL) cells showed weaker expression for clock proteins compared with odontoblast expression. In contrast, within the PDL space, epithelial rests of Malassez (ERM) showed strong expression of all the four clock proteins studied here. Clock proteins were also detected in the nucleus of the osteoblasts and osteoclasts in the alveolar bone (Fig. 4). No clock proteins expression could be detected in cementoblasts or osteocytes (Fig. 4).
Figure 4.
Per2 expression in mouse incisor (A) and molar (B–C) at PN21 stage. Per2 showed distinct expression pattern at the root-analogue versus the root-analogue side in the incisor (A). Per2 was detected in root odontoblasts and ameloblasts (A, black arrowheads). In contrast, Per2 protein expression was low/undetectable in the odontoblasts of the crown-analogue side (A, black arrows). Dental pulp was devoid of staining in both incisors and molars (A–B). Odontoblasts in first molars were strongly stained for Per2 (B, black arrowheads). Per2 showed relatively weak expression in periodontal dental ligament (PDL) cells when compared with odontoblast expression (B–C). Epithelial rests of Malassez (ERM) showed strong expression of Per2 within the PDL space (C, black arrows). Per2 proteins are also detected in the nucleus of the osteoblasts and osteoclasts in the alveolar bone (C, white arrows). No Per2 protein expression is detected in cementoblasts (C, white arrowheads). ERM, epithelial rests of Malassez; od, odontoblasts; Scale bars = 200 μm in A, 100 μm in B, 20 μm in C.
Taken all together, we speculate that the expression of clock proteins in tooth directly correlates with cell differentiation and initiation of matrix production. Indeed, clock proteins expression was clearly up-regulated in ameloblasts and odontoblasts of PN4 teeth. At this stage (PN4), dental pulp staining was low further supporting a key role of clock genes in enamel and dentin formation. We are currently analyzing enamel and dentin formation in knock out models of clock genes to definitively elucidate the potential roles of clock genes in enamel and dentin development.
Of interest, at PN21 incisors a down-regulation of clock proteins expression was observed in the odontoblasts of the crown-analogue side. In contrast, root analogue odontoblasts continue to express clock proteins (Fig. 4). Comparison of the two dentin portions (crown versus root-analogue) at the ultra-structural level has revealed that differences occur in the morphology of the secretory granules of the odontoblast layer (Beertsen and Niehof, 1986). Therefore, differences in clock proteins expression levels may reflect differences in the composition of the non-collagenous matrix between the two dentin portions. In molar teeth clock genes were expressed in both crown and root odontoblasts. Differential gene expression has been described in molar versus incisor odontoblasts (Xing et al., 2007). Our study supports the idea that mouse incisors may have different root patterning mechanisms from molars, potentially due to different genetic signals. More studies are needed to clarify the potential role of clock genes differential expression in the root-analogue versus the crown-analogue of the mouse incisor and the differences of expression between molar and incisor odontoblasts.
In conclusion, our results show that the main clock genes known to control circadian rhythms and related functions in many tissues, Bmal1, Clock, Per1 and Per2 are expressed in teeth. Clock protein expression was found preferentially in matrix secreting cells i.e. ameloblasts, odontoblasts and osteoblasts but not in cementoblasts. These results suggest that the clock genes may be involved in the regulation of ameloblast and odontoblast functions, such as enamel and dentin matrix protein secretion and biomineralization. However, the clock genes studied here may not play a role in early tooth development.
3. Experimental procedures
3.1. RNA isolation and PCR
The animal use was approved by the Animal Welfare Committee of the University of Michigan. The teeth and brain tissue were isolated from day 1 post-natal mice (Charles River, Wilmington, MA). Total RNA was isolated from the freshly excised dental and brain tissues using TRIzol (Invitrogen, Carlsbad, CA), and 2 μg of RNA was reverse transcribed with TaqMan reverse transcription reagents (Applied Biosystems, Branchbury, NJ), following the manufacturer’s recommendations. The resulting cDNA was then amplified by RT-PCR using AmpliTaq Gold DNA Polymerase (Applied Biosystems). RT-PCR amplifications were performed at 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s using specific primers for β-actin, Bmal1, Clock, Per1 and Per2. The design of the primers was based on published mouse cDNA sequences (Table 1). The RT-PCR products were subcloned into pGEM-T Easy vector (Promega, Madison, WI) and RNA expression was confirmed by sequencing.
Table 1.
Primers sequences for RT-PCR
| Gene name | 5′- Sequence -3′ | Product size (bp) | GenBank Number | |
|---|---|---|---|---|
| β-actin | Forward Reverse |
AAGTACCCCATTGAACACGG ATCACAATGCCAGTGGTACG |
257 | NM_031144 |
| Bmal1 | Forward Reverse |
CCAAGAAAGTATGGACACAGACAAA GCATTCTTGATCCTTCCTTGGT |
81 | BC025973 |
| Clock | Forward Reverse |
CAAAATGTCACGAGCACTTAATGC ATATCCACTGCTGGCCTTTGG |
84 | MMAF000998 |
| Per1 | Forward Reverse |
AAACCTCTGGCTGTTCCTACCA AATGTTGCAGCTCTCCAAATACC |
74 | NM_011065 |
| Per2 | Forward Reverse |
ATGCTCGCCATCCACAAGA GCGGAATCGAATGGGAGAAT |
72 | NM_011066 |
3.2. Immunohistochemistry and imaging
For immunohistochemistry, serial sections (7 μm thick) of E13, E17, P4 and P21 mouse head were used. The endogenous peroxidase activity was blocked with 2% hydrogen peroxide in methanol for 20 min. Sections were then blocked for 1 h with 10% goat serum in PBS and incubated for 1 h with Rabbit anti-BMAL1 (1:500, NB300-596, Novus Biologicals, Littleton, CO), or rabbit anti-CLOCK (1:500, PA1-520, Thermo Scientific, Rockford, IL), or rabbit anti-PER1 (1:200, ab3443, Abcam, Cambridge, MA), or rabbit anti-PER2 (1:200, LS-C2836, Lifespan Biosciences, Seattle, WA). After that, the rabbit IgG Vectastain ABC kit (Vector Laboratories, Burlingame, CA) were used following manufacturer’s instruction. All slides were developed in parallel using Vector NovaRED substrate kit (SK-4800, Vector Laboratories) or peroxidase substrate kit DAB (SK-4100, Vector Laboratories), and the reaction was stopped before detection of nonspecific staining in control pre-immune serum-treated sections. Sections were then mounted and photographed on an Olympus microscope. As negative controls, both only primary antibodies and only secondary antibodies were used. Normal rabbit IgG instead of the primary antibodies was also used for determination of nonspecific binding of antibodies.
