Abstract
Recent studies have shown that the transcriptional regulator PLZF controls the development of essentially all of the innate-like features of invariant NKT cells. For example, PLZF-deficient NKT cells do not acquire an “activated” phenotype nor do they acquire the capacity to secrete multiple cytokines upon primary stimulation. The function of a subset of γδ T cells has now also been shown to be dependent upon expression of PLZF. Furthermore, IL-4 produced by PLZF-expressing cells causes some CD8 T cells to acquire innate-like features. Therefore, it is becoming clear that PLZF has a broad impact on the immune response. Here we discuss the current understanding of how expression of PLZF, the innate T cell determinant, is initiated during T cell development.
Introduction
Nearly all hematopoietic cells mature in the bone marrow. In contrast, the cells destined to be T cells leave the bone marrow as multipotent progenitors and home to the thymus where signals from stromal cells are required for commitment to the T (cell) lineage [1]. Once directed into the T lineage, the cells undergo a rigorous selection process that eliminates more than 95% of the candidate T cells. Full maturation requires the expression of a T cell receptor (TCR) that binds self-peptide:MHC complexes with sufficient affinity. At some point during development, T cells are directed into one of several distinct T cell lineages such as CD4 single positive (SP) ‘helper’ cells, CD8 SP ‘killer cells’, or CD4+CD25+ regulatory (Treg) cells. Commitment to these various lineages defines the specialized functions of the cell, which is critical since each cell type plays an essential and distinct role for host defense.
The innate-like, invariant Natural Killer T (NKT) cells are an excellent example of the type of genetic programming that occurs during T cell development in the thymus. Early development of NKT cells appears to be identical to conventional CD4 and CD8 T cells [2]. However, concurrent with positive selection, NKT cells are directed into a lineage that is clearly distinct from conventional T cells in several ways. Here we discuss some elements of the transcriptional network that controls the development of this interesting subset of T cells.
PLZF: the innate T cell determinant
The BTB/POZ-ZF [Broad complex, Tramtrack, Bric à brac (BTB) or poxvirus and zinc finger (POZ)-zinc finger] protein family of transcription factors has been found to control a wide variety of biological processes [3]. BTB-zinc finger (BTB-ZF) family members are defined by the presence of an N-terminal protein-protein interaction domain (BTB/POZ) and C-terminal zinc finger domains [3]. Notably, several BTB-ZF genes have been shown to play critical roles in the development or function of some cells of the hematopoietic system [4–10]. Recent data has shown that the BTB-ZF transcription factor, PLZF (promyelocytic leukemia zinc finger), is essential for the proper development of innate T cells [11,12].
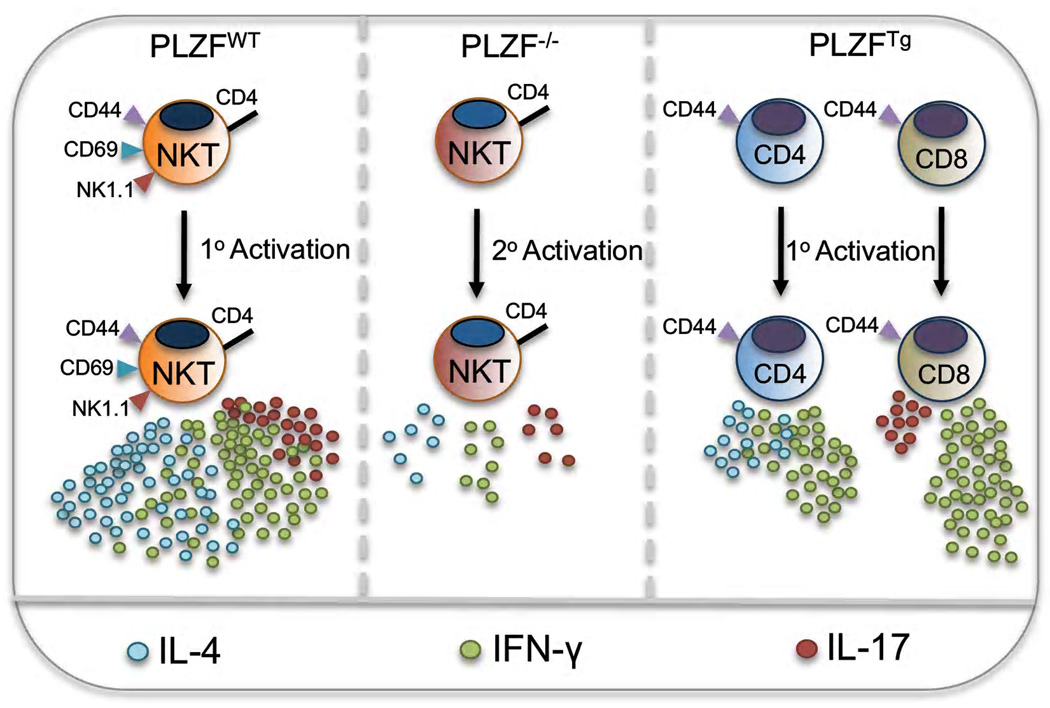
Naïve, conventional T cells must go through activation-induced differentiation, followed by secondary activation by the same or a similar antigen, prior to producing effector cytokines. Therefore, it is days, if not weeks, before there is full participation of conventional T cells in an immune response. In sharp contrast, NKT cells produce vast amounts of a multitude of effector cytokines minutes after activation. Furthermore, they can simultaneously produce both IL-4 and IFN-γ, a feature rarely found among conventional T cells [13]. NKT cells are distinct from naïve T cells in that they constitutively express the activation markers CD44 and CD69. Indeed, acquisition of these markers is a sign of functional maturity in the thymus. In addition to enhanced effector functions, NKT cells are also distinguished by their uneven tissue distribution; mature cells accumulate in the thymus and liver, but are relatively scarce in the lymph nodes. PLZF controls the development of all of these effector functions and characteristics. In the absence of PLZF, NKT cells still develop, but phenotypically and functionally are very similar to conventional naïve CD4 T cells [11,12] (FIGURE 1).
