Abstract
The budding yeast Saccharomyces cerevisiae is an emerging tool for investigating the molecular pathways that underpin several human neurodegenerative disorders associated with protein misfolding. Amyotrophic lateral sclerosis (ALS) is a devastating adult onset neurodegenerative disease primarily affecting motor neurons. The protein TDP-43 has recently been demonstrated to play an important role in the disease, however the mechanisms by which TDP-43 contributes to pathogenesis are unclear. To explore the mechanistic details that result in aberrant accumulation of TDP-43 and to discover potential strategies for therapeutic intervention, we employed a yeast TDP-43 proteinopathy model system. These studies allowed us to determine the regions of TDP-43 required for aggregation and toxicity and to define the effects of ALS-linked mutant forms of TDP-43. We have also been able to harness the power of yeast genetics to identify potent modifiers of TDP-43 toxicity using high-throughput yeast genetic screens. Here, we describe the methods and approaches that we have used in order to gain insight into TDP-43 biology and its role in disease. These approaches are readily adaptable to other neurodegenerative disease proteins.
Introduction
Protein aggregation linked to neurodegenerative disease
As our population continues to age, neurodegenerative diseases will pose an increasing challenge to public health. Identifying and characterizing the specific disease proteins associated with these disorders will provide insight into disease pathogenesis and will aid the development of biomarkers and therapeutic strategies. Several devastating human neurodegenerative diseases are associated with the accumulation of protein aggregates in the brains of affected individuals. These include Alzheimer’s disease [amyloid-beta and tau, (1, 2)], Parkinson’s disease [α–synuclein, (3)], prion diseases [PrP, (4)], polyglutamine diseases [expanded polyglutamine, (5)]; and most recently ALS and frontotemporal lobar degeneration with ubiquitinated inclusions (FTLD-U) [TDP-43, (6)]. Now that we know the identities of the aggregated disease proteins, the next step will be to understand at a mechanistic level how these proteins contribute to disease pathogenesis.
Yeast models of neurodegenerative disease
All cells have to deal with misfolded proteins, from simple yeast cells all the way to complex human neurons. Thus, it is likely that the mechanisms to cope with misfolded proteins as well as the cellular consequences of protein aggregation are conserved from yeast to man. Historically, yeast has been used to study many fundamental eukaryotic cellular pathways, including the cell cycle (7, 8) and the secretory pathway (9–11), but also provides a tractable system to study pathways involved in dealing with misfolded and aggregated proteins (12), including those linked to human disease. Many yeast genes have human homologues and the core cellular pathways are well conserved, meaning that genetic interactions found in yeast are likely to be relevant to human disease (13). The yeast genome is well characterized and easily manipulable (for example, deleting individual genes to probe their function). Thus, using a very simple but powerful genetic system we can define the pathways and genes that are affected by the excess accumulation of a neurodegenerative disease protein and develop innovative approaches to battle neurodegenerative disorders (12).
In the past few years, a number of yeast models of neurodegenerative diseases have been generated by overexpressing the wild type or mutant form of a human disease protein (14–16). A yeast model of the Parkinson’s disease protein α–synuclein has led to discoveries about its pathological properties (14). The yeast α–synuclein model has also provided insight into conserved vesicle trafficking pathways affected by α–synuclein accumulation (17, 18) as well as to suggest novel connections between genetic and environmental contributors to Parkinson’s disease (19). Importantly, results from the yeast studies have been validated in animal and cellular models (17–21). Notably, the relevance of an overexpression model of even the wild type form of the protein is highlighted by 1) the observation that wild type protein accumulates in most cases of disease and the mutant forms are much rarer; and 2) the discovery of duplication and triplications of the α–synuclein gene in rare familial forms of Parkinson’s disease (22–24).
Yeast models have also been created to model polyglutamine (polyQ) disorders (e.g. Huntington’s disease), including expression of a fragment of the huntingtin protein with and without polyQ expansions (15, 25). These studies have recapitulated the polyQ-length dependent aggregation and toxicity as well as to begin to uncover key early pathways affected by polyQ aggregation (26). Because TDP-43 has emerged as a key player in ALS and FTLD-U (see later), we have recently generated a new yeast model to study TDP-43 (16, 27) and describe various experimental approaches used to study critical features of TDP-43 aggregation and toxicity and techniques used to identify modifier genes.
TDP-43 is the major disease protein in ALS and FTLD-U
In 2006, Virginia Lee and colleagues discovered that the TAR DNA binding protein (TDP-43) was the abnormally accumulated protein in amyotrophic lateral sclerosis (ALS), which is also known as Lou Gehrig’s disease, and frontotemporal lobar degeneration with ubiquitinated inclusions (FTLD-U) (6). Subsequent work by a number of groups identified mutations in the TDP-43 gene (TARDBP) in some familial and sporadic cases (28–32). Thus, human genetics and pathology have both converged on TDP-43 as playing a critical role in disease pathogenesis (33). However, very little is known about the cellular pathways affected by TDP-43 in disease. Understanding these critical pathways will be essential for developing effective therapeutic interventions. TDP-43 is a nuclear RNA binding protein that shuttles between the nucleus and the cytoplasm (34, 35). In affected neurons, TDP-43 accumulates in the cytoplasm and is depleted from the nucleus, suggesting that a change in subcellular localization might be important for disease pathogenesis. Despite major efforts to understand the biology of TDP-43 we currently do not know how loss of the normal functions of TDP-43 or perhaps a gain of toxic function might contribute to disease. To address these deficits, we have been exploring TDP-43 aggregation and toxicity using the yeast model system.
