Abstract
Internal physiological states influence behavioral decisions. We have investigated the underlying cellular and molecular mechanisms at the first olfactory synapse for starvation modulation of food search behavior in Drosophila. We found that a local signal by short neuropeptide F (sNPF) and a global metabolic cue by insulin are integrated at specific odorant receptor neurons (ORNs) to modulate olfactory sensitivity. Results from two-photon calcium imaging show that starvation increases presynaptic activity via intraglomerular sNPF signaling. Expression of sNPF and its receptor (sNPFR1) in Or42b neurons is necessary for starvation-induced food search behavior. Presynaptic facilitation in Or42b neurons is sufficient to mimic starvation-like behavior in fed flies. Furthermore, starvation elevates the transcription level of sNPFR1 but not that of sNPF, and insulin signaling suppresses sNPFR1 expression. Thus, starvation increases expression of sNPFR1 to change the odor map, resulting in more robust food search behavior.
Keywords: Drosophila, olfaction, sNPF, gain control, feeding behavior, two-photon imaging, Insulin, PI3K, wortmannin, LY294002
INTRODUCTION
The modulation of behavior by basic physiological need is essential for animal survival. Physiological modulation is often accomplished by release of neuromodulators that alter neuronal excitability or network properties (Destexhe and Marder, 2004). In particular, appetite and satiety modulate feeding behavior in most animals through the actions of neuropeptides. In mammals, the hypothalamus, an important brain region controlling appetite (Berthoud, 2002), integrates hormonal signals such as ghrelin, insulin and leptin from the gut, pancreas and adipose tissues, respectively. Activation of neurons containing neuropeptide Y (NPY) and AgRP in the arcuate nucleus of the hypothalamus augment food intake (for review see (Barsh and Schwartz, 2002)). In insects, two independent homologs of NPY, neuropeptide F (NPF) and short neuropeptide F (sNPF) (Brown et al., 1999; Hewes and Taghert, 2001), promote feeding behavior (Lee et al., 2004; Wu et al., 2003) when broadly overexpressed in neurons. Although much is known about the central control of feeding behavior, little is known about starvation modulation of sensory representation in any animal.
For most animals in their natural environment, feeding begins with a search for the appropriate food source in which the sense of smell plays an indispensible role (Dethier, 1976). While important inroads have been made in identifying neuropeptides that regulate feeding behavior, little is understood about whether or how these hormones/neuropeptides alter olfaction and how that leads to behavioral changes. In rodents, internal state influences olfactory response in the olfactory cortex (Murakami et al., 2005). However, it is not clear whether these metabolic hormones act directly on the olfactory cortex or whether they play a modulatory role in the olfactory bulb where a variety of different neuromodulators influence neural activity.
Insulin is a global metabolic cue that promotes glucose uptake in both vertebrates and invertebrates (Rulifson et al., 2002). In addition to the regulation of blood glucose, insulin signaling is implicated in the modulation of behaviors relating to feeding, reproduction and memory (Gerozissis, 2003) and insulin injection into the hypothalamus reduces food intake in rodents (Woods et al., 1998). However, how insulin signaling fine-tunes defined neural circuits to alter behavior is not well understood. Studies of starvation modulation in the Drosophila nervous system afford an opportunity to investigate an evolutionarily conserved mechanism for energy homeostasis and establish a causal link between neuropeptide modulation and feeding behavior.
We have investigated whether starvation modulates olfactory processing that mediates food-search behavior. We report that starvation alters olfactory representation of food odor at the first olfactory synapse. The neuropeptide, sNPF, which is expressed in Drosophila olfactory receptor neurons (ORNs) (Carlsson et al., 2010; Nassel et al., 2008), mediates this change by facilitating synaptic transmission from select ORNs. Intraglomerular signaling by sNPF is necessary for starvation-dependent enhancement of odor-driven food search behavior. Furthermore, starvation increases the expression level of the sNPF receptor (sNPFR1) by a reduction of insulin signaling. Thus, neuropeptide signaling causes starvation-dependent presynaptic facilitation of sensory transmission, which optimizes olfactory representation for food finding.
RESULTS
Starvation alters olfactory representation and food search behavior
The antennal lobe is the center for early olfactory processing and is a target for many neuromodulators. Within the antennal lobe, ORNs expressing the same odorant receptor genes (Clyne et al., 1999; Vosshall et al., 1999) converge onto a single glomerulus (Vosshall et al., 2000). ORNs make synapses with many local interneurons and the cognate projection neurons (PNs) (Distler and Boeckh, 1997). Output PNs of the antennal lobe transmit olfactory information from glomeruli to higher brain centers such as the lateral horn and mushroom body (Stocker et al., 1990; Vosshall and Stocker, 2007). Although ORNs are the main drivers of PN output (Olsen et al., 2007; Root et al., 2007), interneurons have been shown to control olfactory sensitivity by presynaptic inhibition (Ignell et al., 2009; Olsen and Wilson, 2008; Root et al., 2008) and lateral excitation (Olsen et al., 2007; Root et al., 2007; Shang et al., 2007). Two neuromodulators, serotonin (Dacks et al., 2009) and tachykinin (Ignell et al., 2009), have been shown to alter antennal lobe activity. If starvation modulates antennal lobe neurons, we should observe a change in odor-evoked activity in PNs.
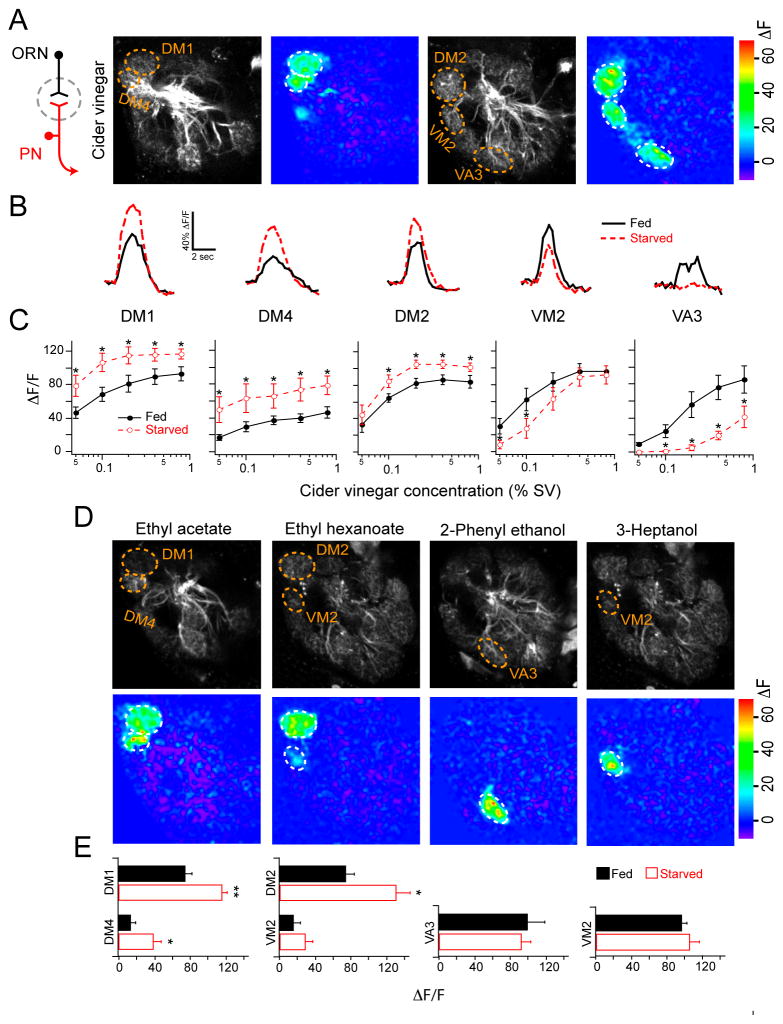
We performed two-photon imaging to measure PN dendritic calcium responses to odor stimulation in fed and starved flies. Flies bearing GH146-Gal4 and UAS-GCaMP transgenes express the calcium sensor GCaMP in many PNs allowing the select measurement of calcium response in PN dendrites (Wang et al., 2003). We first imaged responses to apple cider vinegar (Figure 1A), which is highly attractive to Drosophila and is a complex odor that resembles a natural food source (Semmelhack and Wang, 2009). Cider vinegar excites five glomeruli at the tested concentrations. Starvation significantly enhances odor response in three glomeruli (DM1, DM4 and DM2) but decreases odor response in two glomeruli (VM2 and VA3; Figure 1B, C). It is interesting to note that starvation alters the amplitude of calcium activity without changing the temporal kinetics (Figure 1B). In sharp contrast, our previous study shows that activation of GABAB receptors causes presynaptic inhibition and alters the temporal kinetics of PN calcium activity (Root et al., 2008). Therefore, a change in GABAB receptor signaling is unlikely to account for the starvation dependent change in olfactory response. Rather, our results are more consistent with an excitability change in antennal lobe neurons.
