Abstract
Nearly 2200 genomes encoding some 6 million proteins have now been sequenced. Around 40% of these proteins are of unknown function even when function is loosely and minimally defined as “belonging to a superfamily”. In addition to in silico methods, the swelling stream of high-throughput experimental data can give valuable clues for linking these “unknowns” with precise biological roles. The goal is to develop integrative data-mining platforms that allow the scientific community at large to access and utilize this rich source of experimental knowledge. To this end, we review recent advances in generating whole-genome experimental datasets, where this data can be accessed, and how it can be used to drive prediction of gene function.
What is “function” in the post-genomic era?
With the avalanche of genome sequence data and automated transfer of annotations between those genomes, the definition of function has become increasingly vague. Traditionally, for most biochemists or geneticists, the definition of gene function has been very strict: the corresponding protein has an experimentally defined role with both a molecular and a biological dimension. For an enzyme, for instance, the molecular dimension is fulfilled by discovering the reaction it catalyzes. The biological dimension is fulfilled when the pathway in which the enzyme participates is discovered. Until the molecular role and biological process are both fully understood, the function remains “unknown”.
Traditionally, gene function discovery and/or verification have largely been achieved in this manner, one gene at a time by bench scientists. As of 2008, 59.3% of the genes found in the Escherichia coli genome are affiliated with some type of experimental data [3]. E. coli, however, is the exception rather than the rule, because experimentally characterizing the millions of genes sequenced is so far an impossible task. As a consequence, the vast majority of annotations are bioinformatic-derived predictions. A few of these annotations are based on a combination of bioinformatic evidence, such as metabolic reconstruction, clustering, co-occurrence, or the presence of candidate transcription factor binding sites [4]; but, in most cases, annotations are based solely on sequence similarity to a gene, most likely also annotated in the same way. In addition, it is often difficult to find the experimentally validated progenitor gene and how the annotation was originally acquired [5].
Unfortunately, current bioinformatic-based approaches cannot predict a function for one-third of sequenced genes; moreover, for some gene families at least 60% of the gene predictions are wrong [6]. This difficulty will inevitably become more apparent as newly sequenced genomes emerge containing genes of an ever-increasing phylogenetic distance from those experimentally characterized. Attempts are being made continually to address the need for reliable and accurate gene annotations, such as developing Gold Standard datasets of experimentally verified annotations by the COMputational BRidge to EXperiments (COMBREX: www.combrex.org); nevertheless, the functional annotation dilemma is one of the largest challenges we face in the post-genomic era and threatens to undermine efforts to extract knowledge from genome sequencing efforts.
In order to improve gene annotations, systematic functional verification efforts must be undertaken in combination with the development of better bioinformatic annotation tools. Discovery and verification of gene function have far-reaching impacts as we try to understand every aspect of the cell and discover new ways in which nature provides solutions to emerging issues in our society, including drug discovery and biofuel production. In this review, we focus on recent efforts to directly address gene function discovery in microorganisms through genome-wide high-throughput (HTP) techniques and the benefits and caveats to using these data for gene function prediction and verification (Figure 1). As a general definition, genome-wide experiments include at least 85% of the predicted genes in a given genome [7]. Structural proteomics efforts to link gene with function have been recently reviewed [8] and, as such, will not be discussed here. Instead of an exhaustive review on each type of genome-wide HTP study, we emphasize the aspects of these approaches that can be mined by experimentalists to gain insight into gene function.
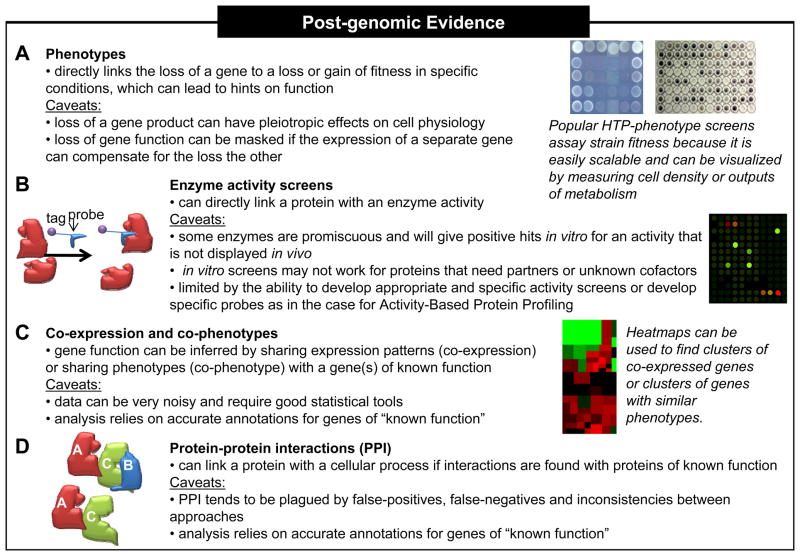
Figure 1.
Post-genomic experiments can give insight into gene function. Owing to genome-wide HTP studies, an ever-increasing number of genes are associated from experimentally derived information, such as (a,c) mutant phenotypes, (b) enzymatic activity, (c) gene expression, and (d) protein-protein interactions (PPI). These experiments can provide different and complementary “clues” about the function of a gene or protein; however, these clues are often reliant on accurate annotation of associated genes (see Caveats ). In a worst case scenario, the gene is found to associate with misannotated genes, which can lead to erroneous predictions and misdirected experiments.