Yeast TDP-43 proteinopathy model
We first sought to determine if we could recapitulate key aspects of TDP-43 biology in yeast cells, including cytoplasmic aggregation and toxicity. To regulate the expression of TDP-43 we placed a single copy of the human gene into a yeast strain under the control of a tightly regulated galactose-inducible promoter. That allowed us to maintain the cells in glucose, to repress TDP-43 expression, and then to rapidly and synchronously induce expression in all cells of the culture. To allow in vivo visualization of the protein in real time, and therefore monitor its propensity to form aggregates, we generated constructs that contain TDP-43 fused to the green fluorescent protein (GFP). To generate these constructs, a TDP-43 Gateway® entry clone was obtained from Invitrogen that contained the full-length human TDP-43 cDNA in the pDONR221 vector. We used this TDP-43 entry clone in Gateway LR reactions to shuttle TDP-43 into Gateway-compatible yeast expression vectors (http://www.addgene.org/yeast_gateway; (36)). To tag TDP-43 with GFP we created new entry clones lacking stop codons (TDP-43nostop) and used these in LR reactions with pAG426Gal-ccdB-GFP to generate the 2-micron (2μ) TDP-43-GFP fusion constructs. To generate the integrating TDP-43-GFP construct the TDP-43nostop entry clone was used in an LR reaction with pAG306Gal-ccdB-GFP. 2μ plasmid constructs (e.g. pAG426Gal-TDP-43-GFP) were subsequently transformed into BY4741 yeast strains (MATa his3 leu2 met15 ura3) and the integrating TDP-43-GFP constructs were transformed into the W303 strain (MATa can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1). For yeast transformation we used the PEG/Lithium acetate method according to standard protocols (37).
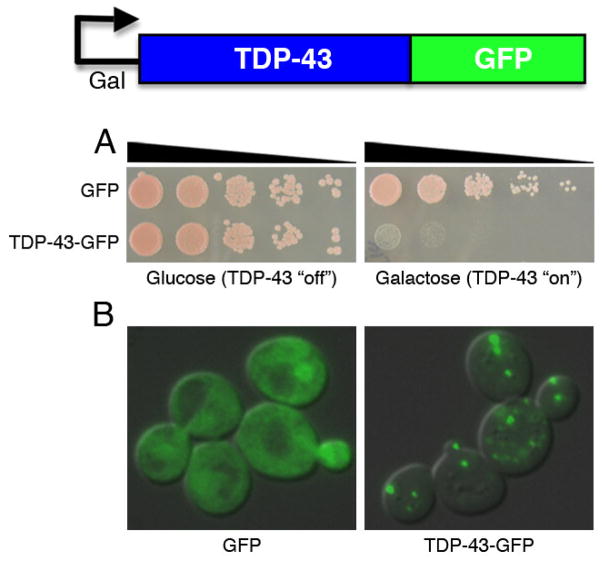
Having generated a panel of TDP-43 yeast expression constructs, we next investigated the effect of TDP-43 expression in living cells. To assess TDP-43 toxicity, we performed spotting assays. For spotting assays, 3 ml of yeast culture were grown overnight at 30 °C in liquid media containing raffinose (SRaf/-Ura) until they reached log or mid-log phase. Growth in raffinose does not activate the galactose-inducible promoter but also does not repress it, thus priming the yeast cells for rapid induction upon addition of galactose. Cultures were subsequently normalized for cell number, serially diluted and spotted onto synthetic solid media containing either glucose or galactose lacking uracil and were grown at 30 °C for 2–3 days. Expression of TDP-43-GFP from a 2-μ plasmid led to inhibition of cell growth compared to cells transformed with a plasmid containing GFP alone (Fig. 1A). Thus, TDP-43 expression in yeast resulted in cellular toxicity.
Figure 1.
Yeast TDP-43 proteinopathy model. A TDP-43 expression construct under the control of a tightly regulated galactose-inducible promoter was introduced into yeast cells. A) Spotting assays using five-fold serial dilutions demonstrates TDP-43 is toxic when expressed at high levels in yeast cells (pAG426Gal-TDP-43-GFP). When cells are spotted on plates containing glucose, TDP-43 expression is repressed and induced in the presence of galactose. B) TDP-43-GFP formed multiple cytoplasmic foci when expressed in yeast cells, whereas GFP alone was diffusely distributed throughout the cytoplasm and nucleus.
We next considered if TDP-43 would form cytoplasmic aggregates in yeast, similar to those observed in human disease and performed experiments to visualize TDP-43-GFP localization. For fluorescence microscopy experiments, we began with a yeast strain harboring an integrated TDP-43-GFP construct. Single colony isolates of the yeast strains were grown to mid-log phase in SRaf/-Ura media at 30°C. Cultures were spun down and resuspended in SGal/-Ura to induce expression of the TDP-43 constructs. Cultures were induced with galactose for 4–6 h, fixed using ethanol and counter-stained with DAPI, to visualize nuclei. At early time points (3–4 h) after induction, GFP alone was diffusely distributed between the cytoplasm and the nucleus, whereas the TDP-43-GFP fusion protein localized mainly to the nucleus with many cells containing 2–3 intranuclear foci of TDP-43 (16). We wondered whether increased expression of TDP-43 would alter its localization. To increase TDP-43 expression levels, we expressed TDP-43-GFP from a 2-μ high copy plasmid. This resulted in altered localization with the majority of cells now containing multiple cytoplasmic foci (Fig. 1B). In yeast cells TDP-43 is initially localized to the nucleus but eventually forms multiple cytoplasmic accumulations. In summary, expressing TDP-43 in yeast cells allowed us to model two key aspects directly related to human disease: cellular toxicity and cytoplasmic aggregation. Thus, having established a yeast TDP-43 model, we are now able to define the molecular players that regulate TDP-43 aggregation and toxicity and therefore gain insight into the pathobiology of TDP-43 in human neurons.
Dissecting the structural features of TDP-43 that confer aggregation and toxicity
Determining which region(s) of TDP-43 drive aggregation and toxicity will be important for designing therapeutic interventions aimed at mitigating these events in human disease. Because full-length TDP-43 aggregates and is toxic in yeast we used this system to define the regions of the protein that confer cytoplasmic aggregation and cellular toxicity. We constructed a panel of TDP-43 truncations and asked which regions of the protein are necessary and sufficient for aggregation and toxicity. To generate C-terminally GFP-tagged TDP-43 constructs, a two-step PCR protocol was used to amplify truncations without a stop codon and incorporate the Gateway attB1 and attB2 sites along with a Kozak consensus sequence. Resulting PCR products were shuttled into pDONR221 using a Gateway BP reaction. The entry clones were then used in LR reactions with pAG426Gal-ccdB-GFP to generate the 2μ TDP-43-GFP truncation constructs. To investigate the domains of TDP-43 required for toxicity and aggregation in yeast, the truncations were individually transformed into the BY4741 strain using the PEG/lithium acetate method and analyzed for toxicity and aggregation using spotting assays and microscopy. These studies revealed that the C-terminal domain of TDP-43 was required for aggregation and toxicity (16). Notably, the majority of disease mutations in TDP-43 are located within the C-terminal region, the domain found in yeast to be required for toxicity. Interestingly, this region was not sufficient for toxicity but required an intact RNA recognition motif (RRM), suggesting that RNA binding is very likely a component of TDP-43 toxicity. Subsequent studies, using point mutations to disable TDP-43 RNA binding, provided further confirmation for a role of RNA binding in mediating TDP-43 toxicity (38).