Figure 1. Olfactory representation in projection neurons is altered by starvation.
A, Two-photon imaging of PN calcium activity in response to cider vinegar stimulation on two optical planes of the antennal lobe in fed flies. Gray-scale images show antennal lobe structure while pseudocolored images reveal odor-evoked activity at 0.4% SV (saturated vapor pressure). B, Representative traces of fluorescence change over time for the five glomeruli excited by cider vinegar at 0.1% SV. C, Peak ΔF/F across a range of cider vinegar concentrations for each glomerulus. D, PN activity of fed flies in response to pure odorants. E, Peak ΔF/F for each glomerulus. D,E, Odors were applied at the following concentrations (%SV): 1% ethyl acetate 1:10,000 in mineral oil, 0.1% ethyl hexanoate 1:10,000 in mineral oil, 0.5% 2-phenyl ethanol, and 0.1% 3-heptanol. C,E, n=5–10 for each condition; error bars show SEM. *P<0.05, **P<0.01; t-test. The flies have GH146-Gal4 and UAS-GCaMP. All starvations were 17–24 hrs.
The apparent starvation-dependent change of olfactory response in the DM1, DM2, DM4, VM2 and VA3 glomeruli could be due to intra- or inter-glomerular mechanisms. We therefore investigated starvation modulation of individual glomeruli with reduced lateral activity. To do this, we imaged PN responses to a panel of four different odorants, each of which excites one or two glomeruli at low concentrations (Figure 1D, E). The responses in DM1 and DM4 to ethyl acetate were significantly enhanced by starvation, while the response of DM2 but not VM2 to ethyl hexanoate is enhanced by starvation. In contrast, the responses of VA3 and VM2 to 2-phenylethanoland and 3-heptanol, respectively, are not modulated by starvation. Therefore, DM1, DM4 and DM2 are more sensitive to odor stimulation in starved animals. However, VA3 and VM2 are not subject to starvation modulation. This result suggests that the apparent suppression of VA3 and VM2 in response to cider vinegar is due to lateral inhibition. We conclude that some antennal lobe neurons are subject to starvation modulation in a glomerular- rather than odor-specific manner, which results in an alteration of the odor map.
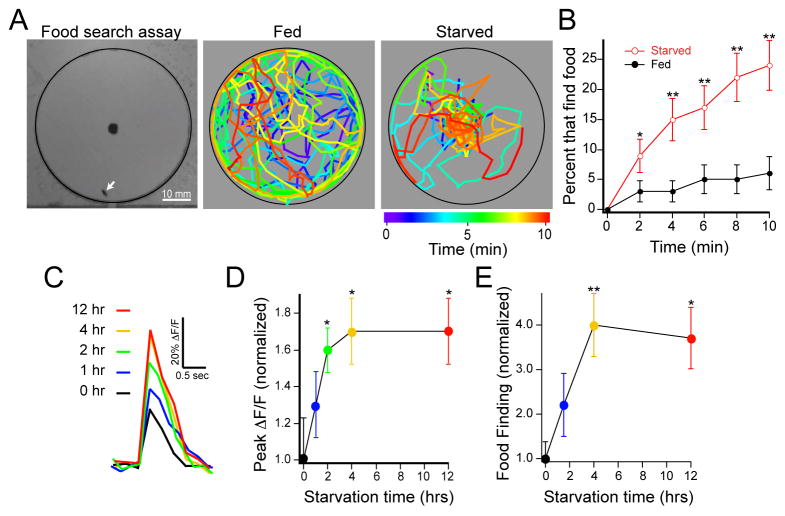
Starvation as an internal state affects feeding behavior (Gelperin, 1971), which begins with an olfaction-dependent search for an appropriate food source. Therefore, we expect that the starvation-dependent change in olfactory representation should be matched by an alteration in behavior. We developed a single fly assay that allows the assessment of starvation modulation on odor-driven food search behavior. We reasoned that latency to find food is a metric of food search. We employed an automatic computer system to monitor the position of individual flies from which we measured the latency required for individuals to reach an odor target. Individual flies were introduced into small arenas each of which contained a food odor, apple cider vinegar, at the center. During the 10 minute observation period, starved flies spend most of the time walking near the food source, whereas fed flies wander in the entire arena with a preference for the perimeter (Figure 2A). The latency of food finding is significantly decreased upon starvation (Figure 2B) and is independent of fly speed (Figure S1). Furthermore, surgical removal of the antennae impairs this behavior (Figure S1). Thus, the sense of smell, mediated by the antennae, is required for food search behavior, and starvation enhances food finding in Drosophila.
Figure 2. Optimum food search behavior and peak olfactory sensitivity are reached within four hours of starvation.
A, A food search assay was used to measure the latency of odor-guided food finding. Grayscale image (left) shows an arena with a food odor, 1% cider vinegar, in the center and a single fly (white arrow). The coordinates of single flies are plotted as a function of time in pseudocolor for a representative fed fly and one starved overnight. B, The latency of food search as the cumulative percentage of flies that find the odor source over time. C,D, Two-photon imaging of PN calcium activity in the DM1 glomerulus in response to electrical stimulation of the olfactory nerve. C, Representative traces of fluorescence change over time from the DM1 glomerulus in flies with varied starvation durations. D, Peak ΔF/F normalized to the average response without starvation. Stimulation was 1 ms in duration, 10 V in amplitude and 4 pulses at 100 Hz. n=5–8 for each starvation condition. Error bars show SEM. *P≤0.05, t-test E, Data from behavioral experiments with varied starvation durations shown as the food finding percentage normalized to that of the fed state. B,E n=53–102 flies for each condition. Error bars show SEM. *P≤0.05, **P≤0.01, z-test for proportions.
What is the time-course of the starvation-dependent change in olfactory activity and food-search behavior? We first varied starvation time and measured calcium activity of PN dendrites in response to precise electrical stimulation of the olfactory nerve. Nerve stimulation decreases variability in calcium response compared to odor stimulation, and allows for finer control of starvation time because the preparation can be prepared faster than the odor preparation. Imaging calcium activity in PN dendrites of the DM1 glomerulus, we found that calcium activity increased with starvation duration up to four hours. Longer starvation duration for twelve hours did not result in more neuronal response (Figure 2C, D). We next varied starvation time and examined the latency of food search behavior. Similar to the starvation-dependent effect on calcium activity, food finding increases up to four hours and is not further increased after 12 hours of starvation (Figure 2E). Thus, the change in antennal lobe activity and food search behavior occurs within four hours of starvation.