Figure 1.
Ectopic expression of PLZF in conventional T cells results in the spontaneous acquisition of memory/effector phenotypes and functions [14,15]. T cells in mice carrying a T cell-specific PLZF transgene were, like NKT cells, found to be nearly all be CD44hi and CD62Llo [14,15]. These cells also were found to produce large amounts of several cytokines upon primary activation [15]. Overall, PLZF expression appears to be necessary, sufficient and cell intrinsic for many of the salient features that characterize innate T cell function and phenotype (FIGURE 1).
Recently, several labs reported that in addition to NKT cells, a subset of mouse γδ T cells expresses high levels of PLZF [16–19]. These Vγ1+Vδ6.3+ T (γδ NKT) cells were also shown to have the innate T cell capacity to co-express IFN-γ and IL-4 [16,18]. In unpublished work, we have also found that PLZF is expressed at high levels in non-invariant, CD1d-restricted T cells as well as in non-CD1d restricted innate T cells. Finally, human NKT cells also express PLZF and, importantly, are dependent upon PLZF for their development [11], and unpublished data D. B. Sant’Angelo). Therefore, rather than use a conglomeration of multiple markers, we propose to simply define the innate T cell lineage as “PLZF-expressing T cells”.
The influence of PLZF-expressing T cells on immune system function is diverse
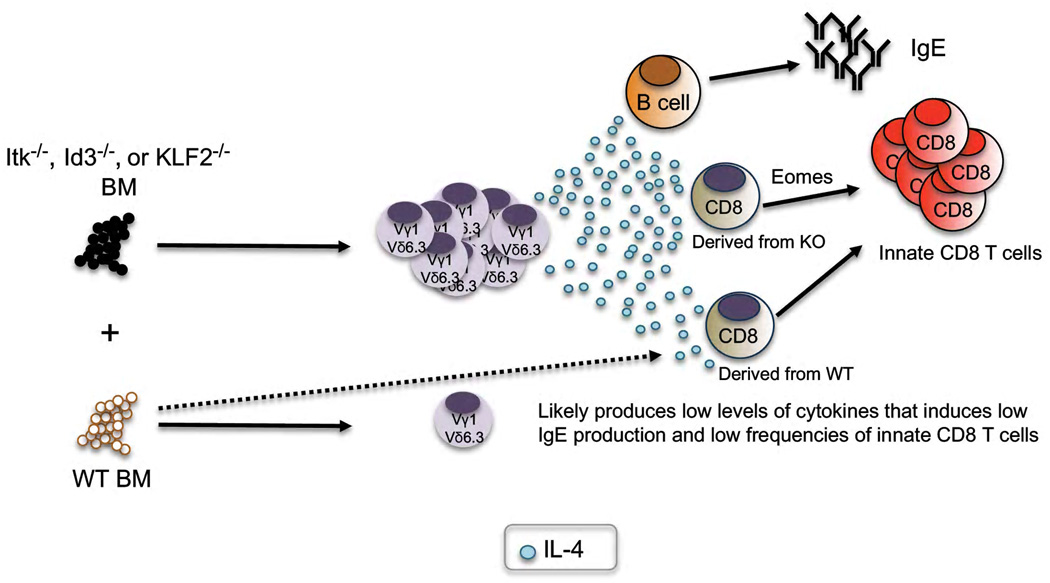
In addition to its cell intrinsic functions, PLZF has recently also been shown to affect the functions of T cells in trans. (FIGURE 2). Several genetic mutations that influence TCR signaling, such as Itk or SLP76, or targets of TCR signaling such as Id3, have been reported to cause the development of an innate-like CD8 T cell population (discussed further below). Initially, the impact of these mutations on the development of these innate CD8 T cells has been reported to be cell intrinsic [20]. For example, Itk-deficient bone marrow used to reconstitute wild-type mice resulted in the development of innate CD8 T cells [20]. Two recent studies, however, convincingly demonstrated that acquisition of the innate T cell characteristics was not intrinsic [21,22], In these experiments, wild-type mice reconstituted with a 50:50 mix of bone marrow carrying the mutation of interest along with wild-type bone marrow. Unexpectedly, the wild-type CD8 T cells also acquired the innate T cell phenotype. The culprit was found to be an expanded population of PLZF-expressing T cells (mainly γδ NKT cells) that developed from the mutant bone marrow (FIGURE 2). The PLZF-expressing cells were found to release low levels of IL-4 in the thymus, which was captured by the IL-4R expressed by some CD8 T cells. These CD8 T cells upregulate eomesodermin (eomes) expression, take on a memory phenotype and produce large amounts of IFN-γ following primary activation. Interestingly, increased Eomes expression is not only a characteristic of the phenotype, but actually appears to be required [23].