The effect of ALS-linked mutations on TDP-43 aggregation and toxicity
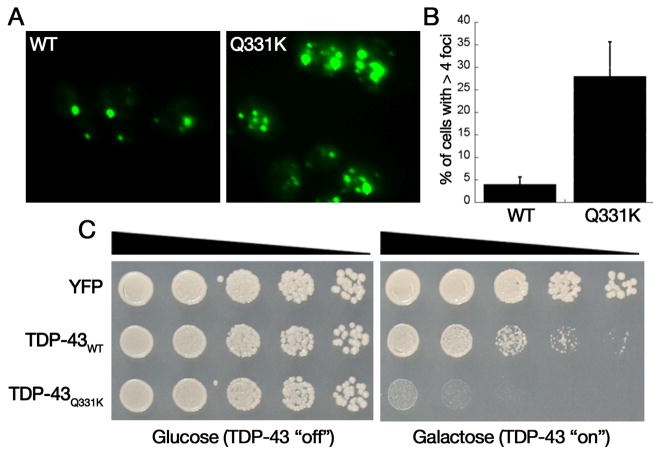
Following the discovery of TDP-43 cytoplasmic accumulations as a hallmark pathological feature of ALS and FTLD-U, several missense mutations in the TDP-43 gene were discovered in ALS and FTLD-U patients (32). However, the mechanism(s) by which these mutations caused disease (for example, gain- or loss-of-function) was unclear. Therefore, we used the yeast model to investigate the effect of these disease-linked mutations on TDP-43. We first considered that the mutations might increase TDP-43 aggregation and/or toxicity. Because the 2μ TDP-43-GFP expression construct produced high levels of expression and resulted in robust aggregation and toxicity, we utilized a new expression system to express TDP-43 at lower levels, enabling us to observe subtle differences between WT and mutant TDP-43. Instead of the 2μ plasmids, we now shuttled TDP-43 into the low copy CEN vectors to generate pAG416GAL-TDP-43-YFP. At this level of expression, WT TDP-43 was only slightly toxic. It is worth noting that the expression of a given protein based in a multi-copy vector (e.g. 2μ) is not always higher than in other expression systems, because yeast cells tend to reduce the expression levels of toxic proteins by reducing the number of copies of the plasmid (14). Nevertheless, using the CEN-based vector system, we were able to distinguish between the effects of WT and ALS-linked TDP-43 mutants. ALS-linked TDP-43 mutant constructs were generated using the QuikChange® site-directed mutagenesis system (Stratagene) with pAG416GAL-TDP-43-YFP as template. The resulting CEN plasmid constructs were transformed into the BY4741 yeast strain and then analyzed for toxicity and aggregation. To assess differences in aggregation between WT and mutant forms of TDP-43, yeast cultures were grown, induced, and processed as described above. After 6 h of induction, several fields of cells were chosen randomly to prevent any bias toward populations of cells with increased amounts of aggregation in addition to obtaining the total number of cells in any given field. At least 200 cells per sample were counted for each replicate. Only cells with >4 foci under the YFP channel were considered as cells with aggregating TDP-43. We used immunoblotting to confirm that WT and mutant TDP-43 proteins were expressed at the same level (See Fig. 2B in (27)). These studies revealed that disease-linked mutations in TDP-43 (especially Q331K) show enhanced aggregation and toxicity in yeast cells (Fig. 2), suggesting that these mutations could cause disease by accelerating TDP-43 aggregation and promoting degeneration. The effects of ALS-linked mutations on TDP-43 aggregation is strikingly similar to those of Parkinson’s disease linked mutations in α–synuclein and Alzheimer’s disease linked mutations in APP (39–41). Importantly, our key results with TDP-43 using this yeast model have been complemented through the use of a number of different cellular and biochemical in vivo and in vitro approaches (42), emphasizing the utility and relevance of modeling key aspects of disease pathogenesis in yeast.
Figure 2.
Accelerated aggregation by mutant TDP-43 linked to ALS. A) Yeast cells expressing YFP-tagged WT or Q331K TDP-43 at low levels. WT cells rarely contain > 4 foci per cell, whereas Q331K cells contain many more aggregates. B) Quantification of increase in aggregation by Q331K performed in triplicate by researcher blinded to identities of samples. C) Spotting assays demonstrate Q331K mutant TDP-43 is much more toxic than WT, despite being expressed at equivalent levels (data not shown).
Yeast genetic screens to define modifiers of TDP-43 toxicity
Having established the yeast TDP-43 model and using it to determine the structural features of TDP-43 required for aggregation and toxicity (16), as well as to explore the role of ALS-linked TDP-43 mutations (27), the next step for the yeast model is to deploy it in large scale genetic screens to identify toxicity modifiers. We still know very little about the mechanisms by which TDP-43 is toxic or those upstream factors that regulate its conversion to a toxic conformation. Unbiased genetic approaches will likely illuminate these critical aspects of TDP-43 pathogenesis. Similar approaches using yeast models and genetic screens have been used to successfully gain insight into mechanisms of Parkinson’s disease and Huntington’s disease (17, 19, 20, 43, 44). Below, we describe two complementary genetic screening approaches that we have recently used with the TDP-43 yeast model to discover genes and pathways that can mitigate toxicity and that might be suitable targets for therapies. The specific genes and pathways that have emerged from these screens, and experiments to validate them in other model systems, will be described elsewhere. However, the approach and methods used to discover these modifier genes will hopefully be useful to others interested in performing similar screens with yeast human disease models.
