SUMMARY
Approximately 35% of the species present in subgingival biofilms are as yet uncultivated, so their role in periodontal pathogenesis is unknown. The aim of the present study was to develop a high throughput method to quantify a wide range of cultivated and uncultivated taxa in subgingival biofilm samples associated with periodontal disease or health. Oligonucleotides targeting the 16S ribosomal DNA gene were designed, synthesized and labeled with digoxigenin. These probes were hybridized with the total nucleic acids of pure cultures or subgingival biofilm samples. Target species included cultivated taxa associated with periodontal health and disease, as well as uncultivated species, such as TM7 sp OT 346, Mitsuokella sp. OT 131 and Desulfobulbus sp. OT 041. Sensitivity and specificity of the probes were determined. A Universal probe was used to assess total bacterial load. Sequences complementary to the probes were used as standards for quantification. Chemiluminescent signals were visualized after film exposure or using a CCD camera. In a pilot clinical study, 266 subgingival plaque samples from eight periodontally healthy people and 11 patients with periodontitis were examined. Probes were specific and sensitivity reached 104 cells. Fusobacterium nucleatum ss polymorphum and Actinomyces gerencseriae were the most abundant cultivated taxa in clinical samples. Among uncultivated/unrecognized species, Mitsuokella sp. OT 131 and Prevotella sp. OT 306 were the most numerous. Porphyromonas gingivalis and Desulfobulbus sp. OT 041 were only detected in patients with periodontitis. Direct hybridization of total nucleic acids using oligonucleotide probes permitted the quantification of multiple cultivated and uncultivated taxa in mixed species biofilm samples.
Keywords: bacteria, biofilms, oral, periodontal, uncultivated
INTRODUCTION
Periodontal diseases are polymicrobial infections that can lead to periodontal inflammation and alveolar bone and tooth loss. One key objective in studying these diseases is the discrimination of host-compatible and pathogenic species. Cultivated species including Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythia were determined to be periodontal pathogens (American Academy of Periodontology, 1996), in part because increased levels of these species were associated with periodontitis and their levels decreased after successful periodontal therapy. Using similar criteria, other species were associated with periodontal health or stability, including members of the genera Actinomyces and Streptococcus.
The oral cavity harbors more than 700 taxa, 35% of which have not yet been cultivated (Chen et al., 2010; Dewhirst et al., 2010). It is likely that the uncultivated segment of the microbiota harbors pathogenic as well as beneficial species. Oral uncultivated taxa have been ‘discovered’ by cloning (Paster et al., 2001b) and further investigated using polymerase chain reaction-based techniques (Brinig et al., 2003), in situ hybridization (Zuger et al., 2007) and more recently microarrays (Colombo et al., 2009) and next generation sequencing (Keijser et al., 2008; Zaura et al., 2009). However, none of these techniques has the ability to quantify the levels of multiple uncultivated species in large numbers of individual biofilm samples simultaneously. Absolute numbers and proportions of organisms in multiple individual biofilm samples are important in distinguishing species associated with periodontal health or disease and to evaluate the effects of periodontal therapy (Haffajee et al., 1998, 2006; Socransky et al., 1998; Socransky and Haffajee, 2005). When attempting to distinguish possible periodontal pathogens from non-pathogenic ‘uncultivable’ species, it is likely that the investigator will have to examine a wide range of candidate taxa in a large number of subgingival biofilm samples from various states of periodontal health or disease. This is because of the rather large variability encountered in the microbial composition of subgingival biofilm samples. Hence, the purpose of the present investigation was to develop a high throughput method to quantify a wide range of uncultivable and cultivable taxa in large numbers of subgingival biofilm samples.
METHODS
Sample preparation
Two types of samples were employed: (i) total nucleic acids extracted from bacterial cells and (ii) total nucleic acids (TNA) extracted from subgingival biofilm samples.
Bacterial cells
As a result of the unavailability of cells from uncultivated/unrecognized bacterial species, cells from cultivated taxa were used as test species for the development of the method and validation purposes in this study.
The majority of strains (Table 1) were grown on trypticase soy agar supplemented with 5% defibrinated sheep blood (Baltimore Biological Laboratories, Cockeysville, MD). Tannerella forsythia was grown on trypticase soy agar supplemented with 5% sheep blood and 10 μg ml−1 N-acetylmuramic acid (Sigma Chemical Co., St Louis, MO). Porphyromonas gingivalis was grown on trypticase soy agar supplemented with 5% sheep blood, 0.3 μg ml−1 menadione (Sigma) and 5 μg ml−1 hemin (Sigma). Eubacterium and Neisseria species were grown on fastidious anaerobic agar (Baltimore Biological Laboratories) with 5% defibrinated sheep blood. Treponema denticola and Treponema socranskii were grown in Mycoplasma broth (Difco Laboratories, Detroit, MI) supplemented with 1 mg ml−1 glucose, 400 μg ml−1 niacinamide, 150 μg ml−1 spermine tetrahydrochloride, 20 μg ml−1 sodium isobutyrate, 1 mg ml−1 L-cysteine, 5 μg ml−1 thiamine pyrophosphate and 0.5% bovine serum. All strains were grown at 35°C under anaerobic conditions (80% N2, 10% CO2, 10% H2).
Table 1.