sNPF signaling in ORNs mediates starvation modulation of food search
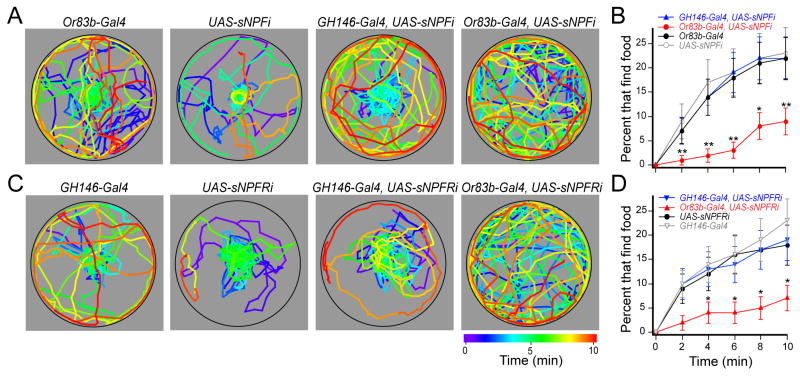
What is the mechanism by which starvation affects odor-guided behavior? The neuropeptide sNPF promotes feeding behavior (Lee et al., 2004) and is expressed in some ORNs (Carlsson et al., 2010; Nassel et al., 2008). We therefore hypothesized that sNPF signaling in ORNs is responsible for the starvation-dependent enhancement of food search behavior. We expressed RNAi to knockdown expression of sNPF (Figure S2A for knockdown verification) in ORNs of flies bearing the Or83b-Gal4 and UAS-sNPF-RNAi transgenes, and as a control we expressed sNPF-RNAi in PNs of flies bearing GH146-Gal4 and UAS-sNPF-RNAi transgenes. We measured the latency of food finding in our behavioral assay and found that indeed starved flies lacking sNPF in ORNs exhibit a significantly longer latency in food finding (Figure 3A, B). Interestingly, starved sNPF knockdown flies behave similarly to fed flies (Figure S2D), suggesting that low sNPF signaling mimics the fed state in the antennal lobe. The difference in latency between sNPF knockdown flies and control flies cannot be attributed to a change in locomotor activity (Figure S2B). Furthermore, flies with a P-element disruption of the first intron of the sNPF gene (sNPFc00448) are similarly impaired in food finding (Figure S2E), although these flies may suffer some growth defect (Lee et al., 2008). Thus, sNPF expression in olfactory receptor neurons mediates the starvation-dependent enhancement of food search behavior.
Figure 3. Starvation-dependent food search requires sNPF signaling in ORNs.
The latency of odor-guided food finding was measured in starved flies with 1% cider vinegar. A, The coordinates of single flies for representative control flies (left two plots) and those expressing sNPF-RNAi (sNPFi) in PNs (third from left) or ORNs (right). B, The latency of food finding. C, The coordinates of two representative control flies (left two plots) and those expressing sNPFR1-RNAi (sNPFRi) in PNs (third from left) or ORNs (right). D, The latency of food finding. n=64–103 flies for each condition. Error bars show SEM. *P<0.05, **P<0.01; z-test for proportions comparing the top three curves to the bottom curve in B,D.
While our findings are in accord with previous work indicating that ORNs express the sNPF peptide (Carlsson et al., 2010; Nassel et al., 2008), the population of neurons that express sNPFR1 (Feng et al., 2003), the receptor for sNPF, is not known. In salamanders, the NPY receptor localizes to sensory neurons of the olfactory epithelium (Mousley et al., 2006), and is thus poised for a feedback modulation. In the mammalian hypothalamus, NPY neurons project from the arcuate nucleus to the lateral hypothalamus (Barsh and Schwartz, 2002; Cowley et al., 1999) and are poised for a feedforward modulation. Thus, two possible mechanisms may account for the observed modulatory effects of the neuropeptide: 1) if sNPFR1 localizes to ORNs, its peptide may modulate starvation-induced behavior through ORN-ORN feedback modulation, or 2) If sNPFR1 localizes to PNs, its peptide may modulate starvation-induced behavior through ORN-PN feedforward modulation. To discriminate between these two possibilities, we expressed RNAi to knockdown sNFPR1 (Figure S2A for knockdown verification) in either the ORNs or PNs. We found that expression of sNPFR1-RNAi in ORNs mimics the effect of the neuropeptide knockdown (Figure 3C, D). In contrast, expression of sNPFR1-RNAi in the PNs has no effect on food search behavior. The difference in latency between sNPFR1 knockdown and control flies cannot be attributed to a change in locomotor activity (Figure S2C). Furthermore, disruption of sNPFR1 by expression of a dominant negative gene (Lee et al., 2008) in ORNs results in a similar decrease in food finding (Figure S2E). Thus, feedback modulation by sNPFR1 expressed in ORNs is necessary for starvation-dependent food search.
Presynaptic activity in ORNs is modulated by sNPF signaling
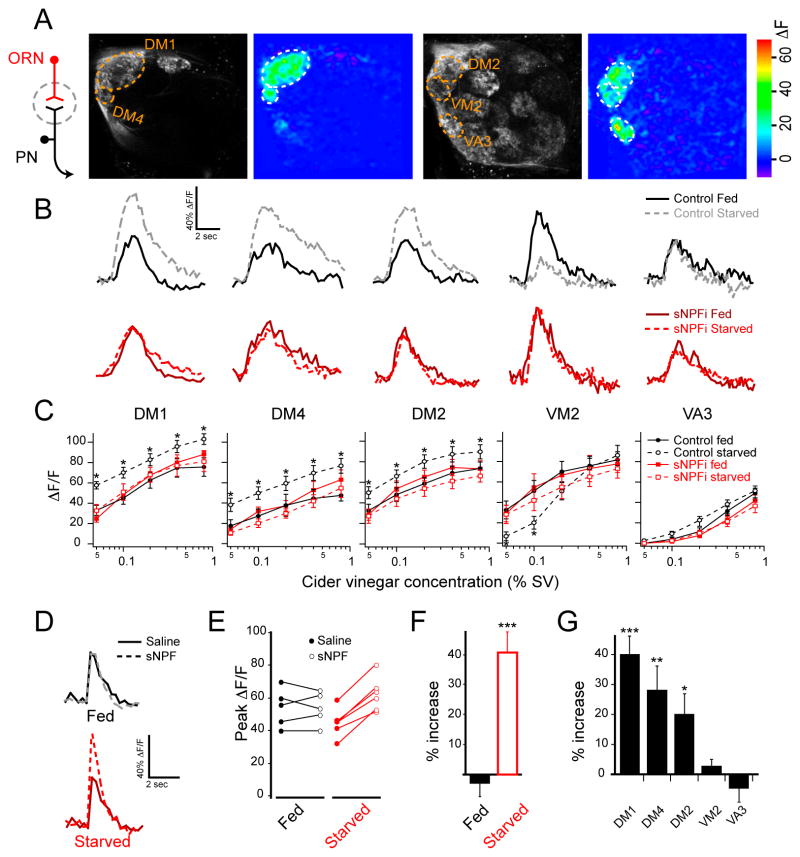
Given that knockdown of sNPF and its receptor in ORNs has a profound effect on starvation-dependent food search behavior, we reasoned that starvation should alter activity in ORN axon terminals. To investigate this, we imaged odor-evoked activity in ORNs in flies that were fed and flies that were starved overnight. Flies bearing the Or83b-Gal4 and UAS-GCaMP transgenes allow the select measurement of calcium activity in ORN axon terminals. We observed that cider vinegar activates the same five glomeruli when comparing ORNs (Figure 4A) to PNs (Figure 1A). Three glomeruli (DM1, DM4 and DM2) exhibit significant increases in calcium activity upon starvation, while the VM2 glomerulus exhibits significant suppression of response at low odor concentration, and the VA3 glomerulus is not affected (Figure 4B,C). Thus, starvation alters olfactory representation in sensory neurons, which is largely consistent with the changes observed in the antennal lobe output PNs.
Figure 4. The sNPF receptor is upregulated upon starvation and mediates presynaptic facilitation in sensory neurons.
A, Two-photon imaging of ORN axon terminal calcium activity in response to cider vinegar stimulation at 0.4% SV in fed flies. B, Representative traces of fluorescence change over time for the five glomeruli excited by 0.1% cider vinegar in control flies (top) and those expressing sNPF-RNAi in ORNs (sNPFi) (bottom). C, Peak ΔF/F across a range of cider vinegar concentrations for each glomerulus. n=10–12 each condition; error bars show SEM. *P<0.05; t-test comparing starved control to fed control. Control flies have Or83b-Gal4 and UAS-GCaMP, and sNPFi flies also have UAS-sNPF-RNAi transgenes. D–G, ORNs axon terminal calcium activity in response to electrical stimulation of the olfactory nerve before and after application of sNPF. D, Representative traces of fluorescence change over time from the DM1 glomerulus of fed and starved flies in saline and after addition of 10μM sNPF. E, Peak ΔF/F before and after sNPF. F, Percent increase in peak ΔF/F after exogenous sNPF addition in DM1. G, Percent increase in peak ΔF/F after sNPF addition in starved flies, for the five glomeruli that respond to cider vinegar. Stimulation was 1 ms in duration, 10 V in amplitude and 16 pulses at 100 Hz. n=5–6; error bars show SEM; *P<0.05, **P<0.01, ***P<0.001, t-test. The flies have Or83b-Gal4 and UAS-GCaMP, and UAS-sNPF-RNAi transgenes.