Plasmid overexpression screen
We have recently performed a yeast plasmid overexpression screen using the TDP-43 proteinopathy model to identify modifiers of TDP-43 toxicity. Since very little is known about why TDP-43 aggregation is toxic in yeast and other cells, we took an unbiased genetic approach. We reasoned that if we could find genes that suppress or enhance toxicity, these would very likely illuminate the key pathways affected by TDP-43 as well as the molecular pathways that control its conversion to a pathological conformation.
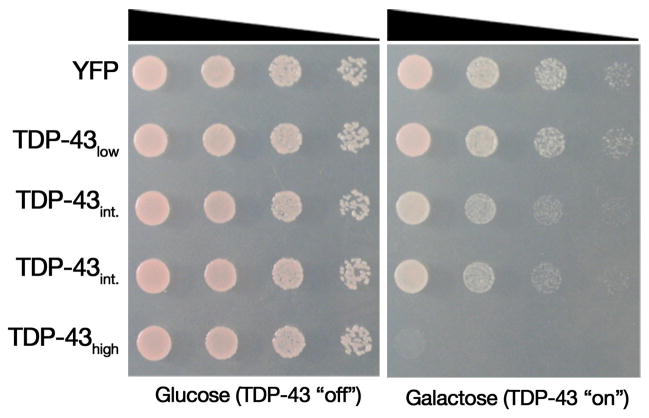
To perform this screen, we generated a yeast strain harboring an integrated galactose-inducible TDP-43 construct (pAG303Gal-TDP-43). Because we wanted to identify both suppressors and enhancers of toxicity using the same screen, we sought a TDP-43 expressing yeast strain with a moderate level of toxicity, such that genes that improved or worsened growth would be readily identifiable (if TDP-43 was too toxic we wouldn’t find enhancers but if it was not toxic enough we wouldn’t find suppressors). Therefore, we analyzed several independent integrants by immunoblot and spotting assays in order to identify the best screening strain, with a moderate level of TDP-43 expression, and a corresponding moderate level of toxicity (Fig. 3). The final strain chosen for the yeast screen was in the W303 strain background (MATa can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 ade2-1 pAG303Gal-TDP-43).
Figure 3.
Yeast strains engineered to express various levels of TDP-43. Spotting assays demonstrate galactose-inducible expression of YFP alone, or low, intermediate, and high levels of TDP-43 expression, which result in a corresponding level of toxicity.
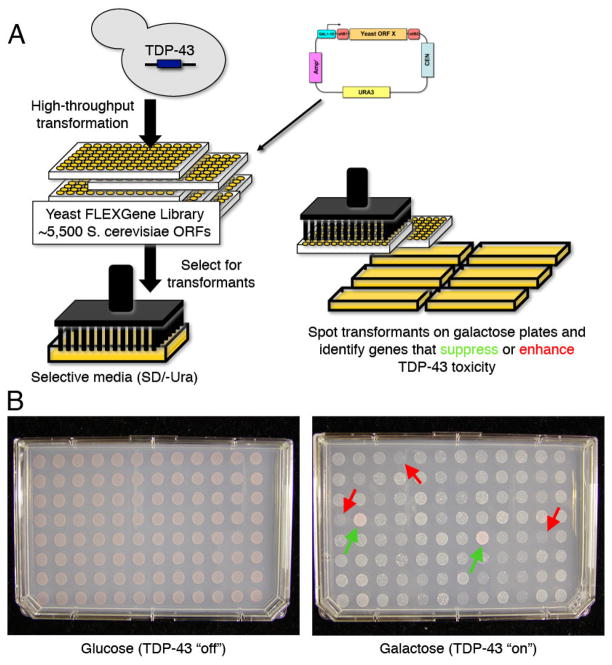
There are several yeast plasmid libraries available, however we chose to use the Yeast FLEXGene ORF library. This library is indeed the “gold standard” in yeast genomic libraries. It is an arrayed library of >5,500 completely sequence-verified yeast genes in Gateway® cloning system-based expression vectors (45). Unlike existing libraries that contain fragments of genes and/or often mutations that complicate interpretation of results, this library contains only full-length genes and is free of mutations. Each yeast ORF in this library has been shuttled into the pBY011 Gateway® yeast expression plasmid (CEN, URA3, galactose-inducible promoter). The library clones are arrayed in 96-well plates; one gene per well (Fig 4A). To perform the TDP-43 modifier screen, we modified the standard PEG/lithium acetate yeast transformation protocol for automated high-throughput transformation in a 96-well format, allowing screening of the entire library by a single student or postdoc, in triplicate, in only a few weeks.
Figure 4.
Yeast plasmid overexpression screen to identify modifiers of TDP-43 toxicity. A) Schematic of yeast genetic screen. Yeast cells harboring an integrated galactose-inducible TDP-43 cassette, were individually transformed with a library of 5,500 yeast open reading frames (ORFs) and spotted onto galactose plates to induce expression of TDP-43 and each gene from the library. B) A representative plate from the yeast screen. Each spot represents a yeast strain expressing TDP-43 along with one gene from the library. Examples of genes that suppressed TDP-43 toxicity (improved growth) are indicated by green arrows and enhancers of toxicity (inhibited growth) are indicated by red arrows.
The Yeast FLEXGene library clones are stored as bacterial glycerol stocks at −80 °C. To isolate plasmid DNA, we inoculate 2.2 ml deep well blocks (Sorenson BioScience) containing 1.7ml of LB-amp, cover with an air-permeable plate seal (Bioexcell Aeraseal, Worldwide Medical Products) and incubate with shaking overnight at 37 °C. The next day, we centrifuge the bacteria and isolate plasmid DNA using the Qiagen BioRobot Universal and a QIAprep 96 Turbo BioRobot Kit (Qiagen). This instrument can process up to four 96-well plates (384 clones) in ~3 hours. Plasmid DNA is eluted into round-bottom 96-well microtiter plates, sealed with aluminum foil seals and stored at −80 °C. To begin the screen, we first aliquot ~1–2 μg of library plasmid DNA (5–10 μl) into 96-well round-bottom microtiter plates using a Qiagen BIOROBOT Rapidplate 96-well pipettor and allowed to dry overnight in a clean bench fume hood (Labconco). We routinely make multiple copies of the library, dry the DNA, and store these plates at room temperature for 1–2 weeks, in order to have them ready for additional rounds of screening. We have found that this protocol works best with ten or fewer 96-well plates, therefore the following yeast transformation protocol is based on ten 96-well plates, but can be scaled up or down accordingly.