Reference bacterial strains used to test specificity of oligonucleotide probes and their American Type Culture Collection numbers
| Aggregatibacter actinomycetemcomitans 1 | Lactobacillus salivarius (11741) |
| Actinomyces georgiae (49285) | Lactobacillus oris (49062) |
| Actinomyces gerencseriae (23860) | Legionella pneumophila (33153) |
| Actinomyces israelii (12102) | Leptotrichia buccalis (14201) |
| Actinomyces meyeri (35568) | Neisseria mucosa (19696) |
| Actinomyces naeslundii I (12104) | Olsenella uli (49627) |
| Actinomyces naeslundii I (43146) | Parvimonas micra (33270) |
| Actinomyces odontolyticus (17929) | Peptostreptococcus anaerobius (27337) |
| Atopobium rimae (49626) | Porphyromonas endodontalis (35406) |
| Bacteroides fragilis (25285) | Porphyromonas gingivalis (33277) |
| Campylobacter concisus (33237) | Prevotella denticola (35308) |
| Campylobacter curvus (35224) | Prevotella intermedia (25611) |
| Campylobacter gracilis (33236) | Prevotella loescheii (15930) |
| Campylobacter rectus (33238) | Prevotella melaninogenica (25845) |
| Campylobacter showae (51146) | Prevotella nigrescens (33563) |
| Campylobacter sputorum ss sputorum (35980) | Prevotella oris (33573) |
| Campylobacter ureolyticus (33387) | Prevotella tannerae (51259) |
| Capnocytophaga gingivalis (33624) | Propionibacterium acnes I 2 |
| Capnocytophaga ochracea (33596) | Propionibacterium acnes II 2 |
| Capnocytophaga sputigena (33612) | Propionibacterium propionicum (14157) |
| Dialister invisus (GBA27) | Rothia dentocariosa (17931) |
| Eikenella corrodens (23834) | Selenomonas artemidis (43528) |
| Enterococcus aerogenes (13048) | Selenomonas infelix (43523) |
| Enterobacter sakazaki (12868) | Selenomonas noxia (43541) |
| Enterococcus faecalis (10100 & 29212) | Selenomonas sputigena (35185) |
| Escherichia coli (10799) | Slackia exigua (700122) |
| Eubacterium brachy (33089) | Staphylococcus aureus (33591) |
| Eubacterium limosum (8486) | Staphylococcus epidermidis (14990) |
| Eubacterium nodatum (33099) | Staphylococcus warneri (27836) |
| Eubacterium saburreum (33271) | Streptococcus anginosus (33397) |
| Eubacterium saphenum (49989) | Streptococcus constellatus (27823) |
| Filifactor alocis (35896) | Streptococcus gordonii (10558) |
| Fusobacterium necrophorum (25286) | Streptococcus intermedius (27335) |
| Fusobacterium nucleatum ss nucleatum (25586) | Streptococcus mitis (49456) |
| Fusobacterium nucleatum ss polymorphum (10953) | Streptococcus mutans (25175) |
| Fusobacterium nucleatum ss vincentii (49256) | Streptococcus oralis (35037) |
| Fusobacterium periodonticum (33693) | Streptococcus parasanguinis (15912) |
| Fusobacterium naviforme (25832) | Streptococcus pneumoniae (49619) |
| Gemella haemolysans (10379) | Streptococcus salivarius (27945) |
| Gemella morbillorum (27824) | Streptococcus sanguinis (10556) |
| Granulicatella adjacens (49175) | Streptococcus vestibularis (49124) |
| Haemophilus aphrophilus (33389) | Tannerella forsythia (43037) |
| Haemophilus influenza (33533) | Veillonella dispar (17748) |
| Haemophilus paraaphrophilus (29242) | Veillonella parvula (10790) |
| Haemophilus segnis (33393) | Treponema denticola (B1) |
| Lactobacillus acidophilus (4356) | Veilonella atypica (17744) |
| Lactobacillus brevis (14869) | Bifidobacterium denticum (27534) |
| Lactobacillus fermentum (14931) | Moraxella catarrhalis (24240) |
All cultivated strains were obtained from the American Type Culture Collection (ATCC number in parenthesis), except for Treponema denticola (B1), which was obtained from The Forsyth Institute.
ATCC strains 43718 and 29523.
ATCC strains 11827 and 11828.
Bacterial cells were harvested from agar plates, placed into 100 μl of RNAse-free TE buffer (10 mM Tris–HCl, 0.1 mM EDTA, pH 7.6) and kept at −80°C until extraction of TNA.
Subgingival plaque samples
Subgingival plaque samples were collected from healthy subjects and from patients with periodontitis. Samples were taken from mesio-buccal sites using 11/12 sterile Gracey curettes (HuFriedy, Chicago, IL) and placed in individual microcentrifuge tubes containing 100 μl RNAse-free TE buffer. Samples were kept at −80°C until TNA extraction.
Extraction of total nucleic acids
Extraction of TNA from all samples was performed using a Masterpure RNA purification kit (Epicentre, Madison, WI). Cells were pelleted by centrifugation at 3500 r.p.m. for 10 min. After the supernatant was discarded, the pellet contained in 25 μl TE was resuspended by vortex mixing. One microliter of proteinase K (50 μg μl−1) and 300 μl of tissue and cell lysis buffer (provided by the manufacturer) were added and the solution was incubated in a 65°C waterbath for 15 min. After 5 min in ice, 175 μl MCP protein precipitation reagent (provided by the manufacturer) was added to each sample. The debris was pelleted by centrifugation at 12,000 r.p.m. for 10 min. The supernatant was transferred to a new tube and 500 μl isopropanol was added. Tubes were mixed by inversion for 2 min and TNA were pelleted by centrifugation at 12,000 r.p.m. for 10 min at 4°C. After carefully pouring off the isopropanol, pellets were rinsed twice with 70% ethanol. Pellets were air dried for 10 min and resuspended in 35 μl TE buffer at 37°C for 10 min. One microliter of ScriptGuard (Epicentre) was added to each sample and samples were kept at −80°C until analysis. The TNA from individual bacterial species were measured with a spectrophotometer (Nanodrop, Wilmington, DE) at 260 nm wavelength. The TNA from bacterial mixtures and from clinical samples were not measured, rather, the entire sample was laid onto a positively charged nylon membrane.
Ninety microliters of 2% glutaraldehyde and 910 μl 6 × SSC (where 1 × SSC = 150 mM NaCl, 15 mM sodium citrate, pH 7.0) were added to each sample. The final solutions were deposited in individual lanes of a Minislot (Immunetics, Cambridge, MA), concentrated onto a nylon membrane (Boehringer Mannheim) by vacuum and fixed onto the membrane by cross-linking using ultraviolet light (Stratalinker 1800, La Jolla, CA) and dried at room temperature. The Minislot device permitted the deposition of 28 different plaque samples in individual ‘lanes’ on a single 15 × 15-cm nylon membrane as well as two control lanes containing standards for quantification of each test species.