Genome-wide phenotype screens
Initial insight into gene function often comes from the discovery of a growth phenotype as a consequence of gene deletion from the chromosome (Figure 1a). Common phenotypic screens look at the effect of the gene deletion on nutritional requirements or on sensitivity or resistance to a chemical agent. In the past ten years, we have seen an influx of experiments aimed at scaling-up phenotype screens and new versions of traditional designs. Screens can involve one gene deletion and thousands of growth conditions [9] or, conversely, thousands of mutants and one or more growth conditions. Whole-genome gain-of-function phenotypic screens as a result of overexpression of a gene in trans can also lead to functional insight [10].
A logical step after obtaining whole-genome sequences is to create strain collections that contain single gene deletions or disruptions (due to transposon insertion) of all or most predicted open reading frames. The first of these studies was in Mycoplasma genitalium shortly after the genome was sequenced [11]. The initial information gained from this effort is an estimate of the essential gene repertoire of that organism under the growth conditions used. By the very definition of “essentiality”, deletion or disruption of essential genes is not feasible.
Once available, experiments characterizing the collection of mutants can be implemented systematically. These genome-wide projects have gained interest rapidly owing to increased feasibility and robustness (Table 1). The time between finishing a genome sequence, building a mutant strain collection, and screening the collection with an array of growth conditions has progressively decreased as new technologies become available and the costs associated with them diminish. This year, a system to knock-down and -up all genes in E. coli in one week, and at $1 USD per gene, has been described [12].
Table 1.
Genome-wide studies
| (a) Bacterial knock-out collections and initial genome-wide phenotype screens | ||
|---|---|---|
| Organism | Screen mutants for: | Refs. |
| Acinetobacter baylyi ADP1 | Carbon source utilization and colony size | [68] |
| Francisella novicida | Host colonization | [69, 70] |
| Francisella tularensis | Host colonization | [71] |
| Mesorhizobium loti | Nodulation deficiency | [72] |
| Pseudomonas aeruginosa PA14 | Abiotic surface attachment | [73] |
| Salmonella enteric serovar Typhi | Defects in growth under standard growth conditions and presence/absence of ox bile | [74] |
| Streptococcus pneumoniae | Fitness under standard growth conditions | [23] |
| Vibrio cholerae | Motility | [75] |
| (b) Genome-wide PPI studies in microbes | ||
| Organism | Strategy | Refs. |
| Escherichia coli | His-tagged bait/prey pull-down | [51] |
| Campylobacter jejuni | HTP yeast two-hybrid (Y2H) screens | [76] |
| Mezorizobium loti | HTP Y2H screens | [77] |
| Saccharomyces cerevisiae | Tandem affinity purification | [78] |
| Synechocystis sp. PCC6803 | HTP Y2H screens | [79] |
| Treponema pallidum | HTP Y2H screens | [80] |
Phenotype screens have advanced from community-based efforts to delete and analyze the function of all uncharacterized genes in a single genome [13] to pooled experiments of barcoded mutant collections that are analyzed with deep sequencing technologies [14]. Screens are not limited to defects in cell proliferation and can be easily applied to myriad experimental designs, such as measuring amino acid levels [15], ATP synthesis [16] or protein localization via HTP microscopy [17].
Model organisms, such as Saccharomyces cerevisiae and E. coli, are leaders in the number of genome-wide phenotype screens owing to the availability of trusted and complete mutant collections. The Saccharomyces Genome Database (SGD; Table 2) maintains an updated list of genome-wide analysis papers, which currently includes 353 journal articles under the heading “large-scale phenotype analysis”. Since 2006, over 20 genome-wide screens have been published that employ the Keio E. coli mutant collection (Keio collection page, EcoTopics: http://www.ecogene.org/topic.php?topic_id=125). In addition to S. cerevisiae and E. coli, genome-wide phenotype screens are feasible for any organism in which gene disruptions can be easily constructed. In recent years, gene knock-out libraries and subsequent genome-wide screens have been performed in a wide range of bacteria (Table 1a). Importantly, to our knowledge, genome-wide mutant collections and phenotype screens are not yet available for any archaeon a – notable deficiency.
Table 2.
