Abstract
Objective
To compare DNA yield from neonatal umbilical cord blood and buccal swab specimens.
Methods
Umbilical cord blood was obtained at birth in a cohort of women enrolled in a preterm labor study. If cord blood was not obtained, neonatal buccal samples were obtained using the Oragene® saliva kits. DNA was extracted from all samples using the QIAamp® extraction kits. DNA concentration and yield were compared between umbilical cord blood and buccal swabs.
Results
DNA concentrations from umbilical cord blood (n=35) was greater than that obtained from buccal swabs (n=20) (total sample: 209.0±110.7 ng/μL vs. 6.9± 6.7 ng/μL respectively, p<0.001; partial sample: n=30 cord blood vs. n=11 buccal, 70.0±51.4 ng/μL vs. 11.3±6.7 ng/μL respectively, p<0.001) and produced more total DNA (total sample: 116.5±70.8 μg vs. 4.2±4.0 μg, p<0.001; partial: 14.0±10.3 μg vs. 1.1±0.7 μg respectively, p<0.001).
Conclusions
Buccal swabs yield less neonatal DNA than umbilical cord blood specimens.
Introduction
Obtaining and storing neonatal genetic samples is an emerging research need. The use of biorepositories has become a critical resource for large scale epidemiological studies, candidate gene and genome-wide association studies(GWAS), and molecular-based biomedical research, bridging the gap between molecular and clinical information1–3. Critical to this endeavor is the ability to easily, conveniently, and cost-effectively acquire high yields of quality DNA from participants4.
Advances in high-throughput genotyping technologies now allow us to assay for thousands of genotypes quickly and with relatively small amounts of DNA (~1–5 ng DNA per assay) 5. However, the samples collected for biorepositories are carefully characterized populations whose finite amount of DNA collected must be maximized to attain increased information, including providing for evaluation of future biomarkers in order to minimize research costs 3, 6. Collection of umbilical cord blood is ideal for obtaining DNA from neonates, but the unpredictable nature of birth makes obtaining these samples difficult at times. While saliva is a robust source of DNA from adults when blood is not available, studies are lacking about the robustness of sample alternatives to umbilical cord blood for the neonate. Review of the literature revealed no studies or data comparing neonatal salivary sample DNA yield to that from umbilical cord blood.
The objective of this study was to compare the DNA yield from neonatal specimens obtained at or just after birth. The hypothesis is that the yield of DNA from neonatal salivary swabs would be equivalent to that from umbilical cord blood and thus would be adequate for genetic studies and biorepositories.
Materials and Methods
Study Population
Our study population consisted of neonates of pregnant women who had been admitted to the hospital with a diagnosis of threatened preterm birth who had received at least one dose of antenatal corticosteroids. The women, admitted to one of two urban hospitals, were recruited to a study and gave consent for obtaining samples for DNA from their newborns. Women consented were at least 23 weeks gestation and greater than or equal to 18 years-old.
Sample Collection and Processing
Blood Samples
Umbilical cord blood samples were obtained immediately following delivery in K2EDTA vacutainers by trained cord blood collectors. Samples were mixed by inversion 8–10 times after being drawn and then stored at −80 °C until DNA isolation.
Saliva Samples
If umbilical cord blood was not acquired at the time of delivery because of an off-hour delivery when the study personnel were not available, a trained member of the research team obtained buccal swabs from the neonate. For buccal cell collection we used Oragene® saliva kit collection with cotton swabs. Oragene® kits are shown to reduce microbial contamination and provide immediate stabilization of samples, allowing it to be stored at room temperature for years without processing or DNA degradation. Oragene® “Saliva collection with cotton swabs or buccal brushes” protocol was followed. Collection protocols call for collector to grip swab without contaminating cotton tip and gently place the cotton tips inside of the infant’s mouth, collecting as much saliva as possible by rubbing the cheeks and moving the tip into the spaces between the check and gums and under the tongue. Once saturated in saliva, tips were cut off of swabs into the base of the Oragene® collection vial. This process was repeated until the 5 cotton tips that come with the collection kit were inside of the collection vial. Immediately upon collection the Oragene® vial cap which contains the Oragene®·DNA solution was placed onto the base and tightened securely. The vial was shaken vigorously and inverted to ensure that the cotton tips were well mixed with the Oragene®·DNA preservation fluid. Kits were then stored at room temperature. Prior to purification the Oragene saliva sample kits were briefly mixed by gentle inversion and incubated for a minimum of 2 hours in an air incubator set to 50 °C. To remove saliva from buccal swabs manufacturer protocols for “DNA Recovery from Saliva Sponges” were followed. First, free liquid was removed and transferred into a 15 mL conical centrifuge tube. A barrel of a 5 mL disposable plastic syringe (with the plunger removed) was placed into the same tube. Sterile, disposable forceps were used to transfer the sponges from the kit collection base into the barrel of the syringe. The syringe barrel containing the sponges in the tube was centrifuged at 200 × g for 10 minutes at 20 °C. The barrel containing the dry sponges was discarded and the tube containing the recovered liquid was capped, labeled and stored at room temperature until purification.