Figure 2.
An increased frequency of PLZF expressing T cells explains another unusual phenotype that has been reported (FIGURE 2). Although Itk-deficient T cells have a reduced capacity to generate Th2 responses, the mice were shown to have increased germinal center activity and increased serum IgE [24]. Recently, however, it was found that by eliminating the PLZF expressing γδ T cells from the Itk-deficient mice by crossing to TCRδ−/− mice reduced the both the serum IgE levels and the frequency of PNA+ B cells back to near wild type mouse levels [17]. Therefore, PLZF-expressing T cells can, in trans, influence the B cell compartment as well.
PLZF has also been reported to influence interferon-mediated innate immunity in mice [25]. In particular, a recent study suggested that NK cell function in PLZF-deficient mice is altered [25]. Sensitive assays conducted in our laboratory have failed to detect PLZF in the vast majority of NK cells. Therefore, it is possible that the interferon-mediated response is also a “trans” effect of PLZF-expressing cells. Finally, Id3 deficient mice have been shown to develop a Sjogren’s Syndrome-like disease autoimmune [26]. We speculate that this disease might also be a consequence of the expanded PLZF-expressing T cells; either by directly causing the disease or by their influence on other cells of the immune system.
TCR signaling in innate T cell development
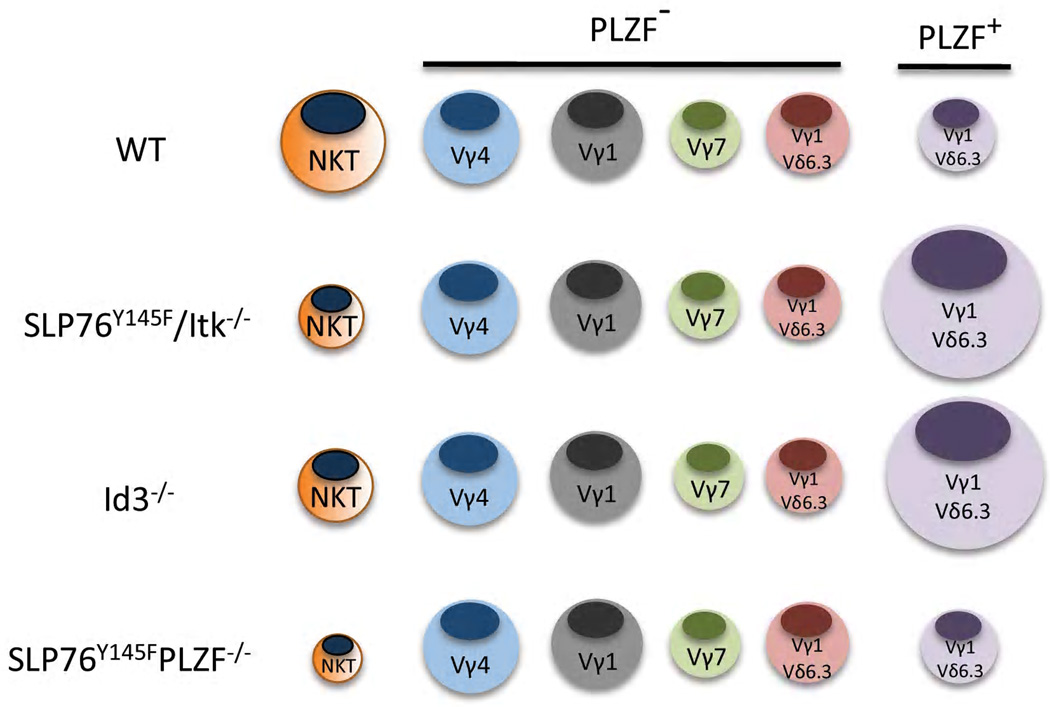
In vitro and in vivo studies support a model whereby αβ/γδ lineage commitment is a result of the relative strength of signal through the TCR. Signaling through the pre-TCR is thought to be weak and potentiates the αβ T cell lineage [27,28]. The relatively strong signaling via the γδ TCR, on the other hand, is thought to secure the γδ T cell fate by initiating a lineage-restricted gene expression program [27,28]. Recent data from several labs, however, forces a reevaluation of the data used to support this model [17–19,24,29,30]. In particular, these studies have shown that mutations in proximal TCR signaling molecules such as LAT, Itk, SLP76, or lack the transcription factor Id3 [29], a downstream target of TCR signals, resulted in a dramatic and selective expansion of PLZF-expressing T cells [17–19,24,29,30]. These cells, which were found to be a rather obscure population of γδ NKT cells, are functionally distinct from the majority of γδ T cells and all express a TCR composed of a Vγ1 and Vδ6.3 gene segments (Figure 3).