Publicly accessible databases that disseminate information from microbial HTP experiments
| Single “-omic” | ||
|---|---|---|
| Dataset type | Database | Website |
| Gene expression | ||
| GEO | http://www.ncbi.nlm.nih.gov/geo/ | |
| Arrayexpress | http://www.ebi.ac.uk/arrayexpress | |
| Stanford Microarray | http://genome-www5.stanford.edu | |
| Database (SMD) Comprehensive Systems-Biology Database (CSB.DB) | http://csbdb.mpimp-golm.mpg.de/ | |
| Enzyme activity | ||
| Structural Proteomics in Torontoa | http://www.utoronto.ca/AlEdwardsLab/eg_list_of_enzymes.html | |
| PPI | ||
| Biological General Repository for Interaction Datasets (BioGRID)b | http://thebiogrid.org/ | |
| IntAct | http://www.ebi.ac.uk/intact/main.xhtml | |
| Agile Protein Interaction DataAnalyzer (APID)c | http://bioinfow.dep.usal.es/apid/index.htm | |
| Molecular INTeraction(MINT) | http://mint.bio.uniroma2.it/mint/Welcome.do | |
| DNA-binding | ||
| Universal PBM Resource for Oligonucleotide Binding Evaluation (UniPROBE) | http://the_brain.bwh.harvard.edu/uniprobe/ | |
| Gene essentiality | ||
| Database of Essential Genes (DEG) | http://www.essentialgene.org/ | |
| Organism-centric that integrate HTP data | ||
| Organism | Database | Website |
| S. cerevisiae | SGD | http://www.yeastgenome.org/ |
| E. coli | eNet | http://ecoli.med.utoronto.ca/ |
| EchoBase | http://www.york.ac.uk/res/thomas/ | |
| EcoliHubd | http://ecolihub.org/ | |
| Ecoliwiki | http://ecoliwiki.net/colipedia/index.php/Welcome_to_EcoliWiki | |
| E. coli Interaction Database (EcID) | http://ecid.bioinfo.cnio.es/ | |
| Ecogene | http://www.ecogene.org/ | |
| Bacillus subtilis | B. subtilis Open Reading Frames (BSORF) | http://bacillus.genome.jp/ |
| Cyanobacteria | CyanoBase | http://genome.kazusa.or.jp/cyanobase |
| Plasmodium falciparum | PlasmoDraft | http://www.lirmm.fr/~dufayard/plasmo_draft_beta/ |
| Neisseria meningitidis | NeMeSys | http://www.genoscope.cns.fr/agc/microscope/expdata/nemesys.php |
| Multi-organism or integration of datasets | ||
| Database | Website | |
| Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) | http://dag.embl-heidelberg.de/newstring_cgi/show_input_page.pl | |
| PAThosystems Resource Integration Center (PATRIC) | http://wwww.patricbrc.org/portal/portal/patric/Home | |
| iProClass | http://pir.georgetown.edu/iproclass/ | |
| The SEED | http://theseed.uchicago.edu/FIG/ | |
Not a database, but does list results of HTP-enzyme screens.
Also contains data from genetic interaction datasets
PPI datasets from BIND (Biomolecular INteraction Network Database), BioGRID, DIP (Depository of Interacting Protiens), HPRD (Human Protein Resource Database), IntAct and MINT are included.
Searches 13 different web resources.
Automation of phenotype screens
The ability to automate phenotypic screens has greatly accelerated the field in recent years. In 2001, Biolog Inc. introduced the Phenotype Microarray (PM), which can be used to compare the growth of a mutant and its isogenic parent in nearly 2,000 growth conditions for roughly $1,200 per strain (reviewed in [18]). Growth is assayed by measuring cellular respiration, which reduces a tetrazolium dye, giving a color change [19]. PM is often a convenient assay to check the validity of systems-level metabolic reconstructions, which has identified discrepancies in annotations and has enabled the discovery of metabolic pathways not represented by current annotations. For instance, using PM to assay substrate utilization by Bacillus subtilis has increased the number of known reactions by 75 [20]. A gap-filling process is used to predict the reactions that can reconcile discrepancies between a metabolic model and PM data. Then predictions can be computationally generated to identify protein candidates responsible for those reactions. Strains carrying deletions in those candidates are then tested for the expected growth defect. For E. coli K-12 MG1655, this approach has led to the functional assignment of eight genes [21].
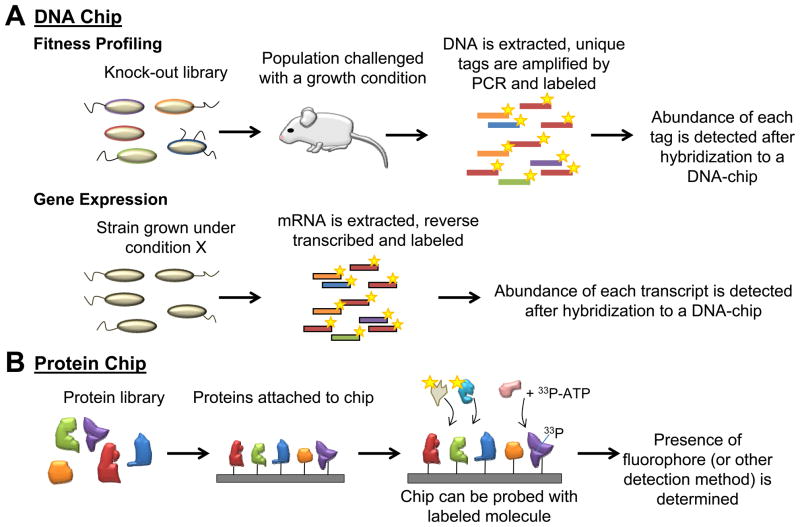
Quantitative fitness profiling of pooled mutants
Fitness profiling, where DNA microarrays or deep sequencing is used to detect mutants in pooled experiments, is a solution to any limitation associated with screening individual mutant strains. If a gene bestows a fitness advantage under a defined growth condition, then its loss will lead to a growth deficit that can be assayed by quantitatively measuring its relative abundance in a population. The yeast deletion collection contains barcodes specific to each allele [22]; therefore, deletion strains can be pooled into a single culture and the abundance of each mutant can be assayed with a DNA microarray before and after an experiment (Figure 2a). Systematic barcodes have also been developed for use in bacteria [21]. These unique tags can then be detected by microarray, similar to the yeast barcode method [19]. In addition to DNA microarrays, deep-sequencing has also been used recently to detect relative abundance of mutants [23]. In this case, the PCR-amplified sequence flanking a transposon or selection cassette can be used for strain detection [24, 25]. As the cost of sequencing decreases, these techniques become increasingly affordable, approaching a few thousand dollars per experiments. As with classical phenotype screens, fitness changes owing to gene loss or over-expression can be assayed. Recently, genome-wide quantitative fitness profiling has been applied to looking at ethanol tolerance [26] and antibiotic susceptibility [27, 28] in E. coli.