DNA Isolation
DNA was extracted from umbilical cord blood samples using the QIAamp® DNA mini, midi, or maxi kits (Qiagen Inc., Valencia, CA). The specific kit used was chosen based on sample volume collected and time period of collection. As with the buccal swab samples, we originally extracted only a portion of the sample collected but later began extracting the full sample to attain maximal DNA yield. Kits are designed to extract the following sample volumes: mini (0.2 mL), midi (1–2 mL), and maxi (3–5 mL). Initially all samples were extracted using the minikit. Subsequent samples were extracted using the kit whose volume capacity best fit the total sample volume collected. Manufacturer spin protocol instructions were followed for all kits with one modification. Our equipment was not capable of reaching 4500 × g for large tube sizes. For steps where the manufacturer protocol called for centrifugation at 4500 × g we used 3100 × g and compensated this difference by using increased spin times. When manufacturer protocols listed steps for highly concentrated DNA those steps were followed. Isolated DNA was transferred into cyrovials and all samples were stored at −80 °C until quantification.
DNA was purified from saliva swab samples using the Oragene® Kit (DNA Genotek) manufacturer recommendations. Two procedures were utilized throughout the course of the study. Initially 500 μL of sample was transferred to a 1.5 mL Eppendorf tube and purified. However, we found DNA yield insufficient using only a portion of the collected sample prompting us to extract the entire sample volume collected from subsequent participant samples. Oragene® saliva kits are designed to collect 2 mL of saliva, which are then added together with 2 mL of Oragene®·DNA present in the cap of each kit resulting in 4 mL of total sample to be extracted. However, using the “saliva collection with cotton swabs or buccal brushes” and “DNA recovery from saliva sponges” protocols, only minimal volumes of saliva were recovered (≤200 μL.) Regardless of sample volume used DNA isolation was done according to manufacturer instructions. Samples were frozen at −80 °C until quantification.
DNA Quantification
Concentration of double-stranded DNA in our samples was determined using a Quant-iT dsDNA Broad Range or High Sensitivity assay Kit and Qubit Flouromter (Invitrogen, Carlsbad, California.) This system utilizes a fluorescent nucleic acid stain to accurately and specifically measure dsDNA at highly sensitive levels.
DNA Quality assessment by gel electrophoresis
The DNA quality was assessed by analyzing the samples for evidence of degradation using gel electrophoresis. Samples of both buccal and umbilical cord blood genomic DNA (180 ng of each sample) were run on an agarose gel (0.9% agarose) and stained with ethidium bromide. The size of the DNA was determined by comparison with a DNA ladder (1 kb; Life Technologies). The largest band in the ladder is ~10 kb. High quality DNA is expected to be mostly >10 kb.
Statistical Analysis
All results were analyzed with SPSS, version 17.0 (SPSS, Inc., Chicago, IL). Non-parametric tests were used to compare mean total yield and concentration of DNA collected from Oragene® buccal swab collection samples and umbilical cord blood samples because the samples were not normally distributed. We utilized the Mann-Whitney U test. We also compared the DNA concentration between extraction protocols for partial sample extraction and complete sample extraction. For the purposes of comparison umbilical cord blood and buccal swab samples were each separated into two groups based on whether the entire blood or buccal cell sample collected or a portion of the entire sample collected was isolated in this study. Additionally, a linear regression analysis was done to compare the possible effect of gestational age at cord blood collection and day of life at buccal swab collection on DNA yield measures.
