Abstract
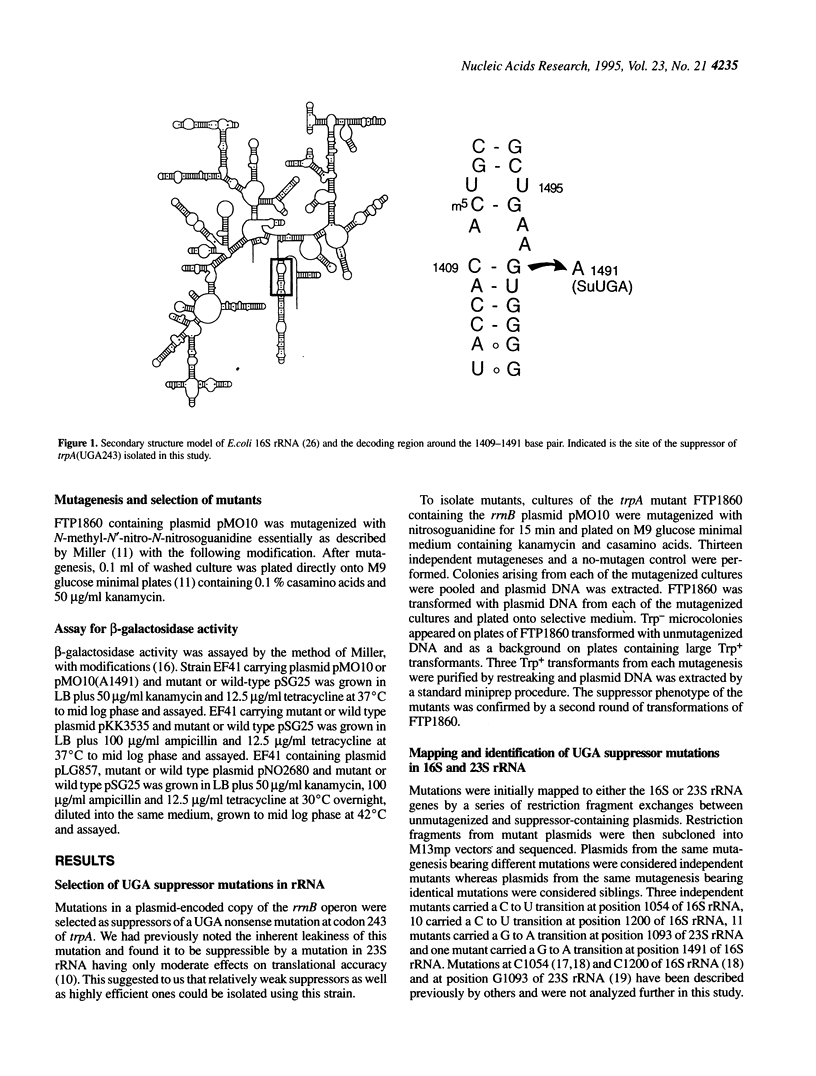
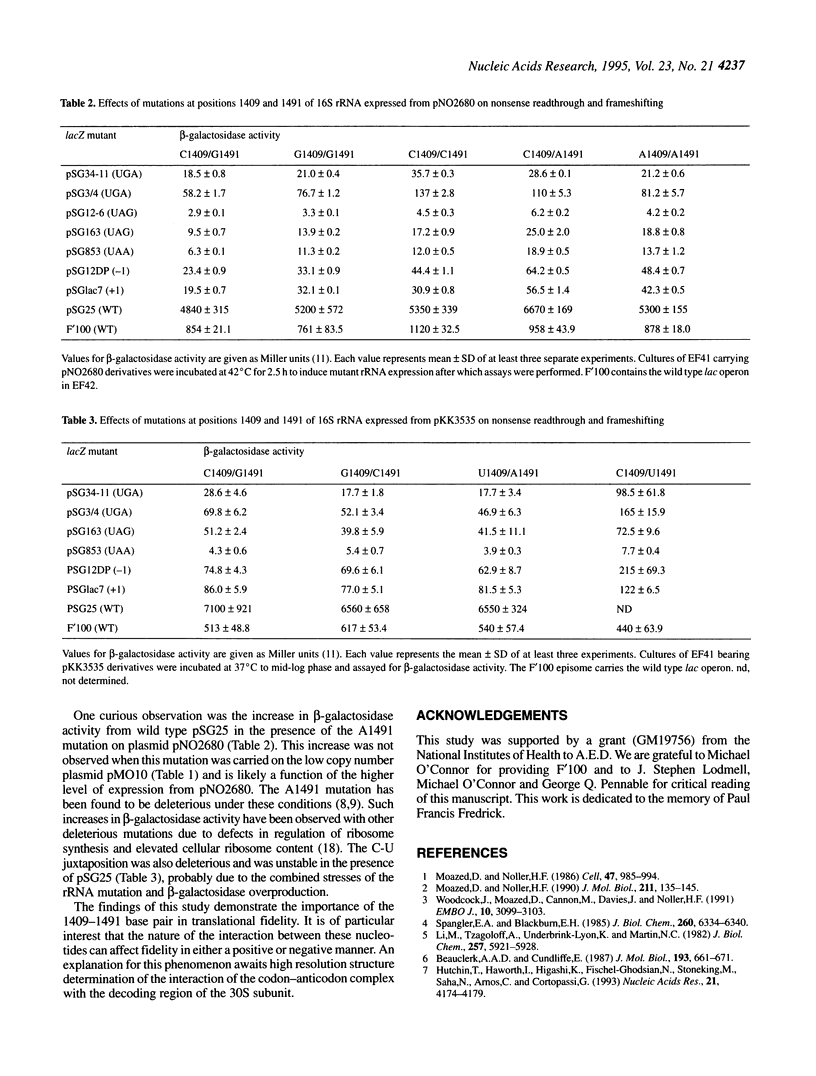
Using a genetic selection for suppressors of a UGA nonsense mutation in trpA, we have isolated a G to A transition mutation at position 1491 in the decoding region of 16S rRNA. This suppressor displayed no codon specificity, suppressing UGA, UAG and UAA nonsense mutations and +1 and -1 frameshift mutations in lacZ. Subsequent examination of a series of mutations at G1491 and its base-pairing partner C1409 revealed various effects on nonsense suppression and frameshifting. Mutations that prevented Watson-Crick base pairing between these residues were observed to increase misreading and frameshifting. However, double mutations that retained pairing potential produced an antisuppressor or hyperaccurate phenotype. Previous studies of antibiotic resistance mutations and antibiotic and tRNA footprints have placed G1491 and C1409 near the site of codon-anticodon pairing. The results of this study demonstrate that the nature of the interaction of these two residues influences the fidelity of tRNA selection.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beauclerk A. A., Cundliffe E. Sites of action of two ribosomal RNA methylases responsible for resistance to aminoglycosides. J Mol Biol. 1987 Feb 20;193(4):661–671. doi: 10.1016/0022-2836(87)90349-4. [DOI] [PubMed] [Google Scholar]
- Brosius J., Ullrich A., Raker M. A., Gray A., Dull T. J., Gutell R. R., Noller H. F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981 Jul;6(1):112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- Ciesiolka J., Gornicki P., Ofengand J. Identification of the site of cross-linking in 16S rRNA of an aromatic azide photoaffinity probe attached to the 5'-anticodon base of A site bound tRNA. Biochemistry. 1985 Aug 27;24(18):4931–4938. doi: 10.1021/bi00339a031. [DOI] [PubMed] [Google Scholar]
- De Stasio E. A., Dahlberg A. E. Effects of mutagenesis of a conserved base-paired site near the decoding region of Escherichia coli 16 S ribosomal RNA. J Mol Biol. 1990 Mar 5;212(1):127–133. doi: 10.1016/0022-2836(90)90309-A. [DOI] [PubMed] [Google Scholar]