To begin the yeast transformation, we inoculate 200 ml YPD with a single colony of our TDP-43 yeast screening strain and grow with shaking overnight at 30 °C until saturated. The next day, we dilute this culture to OD600=0.1 in 2 L of fresh YPD and incubate for a further 4–6 hours, until they have reached OD600=0.6–0.8. We collect the yeast cells by centrifugation (10 minutes at 3,000 RPM in a tabletop centrifuge (Eppendorf 5810R) and wash them once with water and once in 1XTE/LiOAc (0.1M LioAc, 10mM Tris, 1 mM EDTA, pH=8). Following the washes we resuspend the yeast cells in 70 ml 1XTE/LiOAc and incubate with shaking for 30 minutes at 30 °C. To permeabilize the yeast cells we add 0.1 M β-mercaptoethanol and incubate for an additional 30 minutes at 30 °C with shaking. During this incubation period, we boil sheared salmon sperm DNA (2 ml of 10 mg/ml) for 10 minutes, chill on ice for 5 minutes, and add to the yeast/LiOAc mixture. We use the BioRobot RapidPlate to dispense 50 μl of the yeast/LiOAc mixture to each well of the 96-well plates containing plasmid DNA. Note: users without a BioRobot RapidPlate can also use a multi-channel pipettor for this and subsequent steps of the procedure. We incubate the plates for 30 minutes at room temperature without shaking (allowing the yeast cells to settle down into the wells and come in contact with the plasmid DNA. Next we add 125 μl PEG solution (40% PEG/10% DMSO/0.1M LiOAc) to each well and mix thoroughly by pipetting (at least 15–20 times) and then the plates are incubated at room temperature for 30 minutes without shaking. We heat shock the yeast cells for 15 minutes at 42 °C in a dry incubator (do not stack plates). Following heat shock we pellet the cells by spinning the plates in a tabletop centrifuge (2,500 rpm for 5 minutes, Eppendorf 5810R), and then inverting them over a waste container to discard the transformation mixture. Next we wash the yeast cells once with 200 μl of selective media (e.g. SD/-Ura), spin again and discard the media. Finally, we add 200 μl of selective media (e.g. SD/-Ura to select for the library plasmids that confer uracil prototrophy). We incubate the plates at 30 °C for 24–48 hours to allow the transformants to grow.
We are now almost ready to identify toxicity modifers. First we inoculate the transformants into liquid media containing raffinose (SRaf/-Ura). We aliquot 200 μl of SRaf/-Ura into each well of a fresh 96-well plate. Using the BioRobot RapidPlate, we mix the yeast transformants that have been growing for 24–48 hours by pipetting up and down several times, aspirate 5–10 ul of cells, and dispense them into the fresh SRaf/-Ura media. The inoculated SRaf/-Ura plates are incubated overnight at 30 °C without shaking. The next day yeast cells are spotted onto agar plates containing glucose (represses TDP-43 expression) or galactose (induces TDP-43 expression). We use Nunc OmniTrays (Thermo Scientific) for the agar plates and a frogger 96-bolt replicator (V&P Scientific, catalog #VP404) to spot the yeast cells. We incubate the plates at 30 °C in for 2–3 days, photograph colonies using a digital camera (Canon Powershot SD700) and photo copy stand (Kaiser Repro Kid Copy Stand, catalog #KACSKK) and determine which spots grew better (toxicity suppressors) and worse (toxicity enhancers) than controls (Fig. 4B).
We routinely perform the screen in triplicate (making extra copies of the plasmid library when aliquoting DNA is useful for this reason) to ensure that the genetic suppressors and enhancers of TDP-43 toxicity identified are repeatable. The hits that we confirm in triplicate are then individually validated by independent transformation, and a more stringent spotting assay with 5-fold serial dilutions. We perform several additional tests to eliminate non-specific hits. These include toxicity enhancers that are simply toxic in any yeast strain when overexpressed. To eliminate these, we transform each of our candidate toxicity enhancers into a WT yeast strain and test for toxicity. Those genes that are toxic in WT cells are eliminated and we only keep those that are specifically toxic to our TDP-43 strain. We also eliminate suppressors and enhancers that simply down- or up-regulate expression from the galactose-inducible promoter, as these are not likely to be informative about TDP-43 biology, although they do serve as a good indicator that the screen is working. Such genes include MIG1, MIG2, and MIG3 (Multicopy Inhibitor of GAL gene expression) and we routinely recover these in similar galactose promoter-based yeast screens.
Computational approaches can be used to interrogate the screen hits and integrate them with the vast array of available yeast experimental data (e.g. transcriptional profiling, deletion phenotypes, protein-protein interactions, etc.; (20)) The 40 genetic modifiers that we have identified in the TDP-43 overexpression screen included both enhancers (27) and suppressors (13) of toxicity and include a large number of RNA binding proteins and proteins involved in RNA processing, as well as kinases, proteases, and genes of unknown function ((46) and M.A. and A.D.G. unpublished).. We have been encouraged by the ability of these screens to identify genetic modifiers specific to the disease protein. Interestingly, the hits from the TDP-43 proteinopathy model are quite different than a similar screen done in the α–synuclein yeast proteinopathy model (17, 19, 20). Of the 40 hits from the TDP-43 screen, only 1 was shared with the 71 hits from the α–synuclein screen, indicating that the yeast models and screen are specific to the protein biology and not simply overexpression of any aggregation-prone human disease protein.
Deletion screen
As a complementary approach to the plasmid overexpression screen, we have also been using the yeast TDP-43 model to perform a deletion screen to identify toxicity suppressors and enhancers. Yeast deletion screens (also known as synthetic lethal screens) can be used to uncover genetic interactions between seemingly unrelated genes or pathways. Two genes exhibit a “synthetic lethal” interaction when the combination of two non-lethal mutations results in slow growth or cell death (47, 48). This type of interaction can result for genes acting in a single pathway or for genes within two parallel pathways (49). Additionally, these screens can identify alleviating interactions, in which the double mutant grows better than expected based on the growth of each of the single mutants (50). Therefore, we performed a synthetic lethal screen to identify gene deletions that enhance or suppress TDP-43 toxicity (Fig. 5A). A similar approach has been effective at identifying modifiers of α–synuclein or mutant huntingtin toxicity in yeast (43, 44).