Probe preparation
Oligonucleotide probes were prepared for 23 cultivated and 19 uncultivated bacterial taxa. The sequences were 18–22 nucleotides in length and had minimal secondary structure. Each sequence included in this group of probes targeted the 16S ribosomal DNA (rDNA) gene of the species or phylotypes listed in Table 2. The probe panel also included one universal (eubacterial) probe. This sequence was based on a conserved region of the bacterial 16S rDNA gene. All probes used in this study were based on sequences routinely employed in the Human Oral Microbial Identification Microarray (HOMIM). The full list of probe sequences has been published elsewhere (Colombo et al., 2009). One hundre picamoles of each sequence were labeled using a digoxigenin 3' end labeling kit (Roche, Indianapolis, IN).
Table 2.
Taxa for which oligonucleotide probes have been validated
| Cultivated | Uncultivated/Unrecognized |
|---|---|
| Aggregatibacter actinomycetemcomitans 1,2 | Acidaminococcus [G-1] oral taxon 135 |
| Actinomyces gerencseriae 1,2 | Acidaminococcus [G-1] oral taxon 148 |
| Actinomyces odontolyticus 1,2 | Bacteroidetes [G-2] sp. oral taxon 274 |
| Campylobacter concisus 2 | Capnocytophaga sp. oral taxon 326 |
| Campylobacter rectus 1,2 | Capnocytophaga sp. oral taxon 329 |
| Capnocytophaga sputigena 1,2 | Capnocytophaga sp. oral taxon 336 |
| Eikenella corrodens 1,2 | Desulfobulbus sp. oral taxon 041 1 |
| Eubacterium brachy 2 | Mitsuokella sp. oral taxon 131 1 |
| Fusobacterium nucleatum ss. polymorphum 1,2 | Neisseria sp. oral taxon 020 |
| Gemella haemolysans 2 | Peptostreptococcaceae [11][G-7] sp. oral taxon 081 1 |
| Haemophilus parainfluenza | Peptostreptococcaceae [13][G-1] sp. oral taxon 113 |
| Parvimonas micra 1,2 | Porphyromonas sp. oral taxon 279 |
| Porphyromonas endodontalis 2 | Prevotella sp. oral taxon 292 |
| Porphyromonas gingivalis 1,2 | Prevotella sp. oral taxon 306 1 |
| Prevotella denticola 2 | Streptococcus sp. oral taxon 055 |
| Prevotella intermedia 2 | Streptococcus sp. oral taxon 057 |
| Selenomonas noxia 1,2 | Streptococcus sp. oral taxon 066 |
| Shuttleworthia satelles | Tannerella sp. oral taxon 286 1 |
| Streptococcus anginosus 1 | TM7 [G-1] sp. oral taxon 346 1 |
| Streptococcus gordonii 1,2 | |
| Streptococcus mutans 1,2 | |
| Streptococcus parasanguinis 1,2 | |
| Tannerella forsythia 2 | |
| Treponema denticola |
A subset of the probes employed were ‘combination probes’, in that they could not distinguish species/phylotypes. This was the case for S. anginosus/gordonii; C. rectus/concisus and Streptococcus sp. OT055/057/066.
Taxa for which probes were used in the pilot clinical study.
Taxa for which probes were used in the experiments to correlate bacterial cell counts and picomolar levels of complementary sequences.
Hybridization using oligonucleotide probes
Before hybridization, the membranes were pre-wetted in 2 × SSC. The membranes were pre-hybridized in 35 ml of a solution containing 50% formamide, 5 × SSC, 1% casein (Sigma, St Louis MO), 5 × Denhardt’s reagent (Maniatis et al., 1982), 25 mM sodium phosphate (pH 6.5) and 0.5 mg ml−1 yeast RNA (Boehringer Mannheim). The solution was placed into a plastic hybridization bag containing the membrane. The sealed bag was incubated at 42°C for at least 90 min. Each membrane with fixed TNA was placed in a Miniblotter 45 (Immunetics, Cambridge MA), with the “lanes” of TNA at 90° to the channels of the device. A 30 × 45 ‘checkerboard’ pattern was produced
Probes were diluted in a proprietary hybridization buffer (UltraHyb Oligo buffer; Ambion, Austin, TX). The final concentration of the different probes in each lane in the Miniblotter 45 varied from 2 to 60 pM. The digoxigenin-labeled oligonucleotide probes, diluted in UltraHyb Oligo buffer, were placed in individual lanes of the Miniblotter. Empty lanes were filled with hybridization solution. The apparatus was placed in a plastic bag and the membranes were hybridized at 42°C for 80 min. Membranes were washed on a rotator with 250 ml sterile 2 × SSC, 0.5% sodium dodecyl sulfate at 37°C for 1 h. To detect hybrids, membranes were blocked in a maleic acid buffer containing 10% casein and then incubated with a 1 : 2500 dilution of anti-digoxigenin antibody conjugated with alkaline phosphatase (Roche, Indianapolis, IN). The membrane and the solution were placed in a sealed hybridization bag and kept on a rotator for 30 min. The membranes were rinsed with maleic acid buffer for 5 min to remove excess antibody and then washed three times with that buffer for 15 min each time. The membranes were washed with 200 ml ‘buffer 3’ [0.04% MgCl2 and 2.1% diethanolamine (pH 9.5), equal volumes] for 5 min. Finally, 1 ml of a chemiluminescent substrate (CDP Star; Tropix, Bedford, MA) was diluted in 4 ml of ‘buffer 3’. The final solution was deposited onto the membrane surface. The membrane was exposed to an X-ray film in a radiograph cassette for 60 min. The films were scanned and signal intensities of the samples and the standards were measured using TOTAL LAB software (NonLinear USA, Durham, NC). Signals were converted to absolute counts by comparison with standards on the membrane. Failure to detect a signal was recorded as zero.