Figure 2.
Microarray technology has enabled HTP studies that can be aimed at the detection of genome-wide interactions. Simultaneous detection of thousands of knock-out strains, gene transcripts, or protein interactions can be performed. Two main types of microarray chips are shown: (a) DNA and (b) protein. (a) DNA oligonucleotides fixed to the chip can hybridize with labelled DNA generated after growth of cells under specific conditions. (b) Proteins fixed to the chip can interact with a labelled molecule (e.g. metabolite, protein, lipid). Targets of protein kinases can also be detected by kinase-directed 33P-labelling and subsequent radioactivity detection.
The pooling approach has especially been useful for analyzing pathogen–host interactions, where mutant abundance is assayed before and after infection [29–33]. Infecting a host with a pooled library of mutants is more feasible than infecting thousands of hosts with single mutants. Mutants that are required for replication and survival in the host will be underrepresented in the output pool. Since those mutants are of the greatest interest, this approach is referred to as a negative selection screen. Some 27% of Francisella novicida [29] and 53% of Salmonella enterica serovar typhimurium [33] genes identified in negative selection screens were of previously unknown function.
The number of genome-wide mutant collections pales in comparison to the number of genome sequences available, and these mutant collections do not cover all uncharacterized gene families. Also, although helpful, a phenotype alone does not necessarily indicate gene function, because gene deletions can lead to pleiotropic phenotypes that are difficult to interpret. For example, the pleiotropic phenotypes of mutants in the universal sua5/yrdC family could be to the result of a defect in tRNA modification – a function that phenotypic screens had failed to detect [34]. Another drawback in using knock-out libraries to discover gene function is frequent redundancies in gene function. A single gene deletion might not cause an observable phenotype because a second gene can composite for its loss. Metabolic reconstructions can be helpful in pinpointing these cases [35], and genetic interaction studies can help reconcile discrepancies.
Genome-wide genetic interactions
As genetic-interaction experiments are traditionally performed in yeast, it is no surprise that automation of strain construction was first applied to S. cerevisiae with the development of the SGA (Synthetic Genetic Array) analysis [36]. In these experiments, automation enables array-based HTP genetic interaction assays to be performed by systematically combining gene deletions in the same background. Recently, genome-scale genetic interaction screens have also been performed for E. coli [37, 38]; the initial use of the eSGA (E. coli Synthetic Genetic Array) revealed novel genes in iron-sulfur ([Fe-S]) cluster biosynthesis [37], and the initial use of GIANT-coli (Genetic Interaction Analysis Technology) identified proteins of unknown function that might be involved in outer membrane stabilization [38].
Proteomic analyses
To supplement or replace phenotype screens of mutant collections, several groups have focused on HTP screens of enzyme activity (Figure 1b; Box 1). One such genome-wide approach is Activity-Based Protein Profiling (ABPP). ABPP is the application of small molecule probes that are designed to target active sites and label specific classes of proteins [39]. The probe is usually designed so that the labelled protein can be visualized or purified. When combined with mass spectrometry, these proteins can then be identified. The bottleneck in ABPP is the production and testing of probes and ensuring specificity of the probe. β-Lactam probes have been used to identify enzymes involved in the resistance to β-lactam antibiotics of methicillin-resistant Staphylococcus aureus, several of which were previously of unknown function [40].
Box 1. Experimental tools that can be scaled to run genome-wide screens.
Techniques are available to provide information on a large set of genes in a single experiment. The information collected includes measurement of mutant fitness, identification of synthetic lethal gene pairs, co-expression, and protein-protein interactions. By providing more experimental clues, other methods that will prove useful in solving the gene annotation problem are HTP-enzyme assays and metabolite profiling. Currently, scalability of these two techniques remains a major limitation.
Metabolic profiling
Phenotypes. A novel phenotype screen that is now possible because of advances in metabolite extraction and mass spectrometry involves comparing metabolite profiles of a mutant strain and its parent strain [61, 62]. Although not yet used to screen entire knock-out libraries, metabolite profiling of these libraries might allow detection of otherwise-undetectable perturbations of cellular processes.
Metabolite–protein interactions. Instead of tagging a small molecule and identifying the protein that is co-purified (as in ABPP), methods are being developed to tag proteins and then determine the identity of the small molecules that are co-purified [63]. Currently, sensitivity is a limitation, and detection methods are limited to a subset of molecules.
Enzyme activity. Another technique that relies on detection of metabolites is the recently described derivation of metabolic profiling where a protein of unknown function is purified and incubated with a mix of cofactors and metabolites [64]. The substrates and/or products of the enzyme can be deduced from determining the metabolites present after incubation. This technique has directly led to the characterization of three previously hypothetical genes in E. coli [64, 65].
HTP enzyme assays
HTP in vitro screens can be used to probe the activity of purified proteins. This is especially useful for probing the function of genes that are either not associated with a detectable phenotype or are essential. Functional proteomics efforts have focused on developing robust general enzyme assays that connect an unknown protein with a functional sub-class, such as “hydrolase” [66]. In most cases, enzyme assays are limited by the need to analyze purified enzymes and are therefore mainly applied one protein at a time. Exhaustive genome-wide screens might become feasible with ongoing efforts to optimize production of active proteins [67] as well as development of a greater variety of robust enzyme assays.