Results
Ninety-six patient samples were included in this analysis. We were able to collect 65 umbilical cord blood samples (68%) at the time of delivery and collected neonatal buccal swabs from all neonates whose cord blood was unable to be obtained (n=31). We isolated DNA from all samples collected and estimated DNA concentrations and total DNA yield as above. As shown in Table 1, we observed significant differences in both measures between umbilical cord blood and buccal swabs in both the partial and total sample isolation groups. The mean DNA yield in the total sample isolation of umbilical cord blood was 116.5±70.8 μg compared with 4.2±4.0 μg in total sample isolation of Oragene® buccal swabs (p<0.0001). The partial samples yielded total DNA equal to 14.0±10.3 μg for blood versus 1.1±0.7 μg for buccal swabs (p<0.0001). Comparison of DNA concentrations gave similar results, with a mean DNA concentration from total sample isolation of umbilical cord blood and buccal swabs of 209.0±110.7 ng/μL and 6.9±6.7 ng/μL, respectively (p<0.0001). In the partial sample isolation DNA concentrations were 70.0±51.4 ng/μL and 11.3±6.7 ng/μL, respectively (p<0.0001).
Table 1.
DNA concentrations and yield in entire and partial samples of umbilical cord blood and buccal swabs.
| Method of DNA Collection | Umbilical Cord Blood (total sample, n=35) | Oragene® Buccal Swabs (total sample, n=20) | Umbilical Cord Blood (partial sample, n=30) | Oragene® Buccal Swabs (partial sample, n=11) |
|---|---|---|---|---|
| Mean DNA concentration (ng/μl; range) | 209.0 ± 110.7* (7.4–468.5) | 6.9 ± 6.7* (0.6–29.2) | 70.0 ± 51.4* (8.9–204.7) | 11.3 ± 6.7* (4.3–26.2) |
| Average Eluted DNA volume (μl) | 540 | 600 | 200 | 100 |
| Mean total DNA yield (μg; range) | 116.5 ± 70.8* (2.2–281.1) | 4.2 ± 4.0* (0.4–17.5) | 14.0 ± 10.3* (1.8–40.9) | 1.1 ± 0.7* (0.4–2.6) |
Data are means ± standard deviation
P<0.001 for all comparisons of cord blood vs. buccal swab for DNA concentration and total DNA yield. P value also <0.001 comparing mean DNA concentration and total DNA yield between cord blood samples with portion of sample analyzed and entire sample analyzed and between buccal samples with portion of sample and entire sample analyzed.
+Entire sample of umbilical cord blood was between 1–5 ml. Entire sample of buccal swabs was between 2–3 ml. Partial sample of umbilical cord blood utilized 0.2 ml. Partial sample of buccal swabs utilized 0.5 ml.
A secondary point of interest is the difference in DNA yield measures between isolation methods. Due to the claim that Oragene® samples are stabilized upon collection and continue to be stable for up to 20 years at room temperature, we initially completed only partial isolation of the Oragene® buccal swabs samples to act as a safeguard against potential isolation problems. Total DNA yields that were insufficient led to future samples having the total sample collected isolated. As one would expect, mean total DNA yield improved from 1.1±0.7 μg in partial sample isolation to 4.2±4.0 μg in complete sample isolation (p<0.0001). However, mean DNA concentration decreased from 11.3±6.7 ng/μL for partial isolation to 6.9±6.7 ng/μL for full sample isolation (p<0.001) from the buccal swabs.
In addition, the quality of the DNA also appeared to be lower. The analysis of the DNA by gel electrophoresis showed evidence of lower molecular weight DNA and less intense bands >10 kb, both of which are indicative of partial degradation (Figure).
Figure 1.