We next asked if sNPF signaling in ORNs causes the starvation-induced changes in olfactory representation. To investigate this, we imaged ORN response to cider vinegar in starved and fed flies with perturbed sNPF signaling. We found that expression of sNPF-RNAi in the ORNs eliminates the effect of starvation such that the olfactory representation in starved flies lacking sNPF mirrors that of fed control flies (Figure 4C). The overlapping curves between control fed flies and starved RNAi flies suggest that the effect of RNAi is specific to sNPF signaling rather than a potential non-specific effect on neuronal properties. Furthermore, there is no difference between starved and fed sNPF knockdown flies, indicating that sNPF mediates the starvation modulation of ORN activity. In addition, expression of RNAi to knockdown sNPFR1 in ORNs similarly eliminates the effect of starvation (Figure S3A). We further investigated whether abolishing sNPF signaling presynaptically in ORNs eliminates the starvation-dependent enhancement in postsynaptic PNs. To do this, we used flies bearing the GH146-LexA, LexAop-GCaMP, Or83b-Gal4 and UAS-sNPF-RNAi transgenes. Imaging PN calcium activity in the DM1 glomerulus in the absence of presynaptic sNPF, we found that the effect of starvation is abolished such that PN response in starved flies matches that of fed flies (Figure S3B–D). The data suggest that the effect of sNPF-RNAi is not due to a non-specific disruption of synaptic transmission from ORNs. Thus, we conclude that sNPF signaling causes the change in olfactory representation upon starvation.
sNPF signaling mediates presynaptic facilitation
The above results indicate that starvation enhances activity in ORNs by sNPF signaling, suggesting that the neuropeptide could act to facilitate presynaptic activity. To directly test this hypothesis we asked if exogenous application of sNPF affects presynaptic calcium activity in ORN terminals. In order to eliminate the contribution of any potential modulation at ORN cell bodies, we removed the antennae and delivered precise electrical stimulation to one olfactory nerve while imaging calcium activity in the ipsilateral antennal lobe. We expressed sNPF-RNAi in ORNs to eliminate endogenous sNPF, which may occlude the effect of exogenously applied sNPF. Imaging ORN axon terminals, we find that electrical stimulation of the olfactory nerve elicits a calcium transient that is increased upon sNPF application (Figure 4D–G). Interestingly, this increase occurs only in starved flies but not in fed flies, suggesting that sNPFR1 signaling is upregulated upon starvation. We compared the sensitivity to sNPF between the five glomeruli that respond to cider vinegar and found that the DM1, DM2 and DM4 glomeruli exhibit enhanced activity by the neuropeptide, whereas the VM2 and VA3 glomeruli do not (Figure 4G). This result reveals that ORNs terminating in VM2 and VA3 are not modulated by sNPF, which is consistent with the results we obtained with odor stimulation (Figure 1). Therefore, the suppression of calcium activity in VM2 ORNs (Figure 4B) could be a result of lateral presynaptic inhibition (Olsen and Wilson, 2008; Root et al., 2008). Furthermore, the suppression of VA3 PN calcium activity (Figure 1B) could be due to lateral feedforward inhibition (Sachse and Galizia, 2002). Thus, the sNPF peptide and its receptor mediate presynaptic facilitation in starved flies at select glomeruli.
sNPF signaling in DM1 is necessary and sufficient for starvation-dependent food search behavior
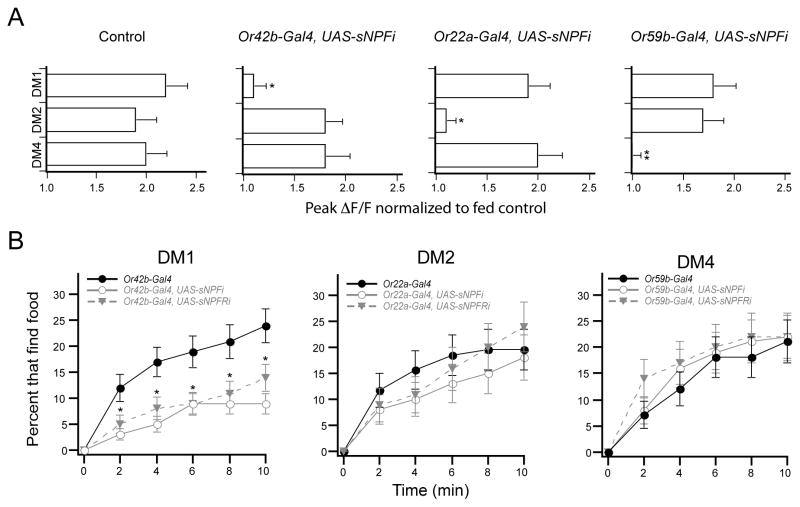
The ORNs of the DM1, DM2 and DM4 glomeruli have the ability to respond to exogenous sNPF, however the endogenous source of the neuropeptide is unclear. The peptide could come from receptor neurons of the same glomerulus or alternatively from neighboring glomeruli. We therefore investigated the inter- vs. intraglomerular source of sNPF by knocking down sNPF expression in specific ORNs and imaging ORN activity in all glomeruli. Flies bearing the Or83b-LexA, LexAop-GCaMP, UAS-sNPF-RNAi and Or-specific-Gal4 transgenes permit the measurement of calcium activity in the axonal termini of many glomeruli, while knockdown of sNPF expression is targeted to one specific glomerulus. We found that knockdown of sNPF expression in Or42b ORNs eliminates starvation modulation in only the cognate DM1 glomerulus without any impact on the ORNs of DM2 or DM4 glomeruli (Figure 5A). Similarly, knockdown of sNPF in Or22a and Or59b ORNs abolished starvation modulation in the ORNs of DM2 and DM4 glomeruli, respectively, without any impact on the other glomeruli (Figure 5A). These results suggest that intraglomerular sNPF peptide is necessary while interglomerular sNPF is not sufficient for starvation modulation of olfactory sensitivity.
Figure 5. sNPF signaling in a single glomerulus is necessary for starvation-dependent food search.
A, Two-photon imaging of ORN axon terminals in flies expressing RNAi to knockdown sNPF expression in the ORNs of individual glomeruli. Peak ΔF/F normalized to the average response from fed control flies to 0.2% SV cider vinegar. n=5–6. *P<0.05, t-test. All flies have Or83b-LexA and LexAop-GCaMP, and where indicated flies also have the Or-specific-Gal4 and UAS-sNPF-RNAi. B, The latency of food finding for starved flies expressing RNAi to knockdown sNPF or sNPFR1 in individual glomeruli. RNAi expression in only the DM1 glomerulus significantly decreases food finding. n=80–195 flies for each condition. *P<0.05, z-test for proportions comparing control to sNPFi and to sNPFRi. Error bars show SEM.
The above results indicate that intraglomerular sNPF signaling selectively increases activity in only three of the five glomeruli activated by cider vinegar. Given that a previous study has found that not all glomeruli contribute equally to odor-guided behavior (Semmelhack and Wang, 2009), we next asked if sNPF signaling in individual glomeruli is necessary for food search behavior. We expressed RNAi to knockdown the peptide or the receptor in the DM1, DM2 and DM4 ORNs, which are modulated by sNPF. We found that knockdown of the neuropeptide or its receptor in DM1 ORNs results in significantly decreased food finding in starved flies (Figure 5B). This difference cannot be attributed to a difference in locomotor activity (Figure S4A). Strikingly, knockdown of the neuropeptide or its receptor in the DM2 or DM4 ORNs has no effect on the starvation-dependent food search behavior (Figure 5B). Expression of the RNAi in the VM2 and VA3 ORNs that are not sensitive to sNPF signaling does not affect food search behavior (Figure S4B). These results indicate that sNPF signaling in a single ORN channel is necessary for the starvation-dependent food search behavior.