Figure 3.
The finding that there is an expansion of γδ T cells in signaling deficient mice has necessitated some revisions of the strength of signal model for γδ T cell development. Lauritsen and coworkers, for example, have suggested that the increase in γδ NKT cells in the absence of Id3 is due to failed negative selection [30]. Some γδ T cells, they suggest, normally received an exceedingly strong TCR signal during development and, as a consequence, are deleted [30]. A clear role for either positive or negative selection in γδ T cell development is, however, unsubstantiated. We, and others, however, have not observed obvious changes in the absolute numbers of other γδ T cell subsets in Id3-deficient mice [18,19]. Therefore, if negative selection were playing a key role, it would follow that it must be specific to the Vγ1Vδ6.3 γδ NKT cell subpopulation and not γδ T cells using other TCRs.
In a fairly direct test of the strength of signaling model for the expansion of γδ NKT cells, we have generated double mutant mice that are deficient for PLZF and carry the SLP76Y145F mutation (unpublished data). If the strength of TCR mediated signals were controlling the development of the γδ NKT cells, the loss of PLZF, which is induced post positive selection, would not be expected to impact the increased frequency of these cells. We found, however, that in the absence of PLZF, the frequency of γδ NKT cells reverted what is found in wild type mice. Therefore, PLZF controls the expansion, not the strength of TCR signals (unpublished data).
It would appear, therefore, that the more salient question is how strength of TCR signaling impacts the development of PLZF expressing T cells. The most obvious explanation, which is that the nature of the TCR signal directly impacts the expression of PLZF, seems unlikely for several reasons. First, the two major types of PLZF-expressing T cells respond differently to alterations in the strength of TCR signals; αβ NKTs being reduced and γδ NKT being increased [17,18,24,31]. Secondly, PLZF expression is not entirely restricted to γδ NKT cells, as nearly 20% of Vγ1 SP and Vδ6.3 SP (most likely Vγ7+Vδ6.3+) express PLZF, albeit at much lower level than found in αβ and γδ NKT cells [16,18]. Next, even within the Vγ1Vδ6.3 γδ NKT cell subpopulation, not all cells express PLZF [16,18]. In the spleen of wild type mice, PLZF is expressed in as low as ~35% to as high as ~60% of the γδ NKT cells [16,18]. There also is a complete absence of PLZF-expressing γδ T cells among intestinal intraepithelial lymphocytes, even among the Vγ1Vδ6.3 TCR population [16,18]. Additionally, sequencing efforts strongly suggest that, similar to the invariant nature of the NKT TCR, these TCRs are identical to each other; even within the potentially variable CDR3 segments [19]. Furthermore, even among Vγ1+Vδ6.3/6.4+ TCR transgenic thymocytes, PLZF only appears to be induced in ~70% of the cells [16].
Overall, we believe that these observations are inconsistent with expected results based a simple strength of TCR signaling model. In summary, we argue that these data suggest that signals generated from the TCR alone cannot induce PLZF expression. Therefore, other undefined signals likely are necessary for the development of PLZF-expressing innate T cells.
If not TCR signals, then what does control innate T cell development?
αβ NKT cells are selected on CD1d expressed on double positive (DP) thymocytes [32]. More recent data showed that αβ NKT cells also require homotypic interactions with SLAM family receptors on DP cells [33,34]. The requirement for SLAM mediated signaling was made evident by previous work that showed that the adaptor molecule SAP was essential for NKT cell development [35,36]. Previous studies also had shown that in the absence of the Src kinase Fyn, which is known to interact with SAP, also resulted in a severe block in αβ NKT cell development [37–39]. FACS analysis and qRT-PCR analysis of early stage αβ NKT cells from SAP-and Fyn-deficient mice, however, showed that PLZF expression could occur independently of these molecules [12,15]. Furthermore, in the absence of SAP or Fyn, γδ NKT cells develop and express PLZF, although PLZF levels are reduced in the absence of SAP [16,18]. As mentioned earlier, ectopic PLZF expression in conventional T cell results in a gain of NKT cell-like functionality [14,15]. Importantly, the ability of PLZF to endow cells with these properties is also independent of SAP and Fyn [15]. Altogether, although these data suggest some differential requirements for SAP and Fyn in αβ- and γδ NKT cells for development, they eliminate the possibility that this pathway is essential for PLZF expression.