Microarray technology is also used for functional proteomics studies and has enabled several assays to be scaled up to the genome-wide level (Figure 2b). Functional protein microarrays combined with expression libraries are used to assay enzyme activity, substrate binding, or protein-protein interactions in a HTP genome-wide manner. For example, proteome chips have been used to identify glycoproteins in yeast [41]; DNA damage recognition proteins in E. coli [42]; and proteins from Streptococcus pyogenes and Steptococcus agalactiae that could bind to three human protein ligands [43].
Experimental association studies
Experimental association studies are defined as experiments aimed at detecting functional links between proteins, which can be inferred from the co-expression of genes or the detection of physical interactions between proteins (Figure 1; Figure 2). The assumption is that proteins that are co-expressed or interact with one another belong to the same pathways or have similar functions in the cell.
Co-expression
Although genome-wide phenotype or enzyme activity screens are gaining popularity, the use of oligonucleotide microarrays to survey mRNA abundance in the cell in a genome-wide manner has become routine (Figure 2A). Consequently, deposition of expression microarray datasets has increased exponentially over the past decade [44]. Owing to the length of time that these techniques have been available, the use and integration of these data are far more advanced than for other types of HTP experimental data.
Recently, transcriptomics studies have benefited from new technologies and approaches, such as HTP sequencing (e.g. RNA-seq) and meta-analysis. RNA-seq involves cDNA synthesis and subsequent sequencing to determine presence and abundance of transcripts (recently reviewed in [45]). Meta-analysis of gene expression includes two approaches. The first is co-expression meta-analysis, which is the analysis of co-expressed genes across species [44]. Confidence that two genes are involved in the same pathway or process is gained by observing that gene A and gene B are co-expressed in species 1, and the homologs of gene A and B are co-expressed in species 2. The second approach is expression meta-analysis, which is the analysis of the expression profiles for a gene family across species [44]. Confidence is gained when the members of a gene family are induced under similar conditions in several species. Analysis of the transcriptional response to hydrogen peroxide in various organisms has identified 18 families of unknown function that were induced in at least two organisms [46]. This type of analysis thereby strengthens the conclusion that these protein families are involved in the cellular response to hydrogen peroxide.
Co-phenotype analysis
Insight into gene function can also come from cluster analysis of phenotypes (Figure 1c). Hierarchical clustering has been traditionally used to analyze gene expression data, but it can also be applied to quantitative phenotype data. Instead of clustering expression values for a set of genes, phenotype profiles for a set of gene deletion strains are used. This approach has been applied to phenotype data from 51 growth conditions in S. cerevisiae [47]. From these phenotype-association data, it was determined that not only do genes with related function cluster together, but also this analysis can be employed as a tool to uncover the function of unknown genes from the known genes that are co-clustered. The authors found that the uncharacterized S. cerevisiae gene YGR122W clusters with genes in the RIM101 pathway, which is involved in sporulation regulation. It was verified that YGR122W is involved in sporulation and part of the RIM101 pathway. Phenotype clustering has also been recently used to enrich the phenotype analysis of Pseudomonas aeruginosa [48] and E. coli [27]. Efforts are being made to widen this type of analysis to the cluster analysis of gene-family phenotypes across species [49].
Protein interactions
Identifying interactions between characterized and uncharacterized proteins is another approach to inferring functional relatedness (Figure 1d). Several experimental approaches are available to analyze protein-protein interactions. Genome-wide protein-protein interactions were first detected using yeast two-hybrid assays [50], and subsequent studies have employed protein microarrays as discussed above and tag-dependent pull-downs coupled with protein sequencing to identify complexes [51]. Recently, large-scale protein-protein interaction data have been collected for several microorganisms (Table 1b).
There’s information out there: how to access it?
There is a wealth of genome-wide HTP data published and the volume will certainly increase with time as these HTP approaches become easier and cheaper. However, at this stage, potentially useful information on gene function is buried in spreadsheets and the supplemental information sections of published reports. Sifting by hand through these resources to extract data on a gene family of interest takes time and is made more difficult by inconsistent nomenclature for the same gene or gene product and by the need to assess the reliability of collated HTP data. Several efforts have therefore been made by the various “-omic” communities to standardize reporting of data [52, 53]. Here, we provide a brief review of publicly accessible databases that integrate HTP data from microbes and then present case studies that illustrate the use of mining genome-wide HTP data to drive gene function prediction and verification.
There are three main types of databases that provide gene- or protein-linked HTP-derived experimental data: single “-omic”, organism-centric and multi-organism/multiple “-omics”. The term “-omic” refers to various post-genomic fields, including transcriptomics (mRNA abundance), proteomics (protein abundance and enzyme activity), interactomics (e.g. protein-protein interactions) and phenomics (phenotypes). Databases containing metabolomic (small molecule abundance) data are not discussed here because no real HTP data analyzing mutants in a genome-scale manner are yet available (Box 1).
Single -omic databases make an effort to provide a comprehensive collection of raw datasets for a single type of data, such as gene expression or protein-protein interactions, from various resources (Table 2). In some cases, such as with GEO (Gene Expression Omnibus), which now provides protein array data in addition to gene expression [54], these databases rely on manual deposition of raw data by experimentalists.