It has been observed that sNPF is also expressed in the mushroom body (Nassel et al., 2008), which suggests that starvation modulation in the central nervous system could be important for food search behavior. We therefore evaluated the contribution of the peripheral modulation by performing gain of function experiments in fed flies to determine if peripheral modulation alone is sufficient to induce starvation-like food search behavior. We first performed imaging experiments to determine if overexpression of sNPFR1 increases odor-evoked calcium activity. We imaged calcium activity in Or42b ORNs in control flies bearing the Or83b-LexA and LexAop-GCaMP transgenes and overexpression flies that also contained the Or42b-Gal4 and UAS-sNPFR1 transgenes. Ectopic expression of sNPFR1 significantly increases ΔF/F in the DM1 glomerulus in fed flies (Figure 6A). Furthermore, this enhanced activity is translated into a shorter latency in food finding behavior in fed flies as ectopic expression of sNPFR1 leads to increased food finding (Figure 6B). The data suggest that sNPFR1 overexpression increases activity of Or42b neurons to produces starvation-like behavior. Is a simple increase in sensitivity sufficient to mimic the behavior? To test this, we artificially increased sensitivity of Or42b neurons by ectopically expressing the bacterial sodium channel (NachBac), which has previously been shown to make Drosophila neurons hyperexcitable (Nitabach et al., 2006). Indeed expression of NachBac in Or42b neurons produced starvation-like food finding in fed flies (Figure 6B). Thus, modulation of activity in the Or42b ORNs is both necessary for, and sufficient to mimic, state-dependent food search behavior. This result also suggests that sNPF is released even in the fed state. Furthermore, the data suggest that modulation of peripheral olfactory activity makes an important contribution to food search behavior.
Figure 6. Overexpression of sNPFR1 is sufficient to enhance activity and food search behavior.
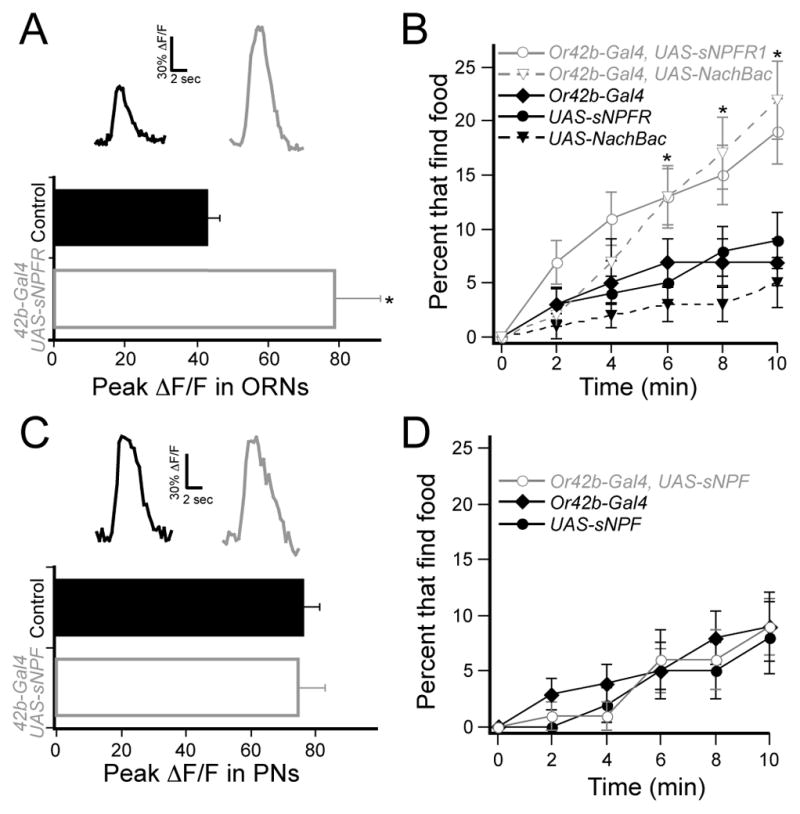
A, Two-photon imaging of ORN axon terminals in the DM1 glomerulus of fed flies in response to 0.2% SV cider vinegar. Control flies have the Or83b-LexA and LexAop-GCaMP transgenes, and experimental flies also bear the Or42b-Gal4 and UAS-sNPFR1 transgenes. n=5–6, *P<0.05, t-test. B, The latency of food finding in fed flies. n=134–168, *P<0.05, z-test for proportions comparing overexpression flies to three controls. C, PN dendritic calcium in the DM1 glomerulus of fed flies in response to 0.2% SV cider vinegar. Control flies have GH146-LexA and LexAop-GCaMP and experimental flies also have Or42b-Gal4 and UAS-sNPF transgenes. n=5–6. D, The latency of food finding in fed flies. n=66–81. Error bars show SEM.
In the above experiments, it appears that sNPF is expressed in fed flies but the receptor is only expressed upon starvation. Therefore, overexpression of the peptide in the fed state should not affect neural activity or behavior. To test this hypothesis, we first overexpressed sNPF in ORNs and imaged the postsynaptic PNs. Indeed overexpression of the peptide did not increase activity in DM1 glomerulus in fed flies (Figure 6C). Next, overexpression of sNPF in Or42b neurons did not alter food-finding behavior in fed flies (Figure 6D). Therefore, an increase in sNPF expression is not sufficient to sensitize ORNs or produce starvation-like food finding in fed flies.
Insulin functions as a satiety signal to suppress sNPFR1 expression
What is the molecular mechanism to increase ORN sensitivity in starved flies to gate appetitive behavior? We first investigated whether this physiological switch involves gene transcription by performing quantitative RT-PCR. We measured the level of sNPF and sNPFR1 transcripts in isolated antennae of fed and starved flies relative to a control gene, rp49 (a ribosomal protein). Interestingly, we found that the level of sNPFR1 mRNA is increased by approximately four-fold upon starvation, while the level of sNPF mRNA does not change (Figure 7B). Although we do not detect a change in sNPF mRNA, we cannot rule out the possibilities of starvation-dependent changes in neuropeptide translation or release. Nevertheless, ectopic expression of sNPFR1 expression is sufficient to induce presynaptic facilitation in fed flies (Figure 6A). Therefore, starvation leads to increased expression of sNPFR1, which is sufficient to cause presynaptic facilitation even in the absence of any starvation-dependent change in sNPF.
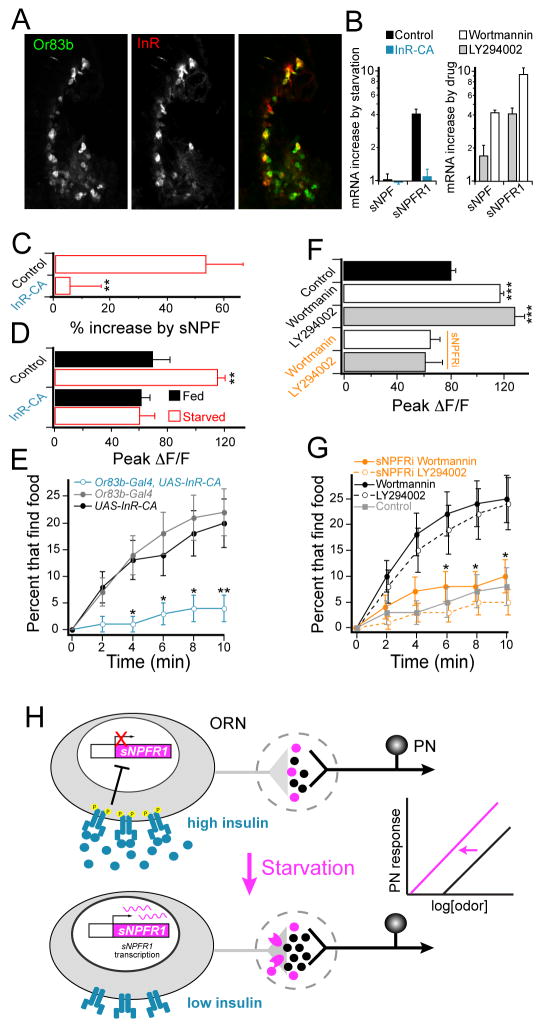
Figure 7. Insulin signaling modulates expression of sNPFR1 and olfactory sensitivity.