- De Stasio E. A., Moazed D., Noller H. F., Dahlberg A. E. Mutations in 16S ribosomal RNA disrupt antibiotic--RNA interactions. EMBO J. 1989 Apr;8(4):1213–1216. doi: 10.1002/j.1460-2075.1989.tb03494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dequard M., Couderc J. L., Legrain P., Belcour L., Picard-Bennoun M. Search for ribosomal mutants in Podospora anserina: genetic analysis of mutants resistant to paromomycin. Biochem Genet. 1980 Apr;18(3-4):263–280. doi: 10.1007/BF00484241. [DOI] [PubMed] [Google Scholar]
- Gourse R. L., Takebe Y., Sharrock R. A., Nomura M. Feedback regulation of rRNA and tRNA synthesis and accumulation of free ribosomes after conditional expression of rRNA genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1069–1073. doi: 10.1073/pnas.82.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory S. T., Lieberman K. R., Dahlberg A. E. Mutations in the peptidyl transferase region of E. coli 23S rRNA affecting translational accuracy. Nucleic Acids Res. 1994 Feb 11;22(3):279–284. doi: 10.1093/nar/22.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Larsen N., Woese C. R. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 1994 Mar;58(1):10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchin T., Haworth I., Higashi K., Fischel-Ghodsian N., Stoneking M., Saha N., Arnos C., Cortopassi G. A molecular basis for human hypersensitivity to aminoglycoside antibiotics. Nucleic Acids Res. 1993 Sep 11;21(18):4174–4179. doi: 10.1093/nar/21.18.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Tzagoloff A., Underbrink-Lyon K., Martin N. C. Identification of the paromomycin-resistance mutation in the 15 S rRNA gene of yeast mitochondria. J Biol Chem. 1982 May 25;257(10):5921–5928. [PubMed] [Google Scholar]
- Martinelli S. D., Sheikh A. Hygromycin- and paromomycin-resistant mutants of Aspergillus nidulans alter translational fidelity. Curr Genet. 1991 Aug;20(3):211–218. doi: 10.1007/BF00326235. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16 S rRNA. J Mol Biol. 1990 Jan 5;211(1):135–145. doi: 10.1016/0022-2836(90)90016-F. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986 Dec 26;47(6):985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Moine H., Dahlberg A. E. Mutations in helix 34 of Escherichia coli 16 S ribosomal RNA have multiple effects on ribosome function and synthesis. J Mol Biol. 1994 Oct 28;243(3):402–412. doi: 10.1006/jmbi.1994.1668. [DOI] [PubMed] [Google Scholar]