Standards for the oligonucleotide probes
The standards for detection using oligonucleotide probes were a mixture of sequences complementary to the oligonucleotide probes designed to detect both the cultivable and as yet uncultivated taxa. The ‘complementary’ sequences were synthesized by SigmaGenosys (Woodlands, TX). The final mixtures of standards had 0.004 or 0.04 pM of each sequence. Ninety microlitres 2% glutaraldehyde and 910 μl 6 × SSC were added to the standards. The final solutions of the standards were deposited as the last two lanes of each membrane.
Relation of complementary sequence concentration to bacterial counts
It was estimated that 0.004 and 0.04 pM complementary probe sequence would approximate 105 and 106 bacterial cells, respectively. As bacterial cells were not available from uncultivated taxa, cultivated bacterial species were employed to test this estimate. Signals were compared from samples containing 10 ng TNA, 106 bacterial cells and 0.04 pM oligonucleotide sequences. These targets were hybridized with probes to the species and the intensities of the signals were compared.
Determination of probe sensitivity
Bacterial suspensions were prepared from pure cultures. Upon harvesting, the cell density of each species was adjusted to an optical density at 600 nm of 1 and 108 cells of each species were pipetted into a microcentrifuge tube. The resulting suspension was serially 10-fold diluted and TNA from each dilution was individually extracted using the Masterpure RNA purification kit. The samples were quantified as described above.
Determination of probe specificity
To determine the specificity of the probes, 10 ng TNA from 96 different bacterial species commonly found in the oral cavity, as well as 20 ‘complementary’ sequences for the uncultivated phylotypes, were laid on the nylon membranes using a Minislot. The membranes were then ‘probed’ in a checkerboard format using all the oligonucleotide probes.
Effect of multiple species in different levels on signal detection
To determine the influence of the presence of different numbers of cells from multiple species on quantification of target species, a series of bacterial mixtures were prepared. Suspensions containing 103 to 106 cells from a range of bacterial species were prepared. The TNAs were extracted from these mixtures and laid on a nylon membrane and quantified as described above.
Pilot clinical study
To assess the feasibility of the proposed method, a small pilot study was conducted. Eight periodontally healthy individuals and 11 patients with periodontitis were selected for study. Periodontally healthy individuals had more than 24 teeth and presented no sites with pocket depth and/or attachment level greater than 3 mm. Patients with periodontitis had at least 20 teeth, at least 5% of sites (measured at up to 168 sites) with pocket depth of 5 mm or greater and attachment level greater than 3 mm. Subjects included in the study had no systemic condition that might influence periodontal disease or dental treatment. Upon enrollment in the study, all subjects signed an informed consent. The informed consent and study protocol were approved by the Institutional Review Board at The Forsyth Institute, where all of the subjects were monitored and sampled. Samples were taken from two randomly selected contralateral quadrants in each subject, providing a total of 14 samples per subject. After removal of supragingival plaque, subgingival plaque samples were individually taken from each mesio-buccal site in the selected quadrants using sterile 11/12 Gracey curettes. Each sample was placed in a microcentrifuge tube containing 100 μl RNAse-free TE buffer. Samples were kept at −80°C until TNA extraction.
Data analysis
Signals were converted to approximate ‘counts’ by comparison with the standards on each membrane. The ‘counts’ were computed by estimating that 0.04 pM of target sequences in the standard was equivalent to approximately 106 cells and that 0.004 pM target sequences approximated 105 cells. Absence of signal detection was recorded as zero. In clinical samples, ‘counts’ for each taxon were averaged within a subject and then across subjects in the periodontitis and periodontally healthy groups, separately. Significance of differences between subject groups was determined using the Mann–Whitney U-test. In this pilot investigation, no effort was made to correct for multiple comparisons.
RESULTS
Sensitivity of detection
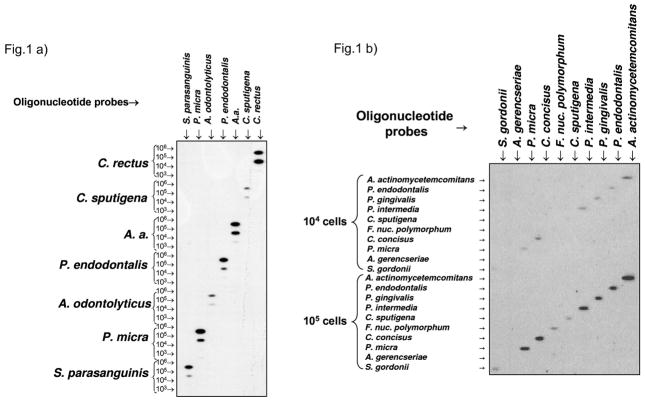
Figure 1 demonstrates the checkerboard format of the assay as well as the sensitivity of detection of the oligonucleotide probes. For all the species tested, signals could be detected on film at the level of 105 bacterial cells and for most species at 104 cells, using a newly acquired CCD camera.
Figure 1.
RNA oligonucleotide quantification technique (ROQT) membrane used to assess probe sensitivity. Total nucleic acids extracted from selected bacterial species were used as targets in the horizontal lanes. Oligonucleotide probes in the vertical lanes were hybridized against these targets. (A) Targets at 106 to 103 bacterial cells. because of higher than ideal concentration of the Campylobacter rectus probe, signals became saturated for both cell concentrations. (B) Targets at 105 to 104 bacterial cells.