A, Antennal tissue with immunoreactivity for the InR and GFP expression under the Or83b promoter. Tissue was stained with anti-GFP (green) and anti-InR (red) antibodies. B, Quantitative RT-PCR analysis of starvation-induced changes in mRNA expression in the antennae of control flies and flies expressing constitutively active InR (InR-CA) in ORNs (left), and that of flies fed PI3K antagonists relative to those fed only sucrose (right). C, Response to electrical stimulation of the olfactory nerve before and after bath application of 10 μM sNPF, as in those Figure 4D–G. n=6–9. D, PN dendritic response to 0.2% SV cider vinegar in the DM1 glomerulus for control flies and those expressing InR-CA in ORNs. n=5–9. C, D, Control flies contain GH146-LexA, LexAop-GCaMP, Or83b-Gal4; InR-CA flies also contain UAS-InR-CA. E, The latency of food search behavior in starved control flies (black and gray) and those expressing InR-CA in ORNs (blue). n=70–90 flies. F, PN dendritic response to 0.2% SV cider vinegar in the DM1 glomerulus for control flies fed sucrose overnight and those fed sucrose plus 25nM wortmannin or 30 μM LY294002. n=5 each. G, The latency of food search behavior in flies fed wortmannin and LY294002, and control flies fed sucrose only. sNPFRi flies (orange) contain both Or83b-Gal4 and UAS-sNPFR1-RNAi, while control flies (black or gray) represent combined data for flies expressing either transgene alone (control groups are not different from each other). n=60–92 flies. H, Model for starvation modulation of olfactory sensitivity. Error bars indicate SEM. *P<0.05, **P<0.01, ***P<0.001, t-test for B, C, D, F, and z-test for E, G.
We next asked what is the metabolic sensor for ORNs to induce expression of sNPFR1? It has been well established that the levels of circulating Drosophila insulin-like peptide plummet in the starvation state (Geminard et al., 2009), and that the downstream signaling from the insulin receptor (InR) has the capacity to control gene expression (Edgar, 2006). Furthermore, expression of an insulin receptor has been observed in ORNs of C. elegans (Chalasani et al., 2010). We therefore asked if ORNs express the insulin receptor, by assaying immunoreactivity with InR antiserum in flies that express GFP in ORNs. Indeed many of the Or83b neurons (Figure 7A) and all of the Or42b neurons (Figure S5A) have InR immunoreactivity indicating that the ORNs projecting to the DM1 glomerulus express InR and therefore could be subject to insulin modulation.
Does InR activity alter the expression of sNPFR1 signaling? We reasoned that ectopic expression of a constitutively active InR (InR-CA) in ORNs should mimic the fed state. We first looked at the starvation-dependent expression of sNPFR1 transcripts and found that starved flies bearing Or83b-Gal4 and UAS-InR-CA do not exhibit an increase in sNPFR1 transcripts measured by qRT-PCR (Figure 7B). Similarly, calcium imaging experiments reveal that expression of InR-CA in ORNs eliminates the sensitivity to exogenous sNPF application in the DM1 glomerulus of starved flies (Figure 7C). In starved control flies, bath application of sNPF enhances the axonal calcium transient evoked by electrical stimulation of the olfactory nerve, while sNPF application had no effect on axonal calcium activity in flies expressing InR-CA in ORNs. This experiment was carried out in the same way as those in Figure 4D–G. These results predict that starvation should not sensitize Or42b ORNs in these flies with the constitutively active InR. Indeed, calcium imaging experiments show that starvation does not increase olfactory response to cider vinegar in DM1 (Figure 7D). Constitutive activation of InR specifically eliminates the starvation-dependent sensitization because the odor response in fed InR-CA flies is not different from fed controls, indicating that the manipulation does not impair these neurons. Measurement of food search behavior indicates that the constitutively active InR in most ORNs (Figure 7E), or selectively in Or42b ORNs (Figure S5B), reduces food finding. Therefore, activation of InR in Or42b neurons prevents starvation-dependent presynaptic facilitation and food search behavior.
We next asked if blockade of InR downstream signals could mimic the effect of starvation in ORNs. Phophatidylinositol 3-kinase (PI3K) is a crucial downstream molecule for insulin control of gene transcription and translation to promote cell growth (Leevers et al., 1996; Weinkove et al., 1999). Chronic blockade of insulin signaling can affect growth, therefore we sought to limit the timing of the perturbation. We hypothesized that pharmacological inhibition of PI3K should mimic the starvation state by preventing InR signaling. Two commonly used anti-tumor drugs, wortmannin and LY294002, have been shown as effective inhibitors of PI3K (Arcaro and Wymann, 1993; Vlahos et al., 1994). Indeed feeding flies overnight with 4% sucrose plus 25 nM wortmannin or 30 μM LY294002 sensitizes olfactory response in the DM1 glomerulus to the same level as that of starved flies and significantly greater than that of flies fed only 4% sucrose (Figure 7F). Do these PI3K antagonists alter ORN sNPFR1 mRNA levels? Indeed qRT-PCR experiments from isolated antennae revealed that feeding flies with wortmannin or LY294002 causes a significant increase in sNPFR1 expression relative to flies fed with 4% sucrose (Figure 7B). Thus, either of these PI3K antagonists causes increased expression of sNPFR1 in ORNs in addition to sensitized olfactory response. Notably, these two PI3K inhibitors appear to increase peptide mRNA level that is not observed in starved flies, however increased expression of sNPF does not change the behavior (Figure 6D). Therefore, we further investigated the link between the drug-induced increase in sNPFR1 and the drug-induced olfactory sensitization with epistatasis experiments. Expression of sNPFR1-RNAi in ORNs eliminates the drug-induced sensitization (Figure 7F), indicating that the sensitization resulting from blocking insulin signaling depends on sNPFR1 expression in ORNs. Lastly, we asked if blocking PI3K induces starvation-like behavior in fed flies. Feeding flies either wortmannin or LY294002 leads to significantly increased food finding in comparison to those expressing sNPFR1-RNAi in Or83b neurons and control flies fed only 4% sucrose (Figure 7G). Expression of sNPFR1-RNAi specifically in Or42b neurons also eliminates the starvation-like effect of wortmannin (Figure S5C). These results demonstrate that reduced insulin signaling is necessary and sufficient for starvation-dependent up-regulation of sNPFR1 and the induction of presynaptic facilitation, indicating that InR in ORNs is the metabolic sensor to trigger appetitive behavior (Figure 7H).
DISCUSSION
We report here that a state of starvation modulates olfactory sensitivity at the first synapse in a form of presynaptic facilitation. Starvation increases sNPFR1 transcription in ORNs, which is both necessary and sufficient for presynaptic facilitation. It has been well established that fluctuation of insulin is a key metabolic cue to maintain energy homeostasis. This study implicates that a low insulin signal via the PI3K pathway increases sNPFR1 expression. Interestingly, a subset of glomeruli exhibit starvation-dependent presynaptic facilitation that depends on intraglomerular sNPF signaling, while selective knockdown of sNPF or sNPFR1 in only the DM1 glomerulus affects food search behavior. This finding corroborates our previous work revealing that the DM1 glomerulus is hardwired for innate odor attraction (Semmelhack and Wang, 2009). Thus, an internal state of starvation, with insulin as a global satiety signal acting on sensory neurons through a local sNPF signal, shifts the odor map. Starvation modulation of the odor map increases the saliency of glomerular activity to match the changing physiological needs of an organism.
The Or42b sensory neurons may be considered as a neural substrate for appetitive choices because they integrate internal and external cues to influence an important innate behavior. In this integration a highly conserved neuropeptide (Hewes and Taghert, 2001) plays an important role in the peripheral olfactory system. A similar presynaptic facilitation mechanism may exist in vertebrates as well. In an aquatic salamander, NPY has been shown to enhance electrical responses of cells in the olfactory epithelium to a food related odorant in starved animals (Mousley et al., 2006). In addition, NPY immunoreactivity has been observed in the olfactory epithelium of mouse (Hansel et al., 2001) and zebrafish (Mathieu et al., 2002). In the nematode C. elegans, elevated activity levels of an NPY-like receptor cause a change in foraging pattern (Macosko et al., 2009). Our study demonstrates that a fluctuating metabolic cue controls sNPFR1 levels in Or42b neurons, which in turn modulates appetitive behavior. However, it remains to be determined whether other ORNs mediate attraction behavior and whether they are subject to sNPF mediated modulation. Given the ubiquitous use of insulin as a metabolic cue, modulation by NPY/sNPF receptors in the early olfactory system could be a conserved mechanism between different animal species.