Figure 5.
Yeast deletion screen to identify modifiers of TDP-43 toxicity. A) Schematic of yeast deletion screen, based on (51). The galactose-inducible TDP-43 expression construct (pAG416Gal-TDP-43) was introduced into MATα strain Y7092 to generate the query strain. This query strain was mated to the yeast haploid deletion collection of non-essential genes (MATa, each gene deleted with KanMX cassette (confers resistance to G418). Mating, sporulation, and mutant selection were performed using a Singer RoToR HDA (Singer Instruments, Somerset, UK). Haploid mutants harboring the TDP-43 expression plasmid were grown in the presence of glucose (TDP-43 expression “off”) or galactose (TDP-43 expression “on”). Following growth at 30 °C for 2 days, plates were photographed and colony sizes measured by ImageJ image analysis software, based on (54). B) A representative plate from the deletion screen. Left is glucose (deletion alone, e.g. xxxΔ) and right is galactose (deletion + TDP-43 expression, e.g. xxxΔ + TDP-43). Each plate contains 384 different strains pinned in duplicate (768 total). The red arrows point to a strong alleviating genetic interaction (toxicity suppressor), in which the gene deletion + TDP-43, grows better than TDP-43- or the deletion alone.
The Boone laboratory in Toronto has developed a high-throughput method to rapidly perform synthetic lethal screens in yeast (referred to as synthetic genetic array (SGA) analysis) (51). To generate our starting “query” strain, we transformed haploid MATα cells (strain Y7092; MATα can1delta::STE2pr-Sp_his5 lyp1delta his3delta1 leu2delta0 ura3delta0 met15delta0) with pAG416Gal-TDP-43, in which TDP-43 expression is under the control of a galactose-inducible promoter. This tightly regulated promoter allows us to keep TDP-43 expression off during routine passage and throughout the various mating and pinning procedures of the screen. The TDP-43 expression cassette in this strain is linked to a dominant selectable marker, URA3, which allows cells to grow in the absence of uracil, as well as the MFA1pr-HIS3 reporter (only expressed in MATa cells).
As detailed below, the number of pinning procedures and the fact that we are working at 768-colony density might seem to be a daunting task. However, we have been using a Singer RoToR HDA robot (http://www.singerinst.co.uk/), designed specifically for yeast synthetic lethal screens. This automated yeast pinning system makes it possible for one student or postdoc to complete a genome-wide synthetic lethal screen (in triplicate) in 3–4 weeks.
To begin the screen, we array our MATα query strain onto four YPD (rich media) agar plates at 768-colony density and grow them overnight at 30 °C. Next, we array our library of ~4,850 MATa viable yeast deletion mutants (Yeast Deletion Collection, Invitrogen), each carrying a gene deletion mutation linked to a G418-resistance marker (KanMX), at 768-colony density (384 individual knockout strains per plate, in duplicate), onto YPD agar plates containing G418, thus creating a deletion mutant array (DMA). The yeast DMA is comprised of 13 plates containing the entire library of non-essential gene deletions. Similar approaches can be used to screen libraries of essential genes (for example, titratable promoter alleles (52) or temperature-sensitive alleles (53)). These plates are also incubated overnight at 30 °C. The next day we mate the query strain with the DMA by first pinning the 768-format query strain onto 13 fresh YPD plates, and then pinning the DMA on top of the query cells. The mating plates are incubated at room temperature for 1 day. We select for diploids by pinning MATa/α zygotes onto glucose-containing synthetic solid media containing G418 and lacking uracil (SD/-Ura+G418) and incubating at 30 °C for two days. The heterozygous diploids that survive this selection are then transferred to enriched sporulation plates (low levels of carbon and nitrogen) and incubated at ~22 °C for 5–7 days. The resulting spores are next transferred to synthetic medium lacking histidine, which selectively germinates the MATa meiotic progeny (because they express the MFA1pr-HIS3 reporter), and incubated at 30 °C for two days. After another round of pinning onto the same plates, to enrich for MATa haploids, the MATa meiotic progeny are transferred, by pinning, to agar plates containing G418 and glucose but lacking uracil, which selects for growth of mutant haploids harboring the TDP-43 plasmid, but keeps its expression off. Following incubation at 30 °C for two days, we digitally photograph each of the 13 plates. Colonies on these plates serve as growth controls (mutant, TDP-43 expression off). Finally, colonies are pinned to agar plates containing G418 and galactose, which turns on TDP-43 expression, and incubated at 30 °C for two days. We photograph these plates and use image analysis software (54) to measure colony size. We compare colony sizes of the mutant+TDP-43 “on” cells to the mutant+TDP-43 “off” cells in order to assess the effect of the double-mutant combinations on growth rate and viability (Fig. 5B). To account for potential differences between cell growth on glucose vs. galactose, we also perform the screen in parallel with a wild-type query strain harboring a galactose-regulated GFP-alone expression cassette.
We perform each screen 3 independent times for a total of 6 replicates (the strains are in duplicate on each plate). Because we are able to analyze each genetic interaction 6 independent times, we are very confident in the interactions that reproduce at least 4 out of 6 times, and most confident in those interactions that repeat 6/6 times. We have also developed custom Excel macros to parse image analysis data and to organize gene lists. The yeast gene deletions that enhance or suppress toxicity are verified by independent transformations and/or random spore analysis, using similar filtering procedures as for the overexpression screen to eliminate deletions that grow poorly on galactose or simply grow much slower than WT cells. Using this approach, we have identified 14 genetic modifiers of TDP-43 toxicity, including 8 alleviating (suppressors) and 6 aggravating (enhancers) interactions. The genes and pathways identified by this screen will hopefully provide insight into the mechanisms underpinning TDP-43 toxicity. The deletion toxicity suppressors represent a particularly attractive class of hits because yeast gene deletions that suppress (alleviate) TDP-43 toxicity might be ideal candidates for drug targets (which typically (though not always) inhibit protein activity, instead of activating).