Organism-centric databases seek to provide a comprehensive collection of experimental and computational data concerning the genes/proteins for a particular genome (Table 2). Many of these databases rely on manual curation and vary greatly in how current and exhaustive they are. Gathering information for a gene family, which can be useful if function is conserved among the members requires extracting the gene names for all homologs, and then using these to search available organism-centric databases. This is a tedious and time-consuming process, even if it is sometimes the only way to gather the maximum amount of data on a specific gene family. Multi-organism and “multi-omics” databases have been developed to start addressing this issue and they are extremely useful in making associations that lead to gene function discoveries (Table 2) [55]. However, none really provide an exhaustive list of HTP data for an entire family; either the database is not up to date with current publications or not all accessible datasets are included.
Mining HTP data to predict function
In general, there is no “magic bullet” to link gene with function. If viewed alone, most post-genomic data for a single gene are meaningless. Clues from a range of post-genomic techniques, both experimental and computational, must be combined to derive or enhance confidence in a hypothesis. Surprisingly, owing to the number and quality of post-genomic databases, mining omics data to discover gene function is far more advanced for eukaryotes than for prokaryotes [56].
However, as more HTP data and better databases become available for prokaryotes, we will surely see more success stories of integrative HTP data mining in microbes, in addition to those described here. Using evidence from microarray data, sequence similarity, gene association, and 3D structure analysis the role of the previously hypothetical E. coli genes yeiC and yeiN in pseudouridine catabolism was predicted and verified [57]. Another example is the use of sequence similarity and observations from published phenotype, expression and protein-protein interaction studies, to predict and validate a role of the the E. coli gene ygfZ in [Fe-S] cluster biosynthesis [58]. More sophisticated approaches to the integration of various datasets for gene function prediction have included applying a statistical method to synthetic lethality data, expression data, mRNA decay rates, and sequence similarity to find genes involved in spindle migration of yeast [59] .
Concluding remarks and future perspectives
With the advent of tools, such as more sophisticated cloning technologies and gene knockout collections, we have seen the influx of HTP techniques that focus on bridging the gap between gene and function. Resources for the functional annotation of genes are being produced and as these approaches become cheaper and easier to perform, data generation will no longer be the limiting factor; instead, access to these datasets and interpretation of that information will become a challenge (Box 2). Currently, the steps required to mine genome-wide data are not trivial and can be daunting [60]. Efforts are needed to ensure that this stream of information is not overlooked and that it becomes another routine tool in linking gene with function.
Box 2. Outstanding data challenges.
Data are generated at an ever-increasing rate, but potential clues for gene function prediction are often lost in the supplementary materials of published reports. To find an exhaustive set of published data on a particular gene, one must dig through databases and individual reports.
-
The major advantage of HTP genome-wide approaches is also their major downfall: these techniques generate a large volume of data, but interpretation of the data and follow-through to validate the predicted functional links between proteins is generally lacking. Mainly, these data are utilized only by the experimentalists who generated it. As similarity searches have become a routine first step towards linking gene and function, the community has an absolute need for tools and databases to truly optimize the usage of experimentally derived genome-wide datasets.
The results of HTP experimental analyses have to be easily minable by experimentalists who can use their expert knowledge to raise functional annotation to the level of precision needed to be biologically relevant. Some results from a HTP experiment may have a greater meaning to experts; these connections between observation and gene function could elude the original investigators. To easily mine this data, it will be necessary to have access to exhaustive and searchable databases that: (i)provide access to multiple data types; (ii) integrate datasets for gene families from all available organisms; (iii) are searchable through similarity searches not only accession numbers; (iv) provide co-expression and co-phenotype analyses; and (v) provide confidence scores for associations.
As of yet, no single HTP method has come remotely close to unravelling the function of all uncharacterized genes in a single genome. Integration of datasets could enhance the knowledge in these individual datasets. However, integration might be a much greater challenge than producing the data, as highlighted in several recent reviews [1, 2].
Acknowledgments
We would like to thank Graciela Lorca for helpful discussions and Andrew Hanson for insightful remarks on the manuscript. This work was supported by the U.S. Department of Energy (grant no. DE–FG02–07ER64498) and by the National Institutes of Health (grant no. R01 GM70641-01) to V. de C.-L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Palsson B, Zengler K. The challenges of integrating multi-omic data sets. Nat Chem Biol. 2010;6:787–789. doi: 10.1038/nchembio.462. [DOI] [PubMed] [Google Scholar]
- 2.Zhang W, et al. Integrating multiple 'omics' analysis for microbial biology: application and methodologies. Microbiology. 2010;156:287–301. doi: 10.1099/mic.0.034793-0. [DOI] [PubMed] [Google Scholar]
- 3.Keseler I, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poptsova M, Gogarten J. Using comparative genome analysis to identify problems in annotated microbial genomes. Microbiology. 2010;156:1909–1917. doi: 10.1099/mic.0.033811-0. [DOI] [PubMed] [Google Scholar]
- 6.Schnoes A, et al. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter A, Sabatini D. Systematic genome-wide screens of gene function. Nat Rev Genet. 2004;5:11–22. doi: 10.1038/nrg1248. [DOI] [PubMed] [Google Scholar]