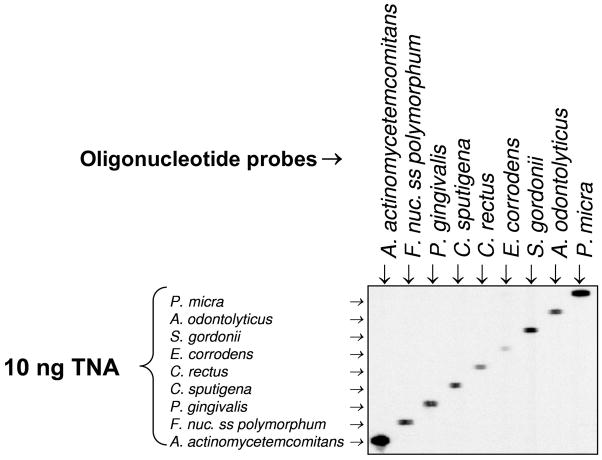
Specificity of detection
Probes for cultivated species were tested for specificity against cultivated species, using 10 ng of TNA as target (Fig. 2). Because of the unavailability of cells from uncultivated taxa, probes from those species were tested against their complementary oligonucleotide sequences. These probes were also tested against TNA extracted from 96 cultivated bacterial species (Table 1). When a signal provided by the hybridization of a test probe with the TNA from any of those species was greater than 10% of the signal provided by the Universal probe for the same TNA, the probe was eliminated from the panel. This was the case for probes for Haemophilus sp. oral taxon (OT) 035, Selenomonas sp. OT 134 and Synergistetes sp. OT 363 [throughout this manuscript, OT or oral taxon designations for uncultivated/unrecognized taxa are provided in accord with the Human Oral Microbiome Database–HOMD (Human Oral Microbiome Database, 2009), when available; GenBank assession numbers can also be found in the HOMD website]. None of the probes for the cultivated taxa showed cross-reactions.
Figure 2.
Assessment of probe specificity. Total nucleic acids (TNA) from selected bacterial species were used as targets in the horizontal lanes. Probes to the same species were hybridized against these targets in the vertical lanes. This figure shows nine of the 23 target taxa tested.
Relation of complementary sequence concentration to bacterial counts
The levels of bacterial taxa in each sample were determined by comparison with standards containing known picomolar levels of sequences complementary to the detection probe sequences. The equivalency between cell numbers and picomolar levels of complementary sequences was individually assessed for 20 cultivated bacterial species. These samples were laid on a nylon membrane and probed using the corresponding specific oligonucleotide probes. It was observed that, on average, 106 bacterial cells were equivalent to 0.068 ± 0.048 pM (mean ± SD).
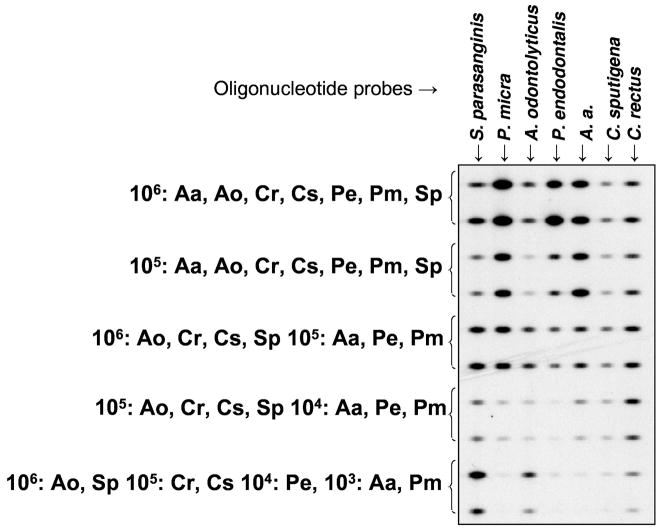
Effect of multiple species at different levels on signal detection
The presence of one or more species in a sample at high levels might affect the detection of other taxa present at lower levels. As there is marked variation in species concentrations in single samples of subgingival biofilms, it was crucial to determine whether the presence of high numbers of cells from a given species would overshadow the presence of less abundant taxa. To evaluate this possibility, mixtures of bacterial species at different levels were prepared. Figure 3 demonstrates that TNA from individual bacterial species could be detected in the range of 104 to 106 cells when in the presence of different levels of cells of other species. For instance, even in the presence of 106 cells of Actinomyces odontolyticus and Streptococcus parasanguinis, 104 cells of Porphyromonas endodontalis and 103 cells of Aggregatibacter actinomycetemcomitans and Parvimonas micra could be detected.
Figure 3.
Effects of different levels of species in mixtures on signal intensity. Aa, Aggregatibacter actinomycetemcomitans; Ao, Actinomyces odontolyticus; Cr, Campylobacter rectus; Cs, Capnocytophaga sputigena; Pe, Porphyromonas endodontalis; Pm, Parvimonas micra; Sp, Streptococcus parasanguinis.
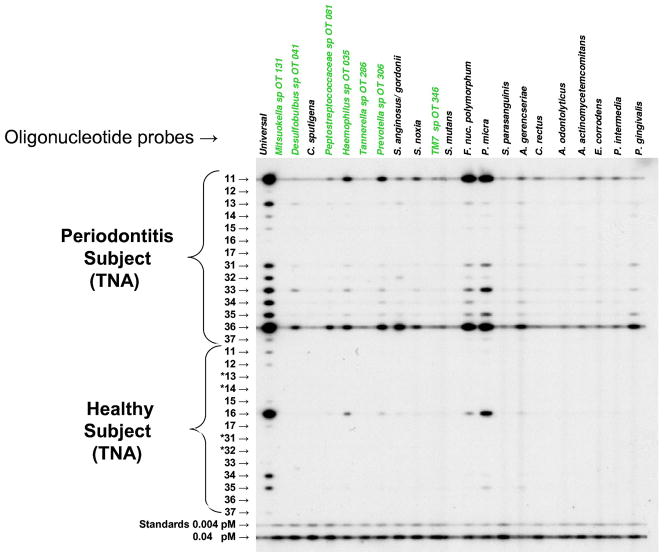
Enumeration of taxa in clinical samples
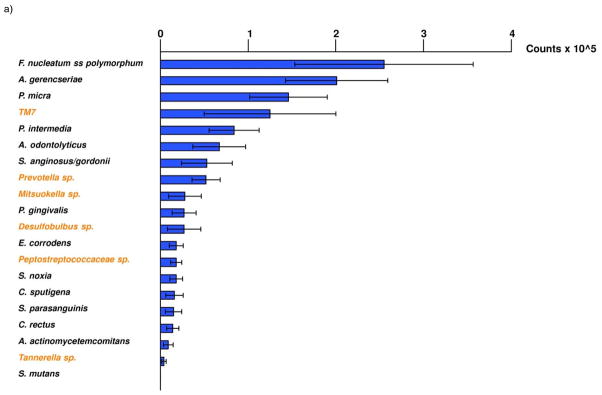
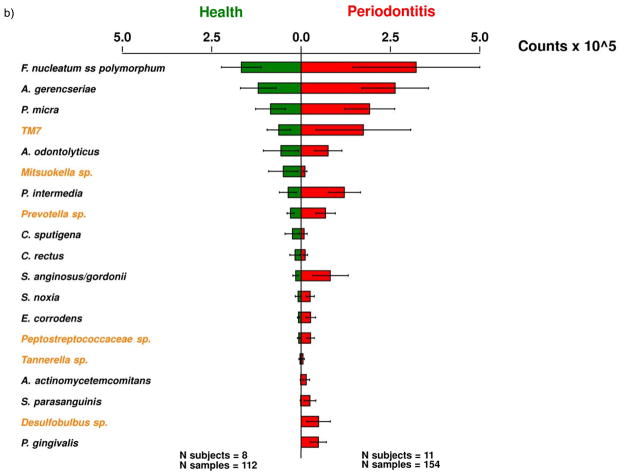
In a pilot clinical study to assess the feasibility of the method, cultivated and uncultivated bacterial taxa could be detected and quantified. Figure 4 shows an example of a typical membrane from this study. The use of a universal (eubacterial) probe gave an estimate of the total bacterial load in each sample. Samples from patients with periodontitis usually showed higher levels of subgingival bacteria based on the universal probe signals than samples from periodontally healthy subjects. Overall, the most commonly detected uncultivated/unrecognized species in the subject population were TM7 sp. OT 346, Prevotella sp. OT 306, Mitsuokella sp. OT 131 (Fig. 5A). In the cultivated segment of the microbiota, the most abundant taxa were Fusobacterium nucleatum ss. polymorphum, Actinomyces gerencseriae and Parvimonas micra. When the data from periodontal health and disease were compared, different mean microbial profiles were observed (Fig. 5B). Patients with periodontitis showed higher mean counts of Prevotella sp. OT 306. Desulfobulbus sp. OT 041 was detected only in the periodontitis group. Mean counts of Prevotella intermedia, a member of the ‘orange complex’ (Socransky et al., 1998), was also elevated in this group. Porphyromonas gingivalis, a periodontal pathogen of the ‘red complex’ could only be detected in the periodontitis group.