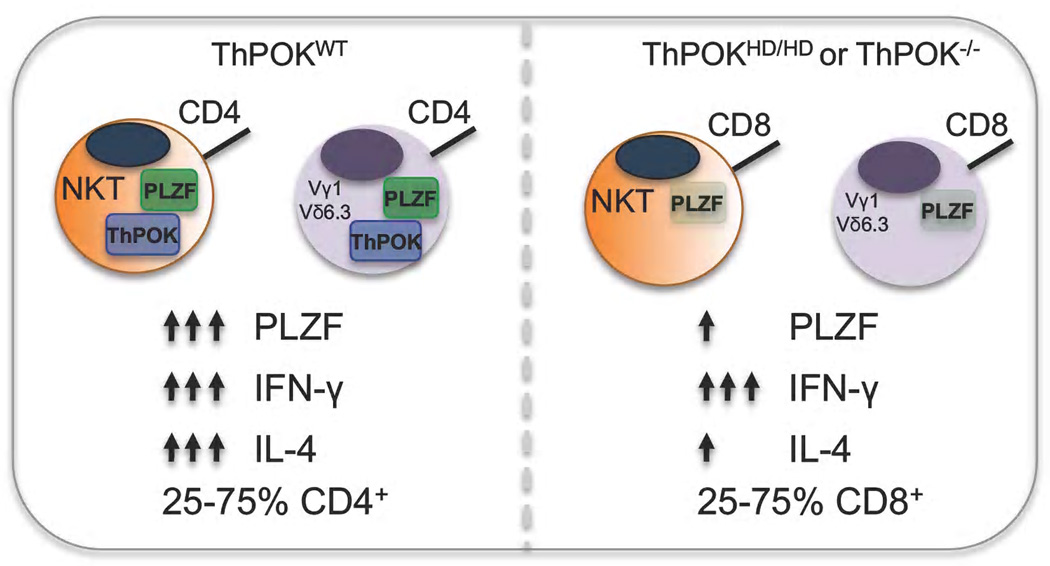
ThPOK, another BTB-ZF transcription factor, has clearly been shown to be necessary and sufficient for the differentiation of αβ CD4 T cells [5]. Recent studies have also defined a role for ThPOK in innate T cell development [18,40–42]. Surprisingly, ThPOK expression was found in all αβ NKT cells, and utilizing ThPOK reporter mice, it was shown that the transcription factor had a heterogeneous expression pattern in γδ T cell populations [18,40,41]. PLZF-expressing γδ NKT cells, however, like αβ NKT cells were found to express high levels of ThPOK; levels that were equivalent to what was found in conventional CD4 T cells [18]. Interestingly, a portion of αβ NKT cells and nearly all of the γδ NKT cells do not express CD4, clearly indicating that ThPOK does not carry out the same function in innate T cells as it does in conventional T cells [18] (Figure 4).
Figure 4.
In the absence of functional ThPOK, defects in the differentiation and function of both αβ-and γδ NKT cells were apparent [18,40]. First, in the absence of ThPOK both cell types now had a substantial percentage of cells that expressed CD8. More interestingly, both cell types also lost the capacity to produce IL-4 upon primary activation. This loss of function, however, might be a consequence of reduced PLZF expression, at least in the γδ NKT cells (Figure 4).
It is interesting that there is a dramatic increase CD4+ γδ NKT cells in TCR signaling mutant-and Id3-deficient mice [17–19,24]. While ThPOK expression is apparent in CD4-negative αβ-and γδ NKT cells, the increased frequency of CD4+ γδ NKT cells in these mice are suggestive of an increase in ThPOK activity. Notably, Park and coworkers reported a decrease in ThPOK mRNA in γδ T cells from Id3-deficient mice [41]. It is tempting to speculate that PLZF and ThPOK cooperate to control some aspects of NKT cell development. While it is clear that ThPOK has an important role in innate T cell development, experiments are needed to directly define how ThPOK impacts the development of these unique cell types.
The alternative pathway for T cell development: thymocyte-thymocyte selected T cells express PLZF
It is clear from the previous two sections that the induction of PLZF in innate T cells requires new transcriptional networks that are intertwined with potentially novel signaling pathways. Presently, there is no clear resolution to the question of how PLZF is induced. Clues to this problem should come from the interesting selection pathway required for NKT cell development that requires interactions with CD1d expressed by DP thymocytes. The interaction with DP thymocytes is obligatory; forced expression of CD1d on thymic epithelial cells does not support NKT cell development [43,44]. In contrast, the development of CD4 T cells was long believed to require unique interactions with thymic epithelial cells. Again, these interactions appeared to be obligatory since MHC class II expressing non-thymic derived epithelial cells were shown to be unable to support T cell development [45]. This concept was, at least partially, refuted by studies showing that MHC class II expression on both mouse [46,47] and human thymocytes [48] was sufficient to support the development of CD4 T cells. Therefore, in mice and humans, there is an alternative pathway for the selection of CD4 T cells.