- Murgola E. J., Hijazi K. A., Göringer H. U., Dahlberg A. E. Mutant 16S ribosomal RNA: a codon-specific translational suppressor. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4162–4165. doi: 10.1073/pnas.85.12.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M., Dahlberg A. E. Mutations at U2555, a tRNA-protected base in 23S rRNA, affect translational fidelity. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):9214–9218. doi: 10.1073/pnas.90.19.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M., Göringer H. U., Dahlberg A. E. A ribosomal ambiguity mutation in the 530 loop of E. coli 16S rRNA. Nucleic Acids Res. 1992 Aug 25;20(16):4221–4227. doi: 10.1093/nar/20.16.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke-Appel J., Jünke N., Brimacombe R., Dukudovskaya S., Dontsova O., Bogdanov A. Site-directed cross-linking of mRNA analogues to 16S ribosomal RNA; a complete scan of cross-links from all positions between '+1' and '+16' on the mRNA, downstream from the decoding site. Nucleic Acids Res. 1993 Jun 25;21(12):2853–2859. doi: 10.1093/nar/21.12.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler E. A., Blackburn E. H. The nucleotide sequence of the 17S ribosomal RNA gene of Tetrahymena thermophila and the identification of point mutations resulting in resistance to the antibiotics paromomycin and hygromycin. J Biol Chem. 1985 May 25;260(10):6334–6340. [PubMed] [Google Scholar]
- Weiss-Brummer B., Hüttenhofer A. The paromomycin resistance mutation (parr-454) in the 15 S rRNA gene of the yeast Saccharomyces cerevisiae is involved in ribosomal frameshifting. Mol Gen Genet. 1989 Jun;217(2-3):362–369. doi: 10.1007/BF02464905. [DOI] [PubMed] [Google Scholar]
- Weiss R. B., Dunn D. M., Atkins J. F., Gesteland R. F. Slippery runs, shifty stops, backward steps, and forward hops: -2, -1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Woodcock J., Moazed D., Cannon M., Davies J., Noller H. F. Interaction of antibiotics with A- and P-site-specific bases in 16S ribosomal RNA. EMBO J. 1991 Oct;10(10):3099–3103. doi: 10.1002/j.1460-2075.1991.tb07863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