Figure 4.
A checkerboard membrane showing hybridization of clinical samples with oligonucleotide probes. Probes for cultivated species (black) and as yet uncultivated species (green) are listed across the top. Each horizontal lane represents the total nucleic acids (TNA) from a sample from the indicated numbered tooth. Standards comprised a mixture of ‘complementary’ sequences from all the test taxa at 0.004 and 0.04 pM, respectively. Teeth marked with an asterisk (*) were absent.
Figure 5.
(A) Mean estimated ‘counts’ ( ×105 ± SEM) of 20 bacterial taxa in 266 subgingival plaque samples obtained from eight periodontally healthy people and 11 patients with periodontitis. The species were ordered according to mean counts. Taxa in black represent cultivated species, whereas those in orange represent uncultivated taxa. (B) Mean estimated ‘counts’ (× 105 ± SEM) of 20 bacterial taxa in 266 samples obtained from eight periodontally healthy people (green) and 11 patients with periodontitis (red). The species were ordered according to mean counts in health. Taxa in black represent cultivated species, while those in orange represent uncultivated taxa.
DISCUSSION
It has long been recognized that many taxa in subgingival plaque were not being cultivated, as there were marked differences between total viable counts (representing cultivated species) and total microscopic counts representing all organisms (Socransky et al., 1963; Newman and Socransky, 1977). This phenomenon has been described as the ‘great plate count anomaly’ (Staley and Konopka, 1985) and it seems to be a constant in bacterial samples originating from different environments, including the oral cavity.
It seems likely that periodontal pathogens are among the uncultivated segment of the microbiota. A number of studies have used cloning to seek novel and uncultivated bacterial species (Paster et al., 2001a; Becker et al., 2002; Dewhirst et al., 2000; Kazor et al., 2003). Cloning, however, is time-consuming, limiting the number of samples and clones that can be analysed at a time, and it does not allow direct quantification. Recently, next generation sequencing (NGS) methods have surpassed the capabilities of cloning and enabled deeper coverage and less labor-intensive sequencing of microbial communities. Even though NGS represents a major advance in the study of oral microbiology, only small numbers of subjects and samples have been reported (Zaura et al., 2009; Li et al., 2010). Furthermore, those studies report data at the genus level, preventing the identification of pathogenic or health compatible species.
Because the differences between periodontal health and disease and before and after therapy are quantitative (Haffajee et al., 1998, 2006; Socransky et al., 1998; Socransky and Haffajee, 2005) and because of the site-specific nature of periodontal diseases, quantification of individual biofilm samples is necessary. Species quantification in samples from periodontal sites with different clinical status in the same or different oral cavities is a powerful first step in discriminating pathogens from host-compatible species. High throughput is another prerequisite. Species counts are highly variable in biofilm samples, requiring the analysis of large numbers of samples from many subjects to detect meaningful differences in species counts. In the present study, the standards for quantification of individual taxa contained 0.004 and 0.04 pM of the sequences complementary to the probes. These levels were estimated based on the molecular weight of the 16S rRNA molecule and the number of copies thought to be present in bacterial cells. Subsequent experiments indicated that 0.04 pM yielded signals equivalent to about 0.44 × 105 bacterial cells.
Ribosomal RNA was selected as the target molecule because it is more abundant in bacterial cells than DNA, and so could enhance the sensitivity of oligonucleotide probes. Additionally, because an actively growing cell has 103 to 104 rRNA molecules (Scheu et al., 1998), rRNA is more associated with cell viability than is DNA (McKillip et al., 1998). Hence, the use of rRNA can provide insights on the relevance of the test species in the ecosystem of interest and avoid biases in the results due to the presence of dead cells (Nocker and Camper, 2006).