- 8.Adams MA, et al. Piecing together the structure-function puzzle: experiences in structure-based functional annotation of hypothetical proteins. Proteomics. 2007;7:2920–2932. doi: 10.1002/pmic.200700099. [DOI] [PubMed] [Google Scholar]
- 9.Viti C, et al. Involvement of the oscA gene in the sulphur starvation response and in Cr(VI) resistance in Pseudomonas corrugata 28. Microbiology. 2009;155:95–105. doi: 10.1099/mic.0.021873-0. [DOI] [PubMed] [Google Scholar]
- 10.Eydallin G, et al. Genome-wide screening of genes whose enhanced expression affects glycogen accumulation in Escherichia coli. DNA Res. 2010;17:61–71. doi: 10.1093/dnares/dsp028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutchison C, et al. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- 12.Warner J, et al. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat Biotechnol. 2010;28:856–862. doi: 10.1038/nbt.1653. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi K, et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith A, et al. Quantitative phenotyping via deep barcode sequencing. Genome Res. 2009;19:1836–1842. doi: 10.1101/gr.093955.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper SJ, et al. High-throughput profiling of amino acids in strains of the Saccharomyces cerevisiae deletion collection. Genome Res. 2010;20:1288–1296. doi: 10.1101/gr.105825.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hara K, et al. Systematic genome-wide scanning for genes involved in ATP generation in Escherichia coli. Metab Eng. 2009;11:1–7. doi: 10.1016/j.ymben.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Fero M, Pogliano K. Automated quantitative live cell fluorescence microscopy. Cold Spring Harb Perspect Biol. 2010;2:a000455. doi: 10.1101/cshperspect.a000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bochner B. Global phenotypic characterization of bacteria. FEMS Microbiol Rev. 2009;33:191–205. doi: 10.1111/j.1574-6976.2008.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bochner B, et al. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 2001;11:1246–1255. doi: 10.1101/gr.186501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh Y, et al. Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J Biol Chem. 2007;282:28791–28799. doi: 10.1074/jbc.M703759200. [DOI] [PubMed] [Google Scholar]
- 21.Reed J, et al. Systems approach to refining genome annotation. Proc Natl Acad Sci U S A. 2006;103:17480–17484. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winzeler E, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 23.van Opijnen T, et al. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith L, et al. Monitoring of gene knockouts: genome-wide profiling of conditionally essential genes. Genome Biol. 2007;8:R87. doi: 10.1186/gb-2007-8-5-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh J, et al. A universal TagModule collection for parallel genetic analysis of microorganisms. Nucleic Acids Res. 2010;38:e146. doi: 10.1093/nar/gkq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodarzi H, et al. Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Mol Syst Biol. 2010;6:378. doi: 10.1038/msb.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girgis H, et al. Genetic architecture of intrinsic antibiotic susceptibility. PLoS One. 2009;4:e5629. doi: 10.1371/journal.pone.0005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamae C, et al. Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J Bacteriol. 2008;190:5981–5988. doi: 10.1128/JB.01982-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss D, et al. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci U S A. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baldwin D, et al. Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect Immun. 2007;75:1005–1016. doi: 10.1128/IAI.01176-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhuri R, et al. Comprehensive identification of Salmonella enterica serovar typhimurium genes required for infection of BALB/c mice. PLoS Pathog. 2009;5:e1000529. doi: 10.1371/journal.ppat.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kizy A, Neely M. First Streptococcus pyogenes signature-tagged mutagenesis screen identifies novel virulence determinants. Infect Immun. 2009;77:1854–1865. doi: 10.1128/IAI.01306-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawley T, et al. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2006;2:e11. doi: 10.1371/journal.ppat.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El Yacoubi B, et al. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Research. 2009;37:2894–2909. doi: 10.1093/nar/gkp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar V, Maranas C. GrowMatch: an automated method for reconciling in silico/in vivo growth predictions. PLoS Comput Biol. 2009;5:e1000308. doi: 10.1371/journal.pcbi.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong A, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 37.Butland G, et al. eSGA: E. coli synthetic genetic array analysis. Nat Methods. 2008;5:789–795. doi: 10.1038/nmeth.1239. [DOI] [PubMed] [Google Scholar]
- 38.Typas A, et al. High-throughput, quantitative analyses of genetic interactions in E. coli. Nat Methods. 2008;5:781–787. doi: 10.1038/nmeth.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cravatt B, Sorensen E. Chemical strategies for the global analysis of protein function. Curr Opin Chem Biol. 2000;4:663–668. doi: 10.1016/s1367-5931(00)00147-2. [DOI] [PubMed] [Google Scholar]
- 40.Staub I, Sieber S. Beta-lactam probes as selective chemical-proteomic tools for the identification and functional characterization of resistance associated enzymes in MRSA. J Am Chem Soc. 2009;131:6271–6276. doi: 10.1021/ja901304n. [DOI] [PubMed] [Google Scholar]
- 41.Kung L, et al. Global analysis of the glycoproteome in Saccharomyces cerevisiae reveals new roles for protein glycosylation in eukaryotes. Mol Syst Biol. 2009;5:308. doi: 10.1038/msb.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C, et al. A proteome chip approach reveals new DNA damage recognition activities in Escherichia coli. Nat Methods. 2008;5:69–74. doi: 10.1038/NMETH1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Margarit I, et al. Capturing host-pathogen interactions by protein microarrays: identification of novel streptococcal proteins binding to human fibronectin, fibrinogen, and C4BP. FASEB J. 2009;23:3100–3112. doi: 10.1096/fj.09-131458. [DOI] [PubMed] [Google Scholar]