Acknowledgments
This research was funded by NIH-NIDCR grant DE018878-01A1 (PP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antoine D, Hillson S, Dean MC. The developmental clock of dental enamel: a test for the periodicity of prism cross-striations in modern humans and an evaluation of the most likely sources of error in histological studies of this kind. Journal of anatomy. 2009;214:45–55. doi: 10.1111/j.1469-7580.2008.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beertsen W, Niehof A. Root-analogue versus crown-analogue dentin: a radioautographic and ultrastructural investigation of the mouse incisor. The Anatomical record. 1986;215:106–118. doi: 10.1002/ar.1092150204. [DOI] [PubMed] [Google Scholar]
- Bei M. Molecular genetics of ameloblast cell lineage. Journal of experimental zoology. 2009;312B:437–444. doi: 10.1002/jez.b.21261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone VM, Stephan FK. Central and peripheral regulation of feeding and nutrition by the mammalian circadian clock: implications for nutrition during manned space flight. Nutrition (Burbank, Los Angeles County, Calif) 2002;18:814–819. doi: 10.1016/s0899-9007(02)00937-1. [DOI] [PubMed] [Google Scholar]
- Dean C. Progress in understanding hominoid dental development. Journal of anatomy. 2000;197(Pt 1):77–101. doi: 10.1046/j.1469-7580.2000.19710077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson GM. Incremental lines of von Ebner in dinosaurs and the assessment of tooth replacement rates using growth line counts. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:14623–14627. doi: 10.1073/pnas.93.25.14623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guatelli-Steinberg D, Larsen CS, Hutchinson DL. Prevalence and the duration of linear enamel hypoplasia: a comparative study of Neandertals and Inuit foragers. Journal of human evolution. 2004;47:65–84. doi: 10.1016/j.jhevol.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Ikeda M. Calcium dynamics and circadian rhythms in suprachiasmatic nucleus neurons. Neuroscientist. 2004;10:315–324. doi: 10.1177/10738584031262149. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Shamanna RK, Kondratova AA, Gorbacheva VY, Antoch MP. Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation. Faseb J. 2006;20:530–532. doi: 10.1096/fj.05-5321fje. [DOI] [PubMed] [Google Scholar]
- Li C, Risnes S. SEM observations of Retzius lines and prism cross-striations in human dental enamel after different acid etching regimes. Archives of oral biology. 2004;49:45–52. doi: 10.1016/s0003-9969(03)00195-x. [DOI] [PubMed] [Google Scholar]
- Lopez Franco GE, Huang A, Pleshko Camacho N, Stone DS, Blank RD. Increased Young’s modulus and hardness of Col1a2oim dentin. Journal of dental research. 2006;85:1032–1036. doi: 10.1177/154405910608501111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Papagerakis P, Yamakoshi Y, Hu JC, Bartlett JD, Simmer JP. Functions of KLK4 and MMP-20 in dental enamel formation. Biological chemistry. 2008;389:695–700. doi: 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EM, Yu J, Sehgal A. Circadian control of eclosion: interaction between a central and peripheral clock in Drosophila melanogaster. Curr Biol. 2003;13:526–533. doi: 10.1016/s0960-9822(03)00167-2. [DOI] [PubMed] [Google Scholar]
- Ohtsuka M, Saeki S, Igarashi K, Shinoda H. Circadian rhythms in the incorporation and secretion of 3H-proline by odontoblasts in relation to incremental lines in rat dentin. Journal of dental research. 1998;77:1889–1895. doi: 10.1177/00220345980770110501. [DOI] [PubMed] [Google Scholar]
- Simmer JP, Papagerakis P, Smith CE, Fisher DC, Rountrey AN, Zheng L, Hu JC. Regulation of dental enamel shape and hardness. Journal of dental research. 2010;89:1024–1038. doi: 10.1177/0022034510375829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- Thesleff I, Sharpe P. Signalling networks regulating dental development. Mechanisms of development. 1997;67:111–123. doi: 10.1016/s0925-4773(97)00115-9. [DOI] [PubMed] [Google Scholar]
- Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Williams JA, Sehgal A. Molecular components of the circadian system in Drosophila. Annual review of physiology. 2001;63:729–755. doi: 10.1146/annurev.physiol.63.1.729. [DOI] [PubMed] [Google Scholar]
- Xing X, Deng Z, Yang F, Watanabe S, Wen L, Jin Y. Determination of genes involved in the early process of molar root development initiation in rat by modified subtractive hybridization. Biochemical and biophysical research communications. 2007;363:994–1000. doi: 10.1016/j.bbrc.2007.09.087. [DOI] [PubMed] [Google Scholar]