Conclusion & Future directions
The methods described above illustrate various ways in which the yeast model system can be used to study the properties of the neurodegenerative disease protein TDP-43. Insight into the features of the TDP-43 protein that are important for aggregation and toxicity, including the effects of disease-linked TDP-43 mutations, will lead to a better understanding of how the protein mislocalizes during disease and what drives aggregation. The genetic modifiers of toxicity discovered using yeast overexpression and deletion screens will hopefully provide novel hypotheses about TDP-43 disease mechanisms and might even help to suggest new drug targets and therapeutic strategies. Obviously, a critical next step involves verifying results from these yeast genetic screens in multiple model systems in order to make fundamental steps towards understanding the pathways that are disturbed due to excess accumulation of the human disease protein. Translating the results from yeast toxicity to neurotoxicity can be accomplished by utilizing other simple model systems including Drosophila, C. elegans and mouse, and is crucial in validating the importance of our findings in yeast and ultimately extending them into the human disease condition.
As one success story so far from the recent TDP-43 modifier screen, we found PBP1 as an overexpression enhancer of TDP-43 toxicity. PBP1 is the yeast homolog of human ataxin 2, the protein mutated in another neurodegenerative disease called spinocerebellar ataxia type-2 (SCA2). We generated transgenic Drosophila expressing TDP-43 and ataxin 2 and observed a similar genetic interaction. We found the two proteins could physically interact in yeast and human cells and that ataxin 2 was mislocalized in motor neurons of ALS patients. Finally, we discovered that intermediate-length polyglutamine repeat expansions in ataxin 2 were significantly associated with increased risk for ALS (38). We continue to characterize the additional hits from these yeast TDP-43 toxicity modifier screens and interrogate their homologs in animal models and human patient samples, with the hopes that additional genes and pathways will provide further insight into the molecular underpinnings of ALS and other TDP-43 proteinopathies.
While we have so far used this yeast TDP-43 model for genetic analyses, in the future we and others could use the yeast model to perform high-throughput screens with small molecule libraries in order to define compounds with activity that could antagonize TDP-43 toxicity. Hits identified from these screens could be optimized using the yeast model, validated with animal models, and then potentially developed as new therapies. A similar strategy has recently been reported for Parkinson’s disease using the yeast α–synuclein model (55, 56). As protein misfolding plays a causal role in many neurodegenerative diseases (57) modeling toxicity of additional human disease proteins in yeast (e.g. tau, Aβ, FUS, polyQ proteins, SOD1, etc.) might give a new perspective into what confers toxicity in many of these disorders.
Acknowledgments
This work was supported in part by a Pilot grant from the University of Pennsylvania Institute on Aging (A.D.G.), an NIH Director’s New Innovator Award 1DP2OD004417-01 (A.D.G.), and 1R01NS065317-01 (A.D.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee VM, Balin BJ, Otvos L, Jr, Trojanowski JQ. Science. 1991;251:675–8. doi: 10.1126/science.1899488. [DOI] [PubMed] [Google Scholar]
- 2.Glenner GG, Wong CW. Biochem Biophys Res Commun. 1984;120:885–90. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 3.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Nature. 1997;388:839–40. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 4.McKinley MP, Bolton DC, Prusiner SB. Cell. 1983;35:57–62. doi: 10.1016/0092-8674(83)90207-6. [DOI] [PubMed] [Google Scholar]
- 5.DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Science. 1997;277:1990–3. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 6.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Science. 2006;314:130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 7.Nurse P. EMBO Rep. 2002;3:204–6. doi: 10.1093/embo-reports/kvf060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartwell LH. Biosci Rep. 2002;22:373–94. doi: 10.1023/a:1020918107706. [DOI] [PubMed] [Google Scholar]
- 9.Stevens T, Esmon B, Schekman R. Cell. 1982;30:439–48. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- 10.Novick P, Ferro S, Schekman R. Cell. 1981;25:461–9. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- 11.Novick P, Field C, Schekman R. Cell. 1980;21:205–15. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 12.Gitler AD. Neurosignals. 2008;16:52–62. doi: 10.1159/000109759. [DOI] [PubMed] [Google Scholar]
- 13.Steinmetz LM, Scharfe C, Deutschbauer AM, Mokranjac D, Herman ZS, Jones T, Chu AM, Giaever G, Prokisch H, Oefner PJ, Davis RW. Nat Genet. 2002;31:400–4. doi: 10.1038/ng929. [DOI] [PubMed] [Google Scholar]
- 14.Outeiro TF, Lindquist S. Science. 2003;302:1772–5. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krobitsch S, Lindquist S. Proc Natl Acad Sci U S A. 2000;97:1589–94. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson BS, McCaffery JM, Lindquist S, Gitler AD. Proc Natl Acad Sci U S A. 2008;105:6439–44. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Liu K, Xu K, Strathearn KE, Liu F, Cao S, Caldwell KA, Caldwell GA, Marsischky G, Kolodner RD, Labaer J, Rochet JC, Bonini NM, Lindquist S. Science. 2006;313:324–8. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gitler AD, Bevis BJ, Shorter J, Strathearn KE, Hamamichi S, Su LJ, Caldwell KA, Caldwell GA, Rochet JC, McCaffery JM, Barlowe C, Lindquist S. Proc Natl Acad Sci U S A. 2008;105:145–50. doi: 10.1073/pnas.0710685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gitler AD, Chesi A, Geddie ML, Strathearn KE, Hamamichi S, Hill KJ, Caldwell KA, Caldwell GA, Cooper AA, Rochet JC, Lindquist S. Nat Genet. 2009;41:308–15. doi: 10.1038/ng.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeger-Lotem E, Riva L, Su LJ, Gitler AD, Cashikar AG, King OD, Auluck PK, Geddie ML, Valastyan JS, Karger DR, Lindquist S, Fraenkel E. Nat Genet. 2009;41:316–23. doi: 10.1038/ng.