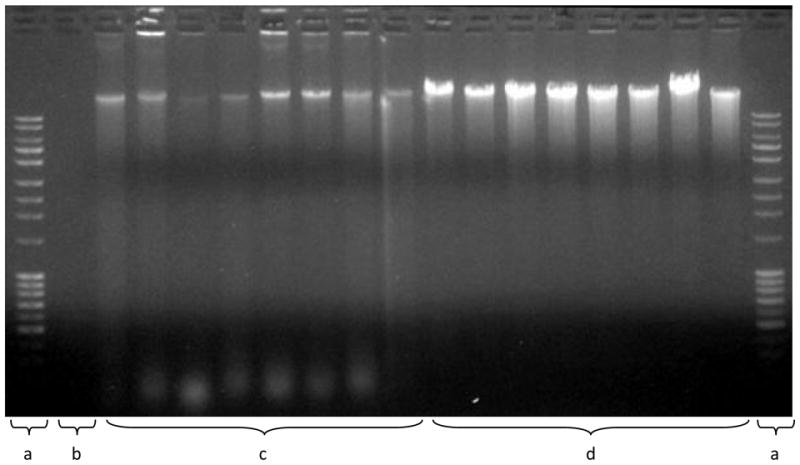
Analysis of DNA fragmentation. The size of the DNA was determined by agarose gel electrophoresis and detected by ethidium bromide staining. Land a) 1 kb Ladder (top band ~10 kb); b) No sample; c) Genomic DNA from buccal swabs (180 ng); d) Genomic DNA from cord blood (180 ng). The smaller molecular weight bands (bottom of the gel) and the lower intensity of the staining of the high molecular weight DNA indicate increased degradation of the DNA from the buccal swabs.
Finally, we completed a linear regression analysis to determine the possible effect of gestational age at the time of cord blood collection and of days of life at the time of buccal swab collection on DNA yield. These two analyses are not comparable due to the fact that days of life is not equivalent to gestational age. Gestational age at the time of cord blood collection does not correlate with total DNA yield (r2=0.006, P=0.681). Days of life at the time of buccal swab collection also does not correlate with total DNA yield (r2=0.119, P=0.328).
Comment
In this study we found statistically significant differences in both total DNA yield (μg) and DNA concentration (ng/μL) between DNA isolated from umbilical cord blood samples and buccal swab samples. We also found that the protocol for partial extraction of an Oragene® buccal swab collection sample did not yield a suitable amount of DNA for a large scale epidemiological study or GWAS analysis and for greatest total yield of DNA (μg) the entire buccal cell collection should be purified. However, for highest concentration of DNA, following manufacturer’s protocols, partial extraction was superior. This is likely due to the fact that sample volumes collected using buccal swabs are not analogous to sample volumes collected from older participants who are able to spit directly into the Oragene® collection container. Manufacturer protocols for swabs and saliva are combined and do not prorate reconstitution volume for initial sample volume. To accommodate swab samples, manufacturer protocols must be edited to decrease reconstitution volume.
Our results for total DNA yield from umbilical cord blood was consistent with typical yield from peripheral blood which is at least 30 μg of DNA/ml of whole blood 7. We obtained mean total DNA yields of 116.5 μg for whole sample isolation (1–5 ml of whole blood) and 14.0 μg for partial sample isolation (0.2 mL of whole blood). Additionally, Qiagen® publishes average DNA yields for their QIAamp® DNA kits stating that DNA yield from whole blood is dependent on leukocyte sample concentration, which was not measured in this study. They show mini, midi, and maxi kit yields ranging from 3–12 μg (based on 0.2 mL whole blood), 3–116 μg (based on 2 mL whole blood), and 16–625 μg (based on 10 mL whole blood), respectively. We found yields in our total sample isolation buccal swab group to match Oragene® kit average yields from cotton swabs of approximately 0.8 μg of DNA per swab. Previous studies measuring DNA by flouremetry have reported variable yields of buccal DNA obtained from children, ranging from 1.7 μg to 28.3 μg8–9. Some of the variation is likely dependent on method of collection, DNA isolation, and DNA quantification. We found yields ranging from 0.4 μg to 17.5 μg with an average total yield of 4.2 μg. We are unaware of other published data using the Oragene® kit in neonates. Our mean concentration of DNA from buccal swabs of 6.9 ng/μL is below what is typically used for genotyping and DNA sequencing. For example, most single gene PCR assays require concentrations of 10 ng/μL. However newer technology platforms such as chip-based arrays require concentrations as high as 60 ng/μL. Buccal sample yields as low as ours would then require either precipitation or amplification to achieve the required DNA concentrations, further limiting the usefulness of these samples for large-scale genetic studies as the samples could be exhausted quickly.
We examined whether there was variability of DNA yield based on gestation age or days of life at the time of cord blood or buccal swab collection because WBC varies with gestational age and one would expect DNA yield to correlate with this variability. However, we found that neither measure of age was significantly associated with DNA yield in this study.