The phenotype of T-CD4 T cells (CD4 T cells selected via thymocyte-thymocyte interactions) immediately suggested that these cells were somehow distinct from E-CD4 T cells (CD4 T cells selected via thymic epithelial cells). In particular, the T-CD4 T cells tended to have an activated phenotype (CD44hi), lower TCR levels and the capacity to produce cytokines upon primary activation [46,47]. The discovery that a subset of T-CD4 T cells were induced to express high levels of PLZF offered a least a partial explanation for the phenotype and function of these cells [48]. More importantly, these cells provided a second clear example of the need for the alternative pathway to induce PLZF expression. Intriguingly, however, only some (~30%) of the T-CD4 T cells are induced to express PLZF. We expect that the differences between the signals received by these two cell populations will be very informative.
But what about the PLZF-expressing γδ NKT cells? Do these cells also require interactions with specific immune cells to induce PLZF expression? In Figure 5, we suggest several possibilities. First, PLZF-expressing γδ T cells, like αβ NKT cells and T-CD4 T cells, might require a DP stage of development, while non-PLZF expressing γδ T cells develop directly from double negative (DN) thymocytes (Figure 5A). Although our published work shows that γδ NKT cells develop and express PLZF in TCRβ-deficient mice [18,49], these mice do still have DP thymocytes, but instead express γδ TCRs. These data would suggest that if γδ NKT cells require a DP stage of development, it likely is somewhat distinct from what is required for αβ NKT cell development. A second idea is that both PLZF-expressing and non-expressing γδ T cells arise from the DN stage, but the precursor cells are in some way distinct. Intriguingly, PLZF expression can be detected at fairly high levels in a subset of DN2a thymocytes (Figure 5B). It is possible, therefore, that expression of PLZF in a precursor cell type “sets the stage” for later development into a PLZF-expressing lineage. Since at this time we cannot discriminate between DN and DP pathways, we must also consider that both are in play and, therefore, signaling required for γδ NKT cells development can occur at either stage (Figure 5C). Overall, we believe that comparison of the developmental requirements for each of these lymphocyte subsets will be of great help in uncovering the signals that induce PLZF expression.
Figure 5.
Concluding Remarks
Overall, it is has become clear that the BTB-ZF family of transcriptional regulators is necessary for fundamental steps during immune system development and function. Our findings show that PLZF has a specific, non-redundant and essential function for the development of a complete immune system. PLZF is an extraordinarily powerful transcription factor. Data shows that the phenotype and function of any T lymphocyte that expresses or is forced to express PLZF is dramatically altered. These findings suggest that PLZF might be useful therapeutically to enhance T cell responses, for example, against tumors. On the other hand, aberrant expression of PLZF in nominally self-reactive T cells may lead to autoimmunity. There is a growing appreciation that numerically minor subsets of T cells can potently modulate immune responses. In particular, the fast and robust response of innate T cells has led to a series of clinical trials designed both to enhance immunity against cancer and to dampen immunity against self. The discovery that PLZF controls the function of these T cell responses should provide new insight into how to control and manipulate this lineage of cells.
Acknowledgements
This work was partially supported by NIH grant R01AI059739 and R01AI083988. E.S.A. is supported by The Ruth L. Kirschstein NRSA F31CA130744. We thank Drs. L. Denzin, E. J. Quann and A. Beaulieu for critical reading of the manuscript and all members of the Sant’Angelo lab for their contributions to understanding the function of PLZF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bhandoola A, von Boehmer H, Petrie HT, Zuniga-Pflucker JC. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 2007;26:678–689. doi: 10.1016/j.immuni.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Das R, Sant'Angelo DB, Nichols KE. Transcriptional control of invariant NKT cell development. Immunol Rev. 2010;238:195–215. doi: 10.1111/j.1600-065X.2010.00962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelly KF, Daniel JM. POZ for effect--POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van den Brink MR, Zelent A, Shigematsu H, Akashi K, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 6.Duy C, Yu JJ, Nahar R, Swaminathan S, Kweon SM, Polo JM, Valls E, Klemm L, Shojaee S, Cerchietti L, et al. BCL6 is critical for the development of a diverse primary B cell repertoire. J Exp Med. 2010;207:1209–1221. doi: 10.1084/jem.20091299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakaguchi S, Hombauer M, Bilic I, Naoe Y, Schebesta A, Taniuchi I, Ellmeier W. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat Immunol. 2010;11:442–448. doi: 10.1038/ni.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda T, Yoshida T, Okada S, Hatano M, Miki T, Ishibashi K, Okabe S, Koseki H, Hirosawa S, Taniguchi M, et al. Disruption of the Bcl6 gene results in an impaired germinal center formation. J Exp Med. 1997;186:439–448. doi: 10.1084/jem.186.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011.. Ref. 11 and 12 introduced PLZF to immunology by showing that the transcription factor was expressed in NKT cells and was necessary for the development of the innate T cell effector funcitons.