The internal state of an organism influences its behavior. There is abundant evidence indicating that the global metabolic cue, insulin, works together with local neuropeptides in specific neural circuits to generate state-dependent behavioral responses. In Drosophila, the tolerance of a noxious food source is suppressed by insulin signaling and enhanced by NPF signaling such that these two peptides exert their opposing effects on the same neurons that mediate the behavior (Wu et al., 2005). In the mammalian hypothalamus, expression of the orexigenic NPY is suppressed in the satiety state via insulin signaling (Mayer and Belsham, 2009; Schwartz et al., 1992). Results from our study indicate that olfactory response in the periphery is reduced in the satiety state, in which insulin suppresses NPFR1 expression to alter neuronal excitability. Insulin's upstream control over sNPFR1 expression, however, appears to be specific to select neuronal types. Previous work in Drosophila has shown that sNPFR1 signaling exerts upstream control of insulin production in the Dilp2 neuroendocrine cells (Lee et al., 2008). In C. elegans, the release of an insulin-like peptide in an interneuron is downstream of a neuropeptide involved in promoting behavioral adaptation to food odors (Chalasani et al., 2010). Thus, different neuronal subtypes may adopt the same neuropeptides for unique and divergent molecular responses. Peptidergic modulation provides a rich repertoire of functional states for the same neural circuit to meet the demand of different internal states.
Central mechanisms to control appetitive behavior, similar to the well-documented modulation of the hypothalamus by NPY, also appear to be important in Drosophila. A recent study demonstrates that appetitive memory requires the NPF receptor in the dopaminergic neurons that innervate specific mushroom body lobes (Krashes et al., 2009). This poses the question: what functions are subserved by starvation modulation of multiple neural substrates? It is interesting to note that sensitization of Or42b ORNs is sufficient to enhance food search behavior in fed flies. Perhaps central modulation by starvation is not necessary for food search behavior. Modulation in the periphery may serve to gate an animals’ sensitivity to specific food odorants, while central modulation may serve to enhance an animal’s ability to remember the relevant cues in finding a particular food source.
EXPERIMENTAL PROCEDURES
Two-photon calcium imaging
GCaMP imaging was performed as previously described (Root et al., 2008; Wang et al., 2003). In odor experiments, a constant airflow of 1 l/min was applied to the antennae via a pipe of 12 mm diameter. Odor onset was controlled by mixing a defined percentage of carrier air with air redirected through odor bottles (presented as %SV, or saturated vapor pressure) as previously described (Root et al., 2008; Semmelhack and Wang, 2009). Nerve stimulation was performed with a glass suction electrode and an S48 stimulator (Grass, Warwick, RI) as previously described (Root et al., 2008; Wang et al., 2003). Starved flies were starved with water for 17–24 hr.
Behavior assay
Single female flies were introduced into chambers that were 60 mm in diameter and 6 mm in height. The chamber was illuminated by 660 nm LEDs. Flies were tracked at 2 Hz with custom software written in Labview (V.8.5, National Instruments), and analysis was performed with Igor Pro (V.6, Wavemetrics, Inc) using a custom macro. Latency is defined as the elapsed time before an individual fly spends more than 5 seconds within a distance of 5 mm from the odor source, which minimizes false positives due to random entry into the odor zone. Apple cider vinegar was diluted 1:100 in 1% low melting temperature agarose and 5 μl were placed in the center of the chamber. Increased cider vinegar concentration leads to increased food finding with a maximum food finding of about 60% at 25% cider vinegar (data not shown). For all of the presented experiments, we used a concentration of cider vinegar that produces moderate food finding such that it could be modulated up or down. In addition, we observed that 17–24 hr starvation and 4–6 hr starvation produced similar results, consistent with the starvation effect measured by calcium imaging (Figure 2e). Therefore, some experiments were carried out with 4–6 hr starvation and others overnight; controls and experimentals were always treated the same.
Pharmacology
sNPF peptide, AQRSPSLRLRF-NH2, 98% purity (Celtek Peptides) was dissolved in saline to a final concentration of 10 μM. Wortmannin and LY294002 (LC Laboratories, Woburn, MA) were dissolved in DMSO at stock concentrations of 10 mM and 50 mM, respectively. Flies were fed overnight in vials containing 4% sucrose solution alone, or that plus 25nM wortmannin or 30μM LY294002.
Quantitative RT-PCR
RNA was isolated from antennae of 50 female flies for each sample. The RNeasy kit (Qiagen) was used to isolate RNA and the reverse transcription was performed using the Retroscript kit (Ambion) with random decamers. This cDNA was subjected to PCR analysis using SYBR green detection on an iCycler thermocycler (Biorad). All values are the average of four replicates, each of which was measured in triplicate and normalized to a rp49 as a control gene.
Immunostaining
Antennal sections were obtained by mounting live fly heads in OCT, freezing in a dry ice ethanol bath, and 14 mm thick section were cut on a cryostat. Slides were immediately fixed with ice-cold 4% paraformaldehyde in 0.1X PBS for 10 min. Staining was performed using standard techniques with chick-anti-GFP (Ab13970, Abcam, Cambridge, MA), rabbit-anti-InR (3021, Cell Signaling Technology, Danvers, MA), at 1:1000, and 1:200 respectively.
Transgenic flies
See supplemental methods for a list of fly stocks.
Supplementary Material
Acknowledgments
We would like to thank R. Axel, M. Gallio, A. Gelperin, S. Kim, and M. Ramaswami for comments on the manuscript. We thank D. Green for assistance with the fly tracking software, E. Troemel for assistance with quantitative RT-PCR, and Jun Hee Lee for helpful discussion of insulin signaling. This work was supported by research grants from the National Institute of Deafness and other Communication Disorders to J.W.W. (R01DC009597, R21DC010458) and fellowship to C.M.R. (1F31DC009511).
Footnotes
Competing Interest Statement: The authors declare they have no competing interest.