In addition to lower concentrations and total DNA yield from buccal swabs, the quality of the DNA was lower, as measured by the DNA fragmentation gel. The increased degradation may lead to more problems (e.g. lower call rates) with some of the genotyping assays.
Traditionally, genomic DNA for large scale epidemiological studies comes from peripheral blood collections 3, 10 because it provides high quality genomic DNA4. However, this method can be invasive, making it unsuitable for use with certain populations such as infants and small children. Requiring an additional needle stick to obtain blood from neonates is difficult and may reduce participation rates11. In neonates, this drawback can be mediated by collecting umbilical cord blood immediately following birth. However, this adds the additional complication of requiring collection staff to be available at all hours to collect blood whenever birthing occurs. As noted in this study, 31/96 (32%) deliveries were not able to obtain cord blood and necessitated alternative methods to collect DNA. Various methods for collecting buccal cells have been utilized recently, either as an alternative or supplement to peripheral blood drawing 4, 12–14.
Despite buccal cells being a more convenient source of DNA to collect versus umbilical cord blood, several limitations exist. These limitations include potentially limited quality and quantity of DNA obtained from buccal specimens4, 9, 13, 15, especially in children13, 16. Mouthwash rinsing is one of the most frequently used 10 alternative methods of collection for buccal cells DNA collection, and may produce DNA of higher quantity and purity than cytobrushes10, 17. However, this method is not suitable for neonates or other small children. Furthermore, DNA degradation and microbial contamination have been shown to be problematic in buccal cell collection protocols16–18. Although previous studies have compared blood and buccal samples in adults19 and buccal cell collection methods in pediatric populations4, the literatures lacks studies evaluating the difference in total yield and concentration of genomic DNA isolated from neonatal populations. Additionally, the total cost of DNA extraction (collection supplies plus laboratory reagents) is similar for umbilical cord and buccal swab collection. Thus, the ability to get the highest quality DNA for banking takes precedence.
There are several limitations of this study including that we did not control for time of day, time from feeding or intubation, week of birth from conception, or medications taken by mom and neonate. These factors may have significant effects on DNA yield from buccal cells. However, none of these factors has ever been evaluated for neonatal buccal swab collections for DNA to our knowledge. A previous study has shown that tooth brushing 1 hour before collection decreases DNA yield significantly in adults12. An added complication is that for neonates that are breast fed there is a possibility that residual breast milk, carrying maternal DNA, would be captured by swabbing. There is also a possibility that DNA present could be from maternal blood or fluid from the delivery process. Future studies will be completed to investigate the effects of these factors on buccal cell DNA. Our prior work has documented small but measurable amounts of maternal DNA in human breast milk.20 However, as most genotyping assays are qualitative, it is unlikely that the very small amount of potential maternal DNA contamination would influence the results of the assay. Other limitations include that trained research team staff collected buccal swabs, rather than a lay person, who would need to be utilized to decrease collection costs or in a study conducted by mail. This may negatively impact DNA yield and quality as proper handling has been shown to be important in minimizing contamination and optimizing DNA yield from buccal cell collections19, 21.
Additionally, we did not use quantitative real time analysis to measure dsDNA concentration, but rather flourometry, which does not discriminate well between human and bacterial DNA. Bacterial DNA has been shown to be a problem in DNA isolated from buccal cells12, 17. Although Oragene® saliva collection has been shown to increase percentage of human DNA isolated is it still limited to an average human DNA yield of 68%18. With the use of more specific quantitation methods we may find further decreased DNA yield. In future studies additional methods of quantitation will be compared, however we hypothesize that this will only further separate the total yield of human genomic DNA in umbilical cord blood samples as compared to neonate buccal swab samples. Finally, we did not measure the ability of the DNA samples to amplify for genetic testing, which is the primary purpose of DNA collection in these studies, or for whole genome amplification, which has been used to increase amount of DNA sample. However, previous studies have shown that DNA collected from both whole blood and saliva has good rates of success in downstream applications in adults 4, 12, 14, 18–19.