- 44.Lu Y, et al. Cross species analysis of microarray expression data. Bioinformatics. 2009;25:1476–1483. doi: 10.1093/bioinformatics/btp247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, et al. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandenbroucke K, et al. Hydrogen peroxide-induced gene expression across kingdoms: a comparative analysis. Mol Biol Evol. 2008;25:507–516. doi: 10.1093/molbev/msm276. [DOI] [PubMed] [Google Scholar]
- 47.Brown JA, et al. Global analysis of gene function in yeast by quantitative phenotypic profiling. Mol Syst Biol. 2006;2:2006.0001. doi: 10.1038/msb4100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pommerenke C, et al. Global genotype-phenotype correlations in Pseudomonas aeruginosa. PLoS Pathog. 2010;6:e1001074. doi: 10.1371/journal.ppat.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, et al. An integrative genomic approach to uncover molecular mechanisms of prokaryotic traits. PLoS Comput Biol. 2006;2:e159. doi: 10.1371/journal.pcbi.0020159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 51.Arifuzzaman M, et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006;16:686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burgoon LD. The need for standards, not guidelines, in biological data reporting and sharing. Nat Biotechnol. 2006;24:1369–1373. doi: 10.1038/nbt1106-1369. [DOI] [PubMed] [Google Scholar]
- 53.Field D, et al. Megascience. 'Omics data sharing. Science. 2009;326:234–236. doi: 10.1126/science.1180598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrett T, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu J, et al. A comparative genomics, network-based approach to understanding virulence in Vibrio cholerae. J Bacteriol. 2009;191:6262–6272. doi: 10.1128/JB.00475-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Crécy-Lagard V, Hanson A. Finding novel metabolic genes through plant-prokaryote phylogenomics. Trends Microbiol. 2007;15:563–570. doi: 10.1016/j.tim.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 57.Preumont A, et al. Molecular identification of pseudouridine-metabolizing enzymes. J Biol Chem. 2008;283:25238–25246. doi: 10.1074/jbc.M804122200. [DOI] [PubMed] [Google Scholar]
- 58.Waller JC, et al. A role for tetrahydrofolates in the metabolism of iron-sulfur clusters in all domains of life. Proc Natl Acad Sci U S A. 2010;107:10412–10417. doi: 10.1073/pnas.0911586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schöner D, et al. Annotating novel genes by integrating synthetic lethals and genomic information. BMC Syst Biol. 2008;2:3. doi: 10.1186/1752-0509-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huttenhower C, Hofmann O. A quick guide to large-scale genomic data mining. PLoS Comput Biol. 2010;6:e1000779. doi: 10.1371/journal.pcbi.1000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishii N, et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- 62.Nakahigashi K, et al. Systematic phenome analysis of Escherichia coli multiple-knockout mutants reveals hidden reactions in central carbon metabolism. Mol Syst Biol. 2009;5:306. doi: 10.1038/msb.2009.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li X, et al. Extensive in vivo metabolite-protein interactions revealed by large-scale systematic analyses. Cell. 2010;143:639–650. doi: 10.1016/j.cell.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saito N, et al. Metabolomics approach for enzyme discovery. J Proteome Res. 2006;5:1979–1987. doi: 10.1021/pr0600576. [DOI] [PubMed] [Google Scholar]
- 65.Saito N, et al. Metabolite profiling reveals YihU as a novel hydroxybutyrate dehydrogenase for alternative succinic semialdehyde metabolism in Escherichia coli. J Biol Chem. 2009;284:16442–16451. doi: 10.1074/jbc.M109.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Proudfoot M, et al. High throughput screening of purified proteins for enzymatic activity. Methods Mol Biol. 2008;426:331–341. doi: 10.1007/978-1-60327-058-8_21. [DOI] [PubMed] [Google Scholar]
- 67.Mureev S, et al. Species-independent translational leaders facilitate cell-free expression. Nat Biotechnol. 2009;27:747–752. doi: 10.1038/nbt.1556. [DOI] [PubMed] [Google Scholar]
- 68.de Berardinis V, et al. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gallagher L, et al. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc Natl Acad Sci U S A. 2007;104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kraemer P, et al. Genome-wide screen in Francisella novicida for genes required for pulmonary and systemic infection in mice. Infect Immun. 2009;77:232–244. doi: 10.1128/IAI.00978-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su J, et al. Genome-wide identification of Francisella tularensis virulence determinants. Infect Immun. 2007;75:3089–3101. doi: 10.1128/IAI.01865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shimoda Y, et al. Construction of signature-tagged mutant library in Mesorhizobium loti as a powerful tool for functional genomics. DNA Res. 2008;15:297–308. doi: 10.1093/dnares/dsn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liberati N, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langridge G, et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 2009;19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cameron D, et al. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci U S A. 2008;105:8736–8741. doi: 10.1073/pnas.0803281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parrish J, et al. A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 2007;8:R130. doi: 10.1186/gb-2007-8-7-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shimoda Y, et al. A large scale analysis of protein-protein interactions in the nitrogen-fixing bacterium Mesorhizobium loti. DNA Res. 2008;15:13–23. doi: 10.1093/dnares/dsm028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Krogan N, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 79.Sato S, et al. A large-scale protein protein interaction analysis in Synechocystis sp. PCC6803. DNA Res. 2007;14:207–216. doi: 10.1093/dnares/dsm021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Titz B, et al. The binary protein interactome of Treponema pallidum--the syphilis spirochete. PLoS One. 2008;3:e2292. doi: 10.1371/journal.pone.0002292. [DOI] [PMC free article] [PubMed] [Google Scholar]