Abstract
The isoschizomeric restriction endonucleases Fnu4HI and BsoFI cleave DNA at 5'-GCdecreasesNGC-3' sequences. Fnu4HI has been shown to be inhibited by 5'-CG-3'methylation in the sequences 5'-GmCGGC-3' or 5'-GCGGmCG-3'. We have now investigated the methylation sensitivity of BsoFI by testing its activity on plasmid DNA 5'-CG-3' methylated with the M.SssI DNA methyltransferase or on synthetic (CGG)n repetitive oligodeoxyribonucleotides which have been partly or completely C methylated. The data demonstrate that BsoFI cannot cleave at its recognition sequence when it is completely 5'-CG-3' methylated. These enzymes have proven to be useful in analyses of the methylation status in (CGG)n repeats of the human genome.
Full text
PDF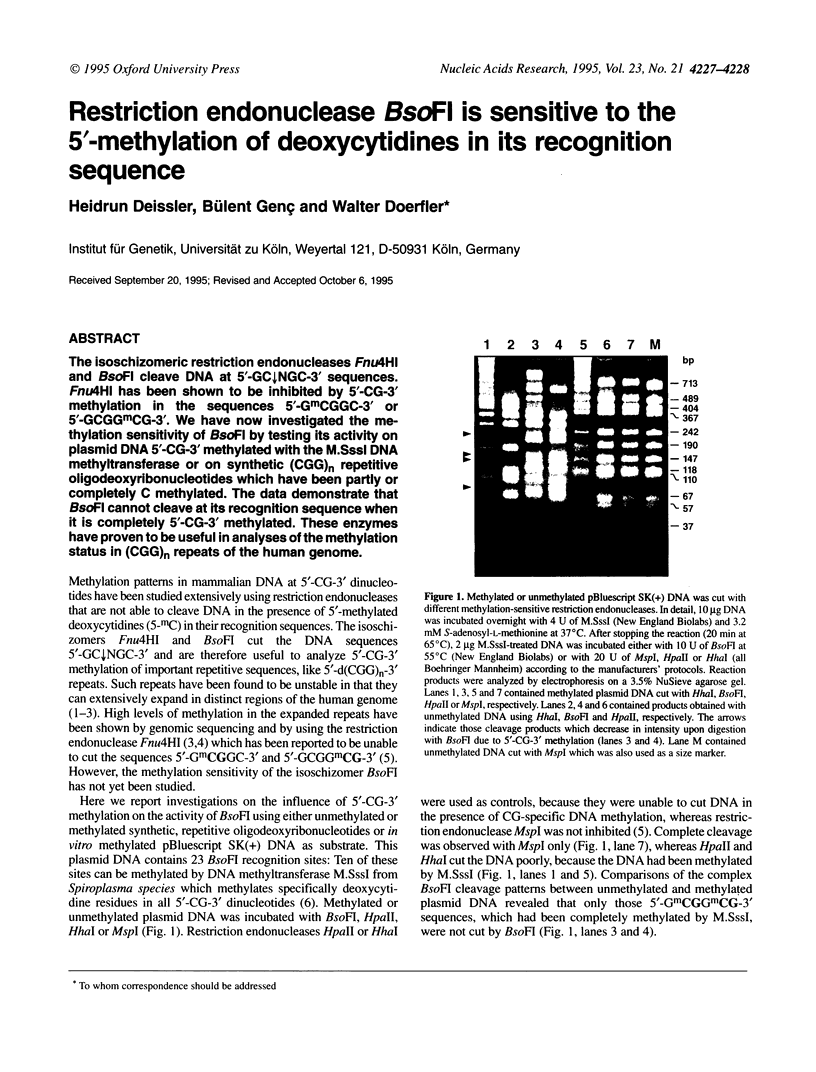

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fu Y. H., Kuhl D. P., Pizzuti A., Pieretti M., Sutcliffe J. S., Richards S., Verkerk A. J., Holden J. J., Fenwick R. G., Jr, Warren S. T. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991 Dec 20;67(6):1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Hansen R. S., Gartler S. M., Scott C. R., Chen S. H., Laird C. D. Methylation analysis of CGG sites in the CpG island of the human FMR1 gene. Hum Mol Genet. 1992 Nov;1(8):571–578. doi: 10.1093/hmg/1.8.571. [DOI] [PubMed] [Google Scholar]
- McClelland M., Nelson M., Raschke E. Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 1994 Sep;22(17):3640–3659. doi: 10.1093/nar/22.17.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlé I., Rousseau F., Heitz D., Kretz C., Devys D., Hanauer A., Boué J., Bertheas M. F., Mandel J. L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991 May 24;252(5009):1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Renbaum P., Abrahamove D., Fainsod A., Wilson G. G., Rottem S., Razin A. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI). Nucleic Acids Res. 1990 Mar 11;18(5):1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk A. J., Pieretti M., Sutcliffe J. S., Fu Y. H., Kuhl D. P., Pizzuti A., Reiner O., Richards S., Victoria M. F., Zhang F. P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991 May 31;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]