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thayanidhi N, Helm JR, Nycz DC, Bentley M, Liang Y, Hay JC. Mol Biol Cell. 2010;21:1850–63. doi: 10.1091/mbc.E09-09-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J, Hulihan M, Waucquier N, Defebvre L, Amouyel P, Farrer M, Destee A. Lancet. 2004;364:1167–9. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 23.Ibanez P, Bonnet AM, Debarges B, Lohmann E, Tison F, Pollak P, Agid Y, Durr A, Brice A. Lancet. 2004;364:1169–71. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 24.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 25.Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU. Proc Natl Acad Sci U S A. 2000;97:7841–6. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duennwald ML, Lindquist S. Genes Dev. 2008;22:3308–19. doi: 10.1101/gad.1673408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. J Biol Chem. 2009;284:20329–39. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. Nat Genet. 2008;40:572–4. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 29.Rutherford NJ, Zhang YJ, Baker M, Gass JM, Finch NA, Xu YF, Stewart H, Kelley BJ, Kuntz K, Crook RJ, Sreedharan J, Vance C, Sorenson E, Lippa C, Bigio EH, Geschwind DH, Knopman DS, Mitsumoto H, Petersen RC, Cashman NR, Hutton M, Shaw CE, Boylan KB, Boeve B, Graff-Radford NR, Wszolek ZK, Caselli RJ, Dickson DW, Mackenzie IR, Petrucelli L, Rademakers R. PLoS Genet. 2008;4:e1000193. doi: 10.1371/journal.pgen.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. Science. 2008;319:1668–72. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Deerlin VM, Leverenz JB, Bekris LM, Bird TD, Yuan W, Elman LB, Clay D, Wood EM, Chen-Plotkin AS, Martinez-Lage M, Steinbart E, McCluskey L, Grossman M, Neumann M, Wu IL, Yang WS, Kalb R, Galasko DR, Montine TJ, Trojanowski JQ, Lee VM, Schellenberg GD, Yu CE. Lancet Neurol. 2008;7:409–16. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pesiridis GS, Lee VM, Trojanowski JQ. Hum Mol Genet. 2009;18:R156–62. doi: 10.1093/hmg/ddp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen-Plotkin AS, Lee VM, Trojanowski JQ. Nat Rev Neurol. 2010 doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ayala YM, Zago P, D’Ambrogio A, Xu YF, Petrucelli L, Buratti E, Baralle FE. J Cell Sci. 2008;121:3778–85. doi: 10.1242/jcs.038950. [DOI] [PubMed] [Google Scholar]
- 35.Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, Lee VM. J Biol Chem. 2008;283:13302–9. doi: 10.1074/jbc.M800342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alberti S, Gitler AD, Lindquist S. Yeast. 2007;24:913–9. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–8. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu M, Padmanabhan A, Clay D, McCluskey L, Elman L, Juhr D, Gruber PJ, Rub U, Auburger G, Trojanowski JQ, Lee VM, Van Deerlin VM, Bonini NM, Gitler AD. Nature. 2010 doi: 10.1038/nature09320. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conway KA, Harper JD, Lansbury PT. Nat Med. 1998;4:1318–20. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- 40.Conway KA, Lee SJ, Rochet JC, Ding TT, Williamson RE, Lansbury PT., Jr Proc Natl Acad Sci U S A. 2000;97:571–6. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lashuel HA, Hartley DM, Petre BM, Wall JS, Simon MN, Walz T, Lansbury PT., Jr J Mol Biol. 2003;332:795–808. doi: 10.1016/s0022-2836(03)00927-6. [DOI] [PubMed] [Google Scholar]
- 42.Barmada SJ, Skibinski G, Korb E, Rao EJ, Wu JY, Finkbeiner S. J Neurosci. 2010;30:639–49. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willingham S, Outeiro TF, DeVit MJ, Lindquist SL, Muchowski PJ. Science. 2003;302:1769–72. doi: 10.1126/science.1090389. [DOI] [PubMed] [Google Scholar]
- 44.Giorgini F, Guidetti P, Nguyen Q, Bennett SC, Muchowski PJ. Nat Genet. 2005;37:526–31. doi: 10.1038/ng1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu Y, Rolfs A, Bhullar B, Murthy TV, Zhu C, Berger MF, Camargo AA, Kelley F, McCarron S, Jepson D, Richardson A, Raphael J, Moreira D, Taycher E, Zuo D, Mohr S, Kane MF, Williamson J, Simpson A, Bulyk ML, Harlow E, Marsischky G, Kolodner RD, LaBaer J. Genome Res. 2007;17:536–43. doi: 10.1101/gr.6037607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, Padmanabhan A, Clay-Falcone D, McCluskey L, Elman L, Juhr D, Gruber PJ, Rub U, Auburger G, Trojanowski JQ, Lee VM, Van Deerlin VM, Bonini NM, Gitler AD. Nature. 2010;466:1069–75. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guarente L. Trends Genet. 1993;9:362–6. doi: 10.1016/0168-9525(93)90042-g. [DOI] [PubMed] [Google Scholar]
- 48.Novick P, Osmond BC, Botstein D. Genetics. 1989;121:659–74. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartman JLt, Garvik B, Hartwell L. Science. 2001;291:1001–4. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- 50.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, Krogan NJ. Cell. 2005;123:507–19. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 51.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. Science. 2001;294:2364–8. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 52.Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, Trochesset M, Morse D, Krogan NJ, Hiley SL, Li Z, Morris Q, Grigull J, Mitsakakis N, Roberts CJ, Greenblatt JF, Boone C, Kaiser CA, Andrews BJ, Hughes TR. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 53.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin ZY, Liang W, Marback M, Paw J, San Luis BJ, Shuteriqi E, Tong AH, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pal C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras AC, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C. Science. 2010;327:425–31. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collins SR, Schuldiner M, Krogan NJ, Weissman JS. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su LJ, Auluck PK, Outeiro TF, Yeger-Lotem E, Kritzer JA, Tardiff DF, Strathearn KE, Liu F, Cao S, Hamamichi S, Hill KJ, Caldwell KA, Bell GW, Fraenkel E, Cooper AA, Caldwell GA, McCaffery JM, Rochet JC, Lindquist S. Dis Model Mech. 2010;3:194–208. doi: 10.1242/dmm.004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeSantis ME, Dersh D. Dis Model Mech. 2010;3:399–400. doi: 10.1242/dmm.005678. [DOI] [PubMed] [Google Scholar]
- 57.Forman MS, Trojanowski JQ, Lee VM. Nat Med. 2004;10:1055–63. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]