Levels of periodontal bacteria in single curette stroke samples of subgingival plaque commonly range from 104 to 107 cells (Socransky and Haffajee, 2005). RNA-oligonucleotide quantification technique (ROQT) is able to consistently detect 105 bacterial cells, and 104 cells of most species (Fig. 1). This is in line with the level of sensitivity obtained using whole genomic DNA probes.
A number of previous studies have investigated uncultivated species (Paster et al., 2001a; Kumar et al., 2005; Li et al., 2006). Kumar et al. (2005) evaluated the subgingival microbiota of periodontal health and disease. The most abundant species belonged to the genera Selenomonas, Streptococcus, Veillonella, Campylobacter and Peptostreptococcus. Fusobacterium species and Actinomyces species, both routinely found by culture and DNA probes (Slots, 1977; Ximenez-Fyvie et al., 2000; Teles et al., 2008) were rarely detected. In contrast, in our study, Fusobacterium nucleatum ss polymorphum and Actinomyces gerencseriae were the most abundant taxa detected. The authors found a robust association of peptostreptococci with periodontitis: Peptostreptococcus strains BS044 (not in HOMD) and CK035 (HOMD: Peptostreptococcus stomatis) were numerous. Targeted DNA approaches have found Parvimonas micra (formerly Peptostreptococcus micros) to be elevated in patients with periodontitis (Nonnenmacher et al., 2004). In the present study, Parvimonas micra was also more prevalent in periodontitis samples. The authors also reported that Desulfobulbus sp. CH031 (not in HOMD) and OT 041 were significantly associated with deep periodontal sites. In our study, although in low numbers, Desulfobulbus sp. OT 041 was only detected in periodontitis samples. Selenomonas clones were not associated with periodontal disease in the Kumar et al. study. In the present study, Selenomonas noxia was present in low numbers in healthy subjects and Selenomonas sp. CS002 (HOMD: Mitsuokella sp. OT 131) was increased in samples from healthy individuals. Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia were rarely detected in periodontitis samples by cloning, which contrasts with investigations using culture, polymerase chain reactions, real-time polymerase chain reactions, immunofluorescence, in situ hybridization, immunocytochemistry, DNA probes and oligonucleotide probes (Tanner et al., 1979; Kigure et al., 1995; Klein and Goncalves, 2003; Nonnenmacher et al., 2004; Yang et al., 2004). In our study, Porphyromonas gingivalis was solely detected in patients with periodontitis and accounted for a significant portion of the total probe count (Fig. 5B).
In our study, Tannerella sp. OT 286 was the least abundant uncultivated taxon present in patients with periodontitis, corroborating findings by others (Zuger et al., 2007). TM7 sp. OT 346 was the most prevalent uncultivated phylotype overall and in the periodontitis group, which is in accord with previous reports (Brinig et al., 2003). The Human Oral Microbe Identification Microrray (HOMIM) has been employed to study the subgingival microbiota of periodontal health and disease (Colombo et al., 2009). The frequency of detection of Porphyromonas gingivalis, Selenomonas noxia, Streptococcus anginosus/gordonii, Actinomyces gerencseriae and TM7 OT 346 was significantly higher in diseased sites and the frequency of Capnocytophaga sputigena was elevated in healthy sites. These results were in line with the levels of taxa found in the present study.
The aim of the present study was to devise a method to detect and quantify uncultivated bacterial species in subgingival biofilm samples in periodontal health and disease. The specificity of the probes used was confirmed by the absence of cross-reactions with any of the 96 bacterial taxa tested, representing the most prominent cultivable oral bacterial taxa. The small clinical study demonstrated the feasibility of the method for its use in clinical trials. The strengths of the proposed method include the absence of pooling, amplification or dilution bias, because an entire individual sample is laid onto the membrane. It allows the quantification of both cultivable and uncultivable bacterial taxa. It is high throughput, in that multiple samples can be analysed for the levels of multiple species at the same time on a single membrane and it is relatively inexpensive. The method also has limitations. The standard curve presented has three data points, which enables quantification of taxa in the 104 to 107 cells range in a given sample. However, additional levels of standards can be added to provide a tighter or more comprehensive standard curve. The data presented above were obtained using film exposure, which has a dynamic range of about two orders of magnitude. We have recently switched to image capture by a CCD camera, reaching a dynamic range of 4.8 orders of magnitude and enhancing the accuracy of the method. Although the format of the ROQT may resemble that of the checkerboard DNA–DNA hybridization technique (Socransky et al., 1994, 2004), it is not meant to represent a ‘more sensitive checkerboard’. The techniques differ in the nature of the probes, their target molecules, their hybridization protocols and the nature of the species sought.
It seems that an ever growing number of taxa can be detected in the oral cavity and more than 35% of them cannot be cultivated. It is likely that only a subset of this segment is relevant to disease development. ROQT provides the ability to examine large numbers of biofilm samples from large numbers of subjects for the levels of uncultivated taxa. This approach can indicate the most numerous, and thereby possibly the most relevant, taxa associated with periodontal diseases, clarifying their potential role in initiation and progression of periodontal infections. By identifying the more relevant uncultivated/unrecognized taxa, this technique will guide the isolation and cultivation of disease-associated uncultivated and unrecognized taxa. Such taxa merit further pursuit by cultivation, to evaluate the pathogenic mechanisms of the selected taxa and to develop more targeted treatment and prevention strategies.
Acknowledgments
This work was supported in part by grants T32-DE-07327, DE-12108 and DE-14242 from the National Institutes of Dental and Craniofacial Research. This work was also supported in part by the Eleanor and Miles Shore 50th Anniversary Fellowship Program for Scholars in Medicine (The Forsyth Institute/Harvard Medical School).