Author Contributions:
C.M.R. and J.W.W. designed experiments, analyzed data and wrote the manuscript. C.M.R, K.I.K and A.J. carried out experiments.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arcaro A, Wymann MP. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem J. 1993;296(Pt 2):297–301. doi: 10.1042/bj2960297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsh GS, Schwartz MW. Genetic approaches to studying energy balance: perception and integration. Nat Rev Genet. 2002;3:589–600. doi: 10.1038/nrg862. [DOI] [PubMed] [Google Scholar]
- Berthoud HR. Multiple neural systems controlling food intake and body weight. Neurosci Biobehav Rev. 2002;26:393–428. doi: 10.1016/s0149-7634(02)00014-3. [DOI] [PubMed] [Google Scholar]
- Brown MR, Crim JW, Arata RC, Cai HN, Chun C, Shen P. Identification of a Drosophila brain-gut peptide related to the neuropeptide Y family. Peptides. 1999;20:1035–1042. doi: 10.1016/s0196-9781(99)00097-2. [DOI] [PubMed] [Google Scholar]
- Carlsson MA, Diesner M, Schachtner J, Nassel DR. Multiple neuropeptides in the Drosophila antennal lobe suggest complex modulatory circuits. J Comp Neurol. 2010;518:3359–3380. doi: 10.1002/cne.22405. [DOI] [PubMed] [Google Scholar]
- Chalasani SH, Kato S, Albrecht DR, Nakagawa T, Abbott LF, Bargmann CI. Neuropeptide feedback modifies odor-evoked dynamics in Caenorhabditis elegans olfactory neurons. Nat Neurosci. 2010;13:615–621. doi: 10.1038/nn.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Cowley MA, Pronchuk N, Fan W, Dinulescu DM, Colmers WF, Cone RD. Integration of NPY, AGRP, and melanocortin signals in the hypothalamic paraventricular nucleus: evidence of a cellular basis for the adipostat. Neuron. 1999;24:155–163. doi: 10.1016/s0896-6273(00)80829-6. [DOI] [PubMed] [Google Scholar]
- Dacks AM, Green DS, Root CM, Nighorn AJ, Wang JW. Serotonin modulates olfactory processing in the antennal lobe of Drosophila. J Neurogenet. 2009;23:366–377. doi: 10.3109/01677060903085722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destexhe A, Marder E. Plasticity in single neuron and circuit computations. Nature. 2004;431:789–795. doi: 10.1038/nature03011. [DOI] [PubMed] [Google Scholar]
- Dethier VG. The hungry fly : a physiological study of the behavior associated with feeding. Cambridge, Mass: Harvard University Press; 1976. [Google Scholar]
- Distler PG, Boeckh J. Synaptic connections between identified neuron types in the antennal lobe glomeruli of the cockroach, Periplaneta americana: II. Local multiglomerular interneurons. J Comp Neurol. 1997;383:529–540. doi: 10.1002/(sici)1096-9861(19970714)383:4<529::aid-cne9>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Edgar BA. How flies get their size: genetics meets physiology. Nat Rev Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- Feng G, Reale V, Chatwin H, Kennedy K, Venard R, Ericsson C, Yu K, Evans PD, Hall LM. Functional characterization of a neuropeptide F-like receptor from Drosophila melanogaster. Eur J Neurosci. 2003;18:227–238. doi: 10.1046/j.1460-9568.2003.02719.x. [DOI] [PubMed] [Google Scholar]
- Gelperin A. Regulation of feeding. Annual Review of Entomology. 1971;16:365–378. [Google Scholar]
- Geminard C, Rulifson EJ, Leopold P. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 2009;10:199–207. doi: 10.1016/j.cmet.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Gerozissis K. Brain insulin: regulation, mechanisms of action and functions. Cell Mol Neurobiol. 2003;23:1–25. doi: 10.1023/A:1022598900246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansel DE, Eipper BA, Ronnett GV. Neuropeptide Y functions as a neuroproliferative factor. Nature. 2001;410:940–944. doi: 10.1038/35073601. [DOI] [PubMed] [Google Scholar]
- Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 2001;11:1126–1142. doi: 10.1101/gr.169901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignell R, Root CM, Birse RT, Wang JW, Nassel DR, Winther AM. Presynaptic peptidergic modulation of olfactory receptor neurons in Drosophila. Proc Natl Acad Sci U S A. 2009;106:13070–13075. doi: 10.1073/pnas.0813004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes MJ, DasGupta S, Vreede A, White B, Armstrong JD, Waddell S. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell. 2009;139:416–427. doi: 10.1016/j.cell.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Kwon OY, Lee JH, Kwon K, Min KJ, Jung SA, Kim AK, You KH, Tatar M, Yu K. Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat Cell Biol. 2008;10:468–475. doi: 10.1038/ncb1710. [DOI] [PubMed] [Google Scholar]
- Lee KS, You KH, Choo JK, Han YM, Yu K. Drosophila short neuropeptide F regulates food intake and body size. J Biol Chem. 2004;279:50781–50789. doi: 10.1074/jbc.M407842200. [DOI] [PubMed] [Google Scholar]
- Leevers SJ, Weinkove D, MacDougall LK, Hafen E, Waterfield MD. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 1996;15:6584–6594. [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, Clardy J, Bargmann CI. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009;458:1171–1175. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M, Tagliafierro G, Bruzzone F, Vallarino M. Neuropeptide tyrosine-like immunoreactive system in the brain, olfactory organ and retina of the zebrafish, Danio rerio, during development. Brain Res Dev Brain Res. 2002;139:255–265. doi: 10.1016/s0165-3806(02)00577-1. [DOI] [PubMed] [Google Scholar]
- Mayer CM, Belsham DD. Insulin directly regulates NPY and AgRP gene expression via the MAPK MEK/ERK signal transduction pathway in mHypoE-46 hypothalamic neurons. Mol Cell Endocrinol. 2009;307:99–108. doi: 10.1016/j.mce.2009.02.031. [DOI] [PubMed] [Google Scholar]
- Mousley A, Polese G, Marks NJ, Eisthen HL. Terminal nerve-derived neuropeptide y modulates physiological responses in the olfactory epithelium of hungry axolotls (Ambystoma mexicanum) J Neurosci. 2006;26:7707–7717. doi: 10.1523/JNEUROSCI.1977-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Kashiwadani H, Kirino Y, Mori K. State-dependent sensory gating in olfactory cortex. Neuron. 2005;46:285–296. doi: 10.1016/j.neuron.2005.02.025. [DOI] [PubMed] [Google Scholar]
- Nassel DR, Enell LE, Santos JG, Wegener C, Johard HA. A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC Neurosci. 2008;9:90. doi: 10.1186/1471-2202-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SR, Bhandawat V, Wilson RI. Excitatory interactions between olfactory processing channels in the Drosophila antennal lobe. Neuron. 2007;54:89–103. doi: 10.1016/j.neuron.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SR, Wilson RI. Lateral presynaptic inhibition mediates gain control in an olfactory circuit. Nature. 2008;452:956–960. doi: 10.1038/nature06864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root CM, Masuyama K, Green DS, Enell LE, Nassel DR, Lee CH, Wang JW. A presynaptic gain control mechanism fine-tunes olfactory behavior. Neuron. 2008;59:311–321. doi: 10.1016/j.neuron.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root CM, Semmelhack JL, Wong AM, Flores J, Wang JW. Propagation of olfactory information in Drosophila. Proc Natl Acad Sci U S A. 2007;104:11826–11831. doi: 10.1073/pnas.0704523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Sachse S, Galizia CG. Role of inhibition for temporal and spatial odor representation in olfactory output neurons: a calcium imaging study. J Neurophysiol. 2002;87:1106–1117. doi: 10.1152/jn.00325.2001. [DOI] [PubMed] [Google Scholar]
- Schwartz MW, Sipols AJ, Marks JL, Sanacora G, White JD, Scheurink A, Kahn SE, Baskin DG, Woods SC, Figlewicz DP, et al. Inhibition of hypothalamic neuropeptide Y gene expression by insulin. Endocrinology. 1992;130:3608–3616. doi: 10.1210/endo.130.6.1597158. [DOI] [PubMed] [Google Scholar]
- Semmelhack JL, Wang JW. Select Drosophila glomeruli mediate innate olfactory attraction and aversion. Nature. 2009;459:218–223. doi: 10.1038/nature07983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Claridge-Chang A, Sjulson L, Pypaert M, Miesenbock G. Excitatory local circuits and their implications for olfactory processing in the fly antennal lobe. Cell. 2007;128:601–612. doi: 10.1016/j.cell.2006.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker RF, Heimbeck G, Gendre N, de Belle JS. Neuroblast ablation in Drosophila P[GAL4] lines reveals origins of olfactory interneurons. J Neurobiol. 1997;32:443–456. doi: 10.1002/(sici)1097-4695(199705)32:5<443::aid-neu1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Stocker RF, Lienhard MC, Borst A, Fischbach KF. Neuronal architecture of the antennal lobe in Drosophila melanogaster. Cell Tissue Res. 1990;262:9–34. doi: 10.1007/BF00327741. [DOI] [PubMed] [Google Scholar]
- Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu Rev Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Weinkove D, Neufeld TP, Twardzik T, Waterfield MD, Leevers SJ. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class I(A) phosphoinositide 3-kinase and its adaptor. Curr Biol. 1999;9:1019–1029. doi: 10.1016/s0960-9822(99)80450-3. [DOI] [PubMed] [Google Scholar]
- Woods SC, Seeley RJ, Porte D, Jr, Schwartz MW. Signals that regulate food intake and energy homeostasis. Science. 1998;280:1378–1383. doi: 10.1126/science.280.5368.1378. [DOI] [PubMed] [Google Scholar]
- Wu Q, Wen T, Lee G, Park JH, Cai HN, Shen P. Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron. 2003;39:147–161. doi: 10.1016/s0896-6273(03)00396-9. [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhao Z, Shen P. Regulation of aversion to noxious food by Drosophila neuropeptide Y- and insulin-like systems. Nat Neurosci. 2005;8:1350–1355. doi: 10.1038/nn1540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.