Our results have important implications for the conduct of large genome wide epidemiological studies. While whole genome amplification is an emerging technology that could lead to infinitely increased quantity of DNA where little was initially extracted, there are still flaws in this technology. First original DNA must be of high quality22–23. To achieve this in buccal cell collections, samples must have been stored, handled, and extracted properly 21, which may be problematic, especially if self-collection is utilized. Additionally microbial contamination and residual salivary enzymes, leading to only portions of human genomic DNA in isolated samples, and DNA degradation are still difficulties of DNA isolation from buccal cells4. These contaminants may compete with the input WGA template, particularly if template concentrations are low4. Finally, several studies have shown that amplification is not always accurate and loss of heterozygosity can occur, often in G-C rich loci24.
If researchers are looking at specific methylation patterns, the method of collection of the DNA will be important. As methylation is tissue specific, the DNA methylation patterns may be different if the DNA is obtained from umbilical cord blood (leukocyte origin) compared to buccal swabs (epithelial origin).
In conclusion, buccal swab collection from neonates is not equivalent to whole umbilical cord blood collection to isolate high yield, quality human genomic DNA for large epidemiological studies. DNA is able to be obtained from buccal swabs of neonates but the quantity and concentrations may be inadequate for large-scale genetic association studies or biorepository use. Even when only a small volume of blood is collected and extracted DNA yields are better than with a full saliva extraction using the Oragene® kit buccal swab protocols. Where umbilical cord blood collection is not possible, buccal swab collection may be an acceptable, albeit inferior, alternative to obtain neonatal DNA.
Clinical Implications.
Compared to umbilical cord blood, DNA extracted from neonatal buccal swabs is of lower concentration and quality.
When banking neonatal DNA for high-throughput genotyping and DNA sequencing studies, umbilical cord blood at the time of delivery may be the preferred sample.
Acknowledgments
Funding for this study from 5K23HD055305 and from the IUPUI Signature Center Grant PREGMED, The Indiana University Center for Pharmacogenetics and Therapeutics Research in Maternal and Child Health.
Footnotes
Reprints not available from authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cambon-Thomsen A, Rial-Sebbag E, Knoppers BM. Trends in ethical and legal frameworks for the use of human biobanks. Eur Respir J. 2007;30:373–82. doi: 10.1183/09031936.00165006. [DOI] [PubMed] [Google Scholar]
- 2.Kauffmann F the Post Genome Respiratory Epidemiology g. Post-genome respiratory epidemiology: a multidisciplinary challenge. Eur Respir J. 2004;24:471–80. doi: 10.1183/09031936.04.00076803. [DOI] [PubMed] [Google Scholar]
- 3.Holland NT, Smith MT, Eskenazi B, Bastaki M. Biological sample collection and processing for molecular epidemiological studies. Mutation Research/Reviews in Mutation Research. 2003;543:217–34. doi: 10.1016/s1383-5742(02)00090-x. [DOI] [PubMed] [Google Scholar]
- 4.Beckett SM, Laughton SJ, Pozza LD, et al. Buccal Swabs and Treated Cards: Methodological Considerations for Molecular Epidemiologic Studies Examining Pediatric Populations. Am J Epidemiol. 2008;167:1260–67. doi: 10.1093/aje/kwn012. [DOI] [PubMed] [Google Scholar]
- 5.Haque K, Pfeiffer R, Beerman M, Struewing J, Chanock S, Bergen A. Performance of high-throughput DNA quantification methods. BMC biotechnology. 2003;3:20. doi: 10.1186/1472-6750-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holland NT, Pfleger L, Berger E, Ho A, Bastaki M. Molecular epidemiology biomarkers--Sample collection and processing considerations. Toxicology and Applied Pharmacology. 2005;206:261–68. doi: 10.1016/j.taap.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 7.Lench N, Stanier P, Williamson R. SIMPLE NON-INVASIVE METHOD TO OBTAIN DNA FOR GENE ANALYSIS. The Lancet. 1988;331:1356–58. doi: 10.1016/s0140-6736(88)92178-2. [DOI] [PubMed] [Google Scholar]