- 13.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 14.Raberger J, Schebesta A, Sakaguchi S, Boucheron N, Blomberg KE, Berglof A, Kolbe T, Smith CI, Rulicke T, Ellmeier W. The transcriptional regulator PLZF induces the development of CD44 high memory phenotype T cells. Proc Natl Acad Sci U S A. 2008;105:17919–17924. doi: 10.1073/pnas.0805733105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kovalovsky D, Alonzo ES, Uche OU, Eidson M, Nichols KE, Sant'Angelo DB. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. J Immunol. 2010;184:6746–6755. doi: 10.4049/jimmunol.1000776.. Here it is shown that the PLZF functions in a cell intrinsic manner that is not dependent upon signalling mediated via SAP or Fyn.
- 16. Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, Pandolfi PP, Bendelac A, von Boehmer H. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci U S A. 2009;106:12453–12458. doi: 10.1073/pnas.0903895106.. This study was among the first to show that there is a subset of γδ T cells that expresses PLZF and the transgenic expression of the Vγ1.1Vδ6.3 TCR results in an increased frequency of PLZF-expressing γδ T cells.
- 17. Felices M, Yin CC, Kosaka Y, Kang J, Berg LJ. Tec kinase Itk in gammadeltaT cells is pivotal for controlling IgE production in vivo. Proc Natl Acad Sci U S A. 2009;106:8308–8313. doi: 10.1073/pnas.0808459106.. Along with Ref. 23, this paper shows that PLZF-expressing γδ T cells are shown to cause the spontaneous generation of germinal centers and increased serum levels of IgE.
- 18. Alonzo ES, Gottschalk RA, Das J, Egawa T, Hobbs RM, Pandolfi PP, Pereira P, Nichols KE, Koretzky GA, Jordan MS, et al. Development of promyelocytic zinc finger and ThPOK-expressing innate gamma delta T cells is controlled by strength of TCR signaling and Id3. J Immunol. 2010;184:1268–1279. doi: 10.4049/jimmunol.0903218.. The development of PLZF-expressing γδ NKT cells is directly linked to functional interactions of Itk with SLP76 and the expression of Id3. Furthermore, this is the first paper to demonstrate that ThPOK is expressed in γδ T cells and that ThPOK is required for the full function of γδ NKT cells.
- 19. Verykokakis M, Boos MD, Bendelac A, Adams EJ, Pereira P, Kee BL. Inhibitor of DNA binding 3 limits development of murine slam-associated adaptor protein-dependent "innate" gammadelta T cells. PLoS One. 2010;5:e9303. doi: 10.1371/journal.pone.0009303.. Here it is shown that the TCR expressed by γδ NKT cells has very limited diveristy suggesting that these cells might be selected via a ligand in the thymus.
- 20.Broussard C, Fleischacker C, Horai R, Chetana M, Venegas AM, Sharp LL, Hedrick SM, Fowlkes BJ, Schwartzberg PL. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Verykokakis M, Boos MD, Bendelac A, Kee BL. SAP protein-dependent natural killer T-like cells regulate the development of CD8(+) T cells with innate lymphocyte characteristics. Immunity. 2010;33:203–215. doi: 10.1016/j.immuni.2010.07.013.. These two studies showed that the increased frequency of PLZF-expressing T cells that develop either in the absence of Itk or Id3 result in conventional CD8 T cells acquiring an innate-like phenotype.
- 23.Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31:122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi Q, Xia M, Hu J, Hicks E, Iyer A, Xiong N, August A. Enhanced development of CD4+ gammadelta T cells in the absence of Itk results in elevated IgE production. Blood. 2009;114:564–571. doi: 10.1182/blood-2008-12-196345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu D, Holko M, Sadler AJ, Scott B, Higashiyama S, Berkofsky-Fessler W, McConnell MJ, Pandolfi PP, Licht JD, Williams BR. Promyelocytic leukemia zinc finger protein regulates interferon-mediated innate immunity. Immunity. 2009;30:802–816. doi: 10.1016/j.immuni.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Dai M, Zhuang Y. A T cell intrinsic role of Id3 in a mouse model for primary Sjogren's syndrome. Immunity. 2004;21:551–560. doi: 10.1016/j.immuni.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 27.Hayes SM, Li L, Love PE. TCR signal strength influences alphabeta/gammadelta lineage fate. Immunity. 2005;22:583–593. doi: 10.1016/j.immuni.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Haks MC, Lefebvre JM, Lauritsen JP, Carleton M, Rhodes M, Miyazaki T, Kappes DJ, Wiest DL. Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immunity. 2005;22:595–606. doi: 10.1016/j.immuni.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Ueda-Hayakawa I, Mahlios J, Zhuang Y. Id3 restricts the developmental potential of gamma delta lineage during thymopoiesis. J Immunol. 