References
- American Academy of Periodontology. Consensus report. Periodontal diseases: pathogenesis and microbial factors. Ann Periodontol. 1996;1:926–932. doi: 10.1902/annals.1996.1.1.926. [DOI] [PubMed] [Google Scholar]
- Becker MR, Paster BJ, Leys EJ, et al. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol. 2002;40:1001–1009. doi: 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinig MM, Lepp PW, Ouverney CC, Armitage GC, Relman DA. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Appl Environ Microbiol. 2003;69:1687–1694. doi: 10.1128/AEM.69.3.1687-1694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010:10. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo AP, Boches SK, Cotton SL, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, et al. The Human Oral Microbiome. J Bacteriol. 2010 Jul 23; doi: 10.1128/JB.00542-10. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Tamer MA, Ericson RE, et al. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol Immunol. 2000;15:196–202. doi: 10.1034/j.1399-302x.2000.150308.x. [DOI] [PubMed] [Google Scholar]
- Haffajee AD, Cugini MA, Tanner A, et al. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J Clin Periodontol. 1998;25:346–353. doi: 10.1111/j.1600-051x.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- Haffajee AD, Teles RP, Socransky SS. The effect of periodontal therapy on the composition of the subgingival microbiota. Periodontol 2000. 2006;42:219–258. doi: 10.1111/j.1600-0757.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- [Accessed June 2010];Human Oral Microbiome Database. 2009 Available at: www.homd.org.
- Kazor CE, Mitchell PM, Lee AM, et al. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J Clin Microbiol. 2003;41:558–563. doi: 10.1128/JCM.41.2.558-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijser BJ, Zaura E, Huse SM, et al. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res. 2008;87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- Kigure T, Saito A, Seida K, Yamada S, Ishihara K, Okuda K. Distribution of Porphyromonas gingivalis and Treponema denticola in human subgingival plaque at different periodontal pocket depths examined by immunohistochemical methods. J Periodontal Res. 1995;30:332–341. doi: 10.1111/j.1600-0765.1995.tb01284.x. [DOI] [PubMed] [Google Scholar]
- Klein MI, Goncalves RB. Detection of Tannerella forsythensis (Bacteroides forsythus) and Porphyromonas gingivalis by polymerase chain reaction in subjects with different periodontal status. J Periodontol. 2003;74:798–802. doi: 10.1902/jop.2003.74.6.798. [DOI] [PubMed] [Google Scholar]
- Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CL, Liang JP, Jiang YT. Association of uncultivated oral phylotypes AU126 and X112 with periodontitis. Oral Dis. 2006;12:371–374. doi: 10.1111/j.1601-0825.2005.01205.x. [DOI] [PubMed] [Google Scholar]
- Li L, Hsiao WW, Nandakumar R, et al. Analyzing endodontic infections by deep coverage pyrosequencing. J Dent Res. 2010;89:980–984. doi: 10.1177/0022034510370026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- McKillip JL, Jaykus L, Drake M. rRNA Stability in heat-killed and UV-irradiated enterotoxigenic Staphylococcus aureus and Escherichia coli 0157:H7. Appl Environ Microbiol. 1998;64:4264–4268. doi: 10.1128/aem.64.11.4264-4268.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M, Socransky SS. Predominant cultivable microbiota in periodontosis. J Periodontal Res. 1977;12:120–128. doi: 10.1111/j.1600-0765.1977.tb00114.x. [DOI] [PubMed] [Google Scholar]
- Nocker A, Camper AK. Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl Environ Microbiol. 2006;72:1997–2004. doi: 10.1128/AEM.72.3.1997-2004.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonnenmacher C, Dalpke A, Mutters R, Heeg K. Quantitative detection of periodontopathogens by real-time PCR. J Microbiol Methods. 2004;59:117–125. doi: 10.1016/j.mimet.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001a;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Boches SK, Galvin JL, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001b;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheu PM, Berghof K, Stahl U. Detection of pathogenic and spoilage microorganisms in food with the polymerase chain reaction. Food Microbiol. 1998;15:13–31. [Google Scholar]
- Slots J. Microflora of the healthy gingival sulcus in man. Scand J Dent Res. 1977;85:247–254. doi: 10.1111/j.1600-0722.1977.tb00560.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Gibbons R, Dale ABL, Rosenthal E, Macdonald J. The microbiota of the gingival crevice area of man. I. Total microscopic and viable counts and counts of specific organisms. Arch Oral Biol. 1963;8:275–280. doi: 10.1016/0003-9969(63)90019-0. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135–187. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Smith C, et al. Use of checkerboard DNA-DNA hybridization to study complex microbial ecosystems. Oral Microbiol Immunol. 2004;19:352–362. doi: 10.1111/j.1399-302x.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Smith C, Martin L, Paster BJ, Dewhirst FE, Levin A. "Checkerboard" DNA-DNA hybridization. Biotechniques. 1994;17:788–792. [PubMed] [Google Scholar]
- Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- Tanner A, Haffer C, Bratthall G, Visconti R, Socransky SS. A study of the bacteria associated with advancing periodontitis in man. J Clin Periodontol. 1979;6:278–307. doi: 10.1111/j.1600-051x.1979.tb01931.x. [DOI] [PubMed] [Google Scholar]
- Teles FR, Haffajee AD, Socransky SS. The reproducibility of curet sampling of subgingival biofilms. J Periodontol. 2008;79:705–713. doi: 10.1902/jop.2008.070424. [DOI] [PubMed] [Google Scholar]
- Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J Clin Periodontol. 2000;27:648–657. doi: 10.1034/j.1600-051x.2000.027009648.x. [DOI] [PubMed] [Google Scholar]
- Yang HW, Huang YF, Chou MY. Occurrence of Porphyromonas gingivalis and Tannerella forsythensis in periodontally diseased and healthy subjects. J Periodontol. 2004;75:1077–1083. doi: 10.1902/jop.2004.75.8.1077. [DOI] [PubMed] [Google Scholar]
- Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy "core microbiome" of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuger J, Luthi-Schaller H, Gmur R. Uncultivated Tannerella BU045 and BU063 are slim segmented filamentous rods of high prevalence but low abundance in inflammatory disease-associated dental plaques. Microbiology. 2007;153:3809–3816. doi: 10.1099/mic.0.2007/010926-0. [DOI] [PubMed] [Google Scholar]