- 8.Witso E, Stene LC, Paltiel L, Joner G, Ronningen KS. DNA extraction and HLA genotyping using mailed mouth brushes from children. Pediatric diabetes. 2002;3:89–94. doi: 10.1034/j.1399-5448.2002.30205.x. [DOI] [PubMed] [Google Scholar]
- 9.Zheng S, Ma X, Buffler PA, Smith MT, Wiencke JK. Whole genome amplification increases the efficiency and validity of buccal cell genotyping in pediatric populations. Cancer Epidemiol Biomarkers Prev. 2001;10:697–700. [PubMed] [Google Scholar]
- 10.Steinberg K, Beck J, Nickerson D, et al. DNA banking for epidemiologic studies: A review of current practices. Epidemiology. 2002;13:246–54. doi: 10.1097/00001648-200205000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Dlugos DJ, Scattergood TM, Ferraro TN, Berrettinni WH, Buono RJ. Recruitment rates and fear of phlebotomy in pediatric patients in a genetic study of epilepsy. Epilepsy Behav. 2005;6:444–6. doi: 10.1016/j.yebeh.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Feigelson HS, Rodriguez C, Robertson AS, et al. Determinants of DNA yield and quality from buccal cell samples collected with mouthwash. Cancer Epidemiol Biomarkers Prev. 2001;10:1005–8. [PubMed] [Google Scholar]
- 13.Saftlas AF, Waldschmidt M, Logsden-Sackett N, Triche E, Field E. Optimizing Buccal Cell DNA Yields in Mothers and Infants for Human Leukocyte Antigen Genotyping. Am J Epidemiol. 2004;160:77–84. doi: 10.1093/aje/kwh171. [DOI] [PubMed] [Google Scholar]
- 14.Satia A, King IB, Abouta JS, et al. Buccal Cell DNA Yield, Quality, and Collection Costs. Cancer Epidemiology Biomarkers & Prevention. 2002;11:1130–33. [PubMed] [Google Scholar]
- 15.Neuhaus T, Geisen G, Bolt HM, et al. Reliability of non-invasively acquired human genomic DNA as a substrate for real-time PCR-assisted analysis of genetic polymorphisms. Archives of toxicology. 2004;78:390–6. doi: 10.1007/s00204-004-0554-3. [DOI] [PubMed] [Google Scholar]
- 16.Walker AH, Najarian D, White DL, Jaffe JF, Kanetsky PA, Rebbeck TR. Collection of genomic DNA by buccal swabs for polymerase chain reaction-based biomarker assays. Environmental health perspectives. 1999;107:517–20. doi: 10.1289/ehp.99107517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Closas M, Egan KM, Abruzzo J, et al. Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. Cancer Epidemiol Biomarkers Prev. 2001;10:687–96. [PubMed] [Google Scholar]
- 18.Rylander-Rudqvist T, Hakansson N, Tybring G, Wolk A. Quality and quantity of saliva DNA obtained from the self-administrated oragene method--a pilot study on the cohort of Swedish men. Cancer Epidemiol Biomarkers Prev. 2006;15:1742–5. doi: 10.1158/1055-9965.EPI-05-0706. [DOI] [PubMed] [Google Scholar]
- 19.Feigelson HS, Rodriguez C, Welch R, et al. Successful Genome-Wide Scan in Paired Blood and Buccal Samples. Cancer Epidemiology Biomarkers & Prevention. 2007;16:1023–25. doi: 10.1158/1055-9965.EPI-06-0859. [DOI] [PubMed] [Google Scholar]
- 20.Haas DM, Daum M, Skaar T, Philips S, Miracle D, Renbarger JL. Human Breast Milk as a Source of DNA for Amplification. J Clin Pharmacol. 2010 doi: 10.1177/0091270010370847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergen AW, Qi Y, Haque KA, et al. Effects of electron-beam irradiation on whole genome amplification. Cancer Epidemiol Biomarkers Prev. 2005;14:1016–9. doi: 10.1158/1055-9965.EPI-04-0686. [DOI] [PubMed] [Google Scholar]
- 22.Bergen AW, Qi Y, Haque KA, Welch RA, Chanock SJ. Effects of DNA mass on multiple displacement whole genome amplification and genotyping performance. BMC biotechnology. 2005;5:24. doi: 10.1186/1472-6750-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun G, Kaushal R, Pal P, et al. Whole-genome amplification: relative efficiencies of the current methods. Legal medicine (Tokyo, Japan) 2005;7:279–86. doi: 10.1016/j.legalmed.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Bergen AW, Haque KA, Qi Y, et al. Comparison of yield and genotyping performance of multiple displacement amplification and OmniPlex whole genome amplified DNA generated from multiple DNA sources. Human mutation. 2005;26:262–70. doi: 10.1002/humu.20213. [DOI] [PubMed] [Google Scholar]