2009;182:5306–5316. doi: 10.4049/jimmunol.0804249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lauritsen JP, Wong GW, Lee SY, Lefebvre JM, Ciofani M, Rhodes M, Kappes DJ, Zuniga-Pflucker JC, Wiest DL. Marked induction of the helix-loop-helix protein Id3 promotes the gammadelta T cell fate and renders their functional maturation Notch independent. Immunity. 2009;31:565–575. doi: 10.1016/j.immuni.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jordan MS, Smith JE, Burns JC, Austin JE, Nichols KE, Aschenbrenner AC, Koretzky GA. Complementation in trans of altered thymocyte development in mice expressing mutant forms of the adaptor molecule SLP76. Immunity. 2008;28:359–369. doi: 10.1016/j.immuni.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jordan MA, Fletcher JM, Pellicci D, Baxter AG. Slamf1, the NKT cell control gene Nkt1. J Immunol. 2007;178:1618–1627. doi: 10.4049/jimmunol.178.3.1618. [DOI] [PubMed] [Google Scholar]
- 34.Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, Mamchak AA, Terhorst C, Bendelac A. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27:751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, Tangye SG, Schwartzberg PL, Koretzky GA, Stein PL. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung B, Aoukaty A, Dutz J, Terhorst C, Tan R. Signaling lymphocytic activation molecule-associated protein controls NKT cell functions. J Immunol. 2005;174:3153–3157. doi: 10.4049/jimmunol.174.6.3153. [DOI] [PubMed] [Google Scholar]
- 37.Gadue P, Morton N, Stein PL. The Src family tyrosine kinase Fyn regulates natural killer T cell development. J Exp Med. 1999;190:1189–1196. doi: 10.1084/jem.190.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, Howie D, Sumegi J, Terhorst C, Eck MJ. SAP couples Fyn to SLAM immune receptors. Nat Cell Biol. 2003;5:155–160. doi: 10.1038/ncb920. [DOI] [PubMed] [Google Scholar]
- 39.Latour S, Roncagalli R, Chen R, Bakinowski M, Shi X, Schwartzberg PL, Davidson D, Veillette A. Binding of SAP SH2 domain to FynT SH3 domain reveals a novel mechanism of receptor signalling in immune regulation. Nat Cell Biol. 2003;5:149–154. doi: 10.1038/ncb919. [DOI] [PubMed] [Google Scholar]
- 40.Engel I, Hammond K, Sullivan BA, He X, Taniuchi I, Kappes D, Kronenberg M. Co-receptor choice by V alpha14i NKT cells is driven by Th-POK expression rather than avoidance of CD8-mediated negative selection. J Exp Med. 2010;207:1015–1029. doi: 10.1084/jem.20090557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park K, He X, Lee HO, Hua X, Li Y, Wiest D, Kappes DJ. TCR-mediated ThPOK induction promotes development of mature (CD24-) gammadelta thymocytes. EMBO J. 2010;29:2329–2341. doi: 10.1038/emboj.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Carr T, Xiong Y, Wildt KF, Zhu J, Feigenbaum L, Bendelac A, Bosselut R. The sequential activity of Gata3 and Thpok is required for the differentiation of CD1d-restricted CD4+ NKT cells. Eur J Immunol. 2010;40:2385–2390. doi: 10.1002/eji.201040534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei DG, Lee H, Park SH, Beaudoin L, Teyton L, Lehuen A, Bendelac A. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J Exp Med. 2005;202:239–248. doi: 10.1084/jem.20050413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schumann J, Pittoni P, Tonti E, Macdonald HR, Dellabona P, Casorati G. Targeted expression of human CD1d in transgenic mice reveals independent roles for thymocytes and thymic APCs in positive and negative selection of Valpha14i NKT cells. J Immunol. 2005;175:7303–7310. doi: 10.4049/jimmunol.175.11.7303. [DOI] [PubMed] [Google Scholar]
- 45.Anderson G, Partington KM, Jenkinson EJ. Differential effects of peptide diversity and stromal cell type in positive and negative selection in the thymus. J Immunol. 1998;161:6599–6603. [PubMed] [Google Scholar]
- 46.Li W, Kim MG, Gourley TS, McCarthy BP, Sant'Angelo DB, Chang CH. An alternate pathway for CD4 T cell development: thymocyte-expressed MHC class II selects a distinct T cell population. Immunity. 2005;23:375–386. doi: 10.1016/j.immuni.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Choi EY, Jung KC, Park HJ, Chung DH, Song JS, Yang SD, Simpson E, Park SH. Thymocyte-thymocyte interaction for efficient positive selection and maturation of CD4 T cells. Immunity. 2005;23:387–396. doi: 10.1016/j.immuni.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 48. Lee YJ, Jeon YK, Kang BH, Chung DH, Park CG, Shin HY, Jung KC, Park SH. Generation of PLZF+ CD4+ T cells via MHC class II-dependent thymocyte-thymocyte interaction is a physiological process in humans. J Exp Med. 2010;207:237–246. doi: 10.1084/jem.20091519.. This paper shows that CD4 T cells could be selected by MHC class II expressed on human thymocytes and that some of these cells express PLZF and have innate-like T cell phenoytpes.
- 49.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]