Abstract
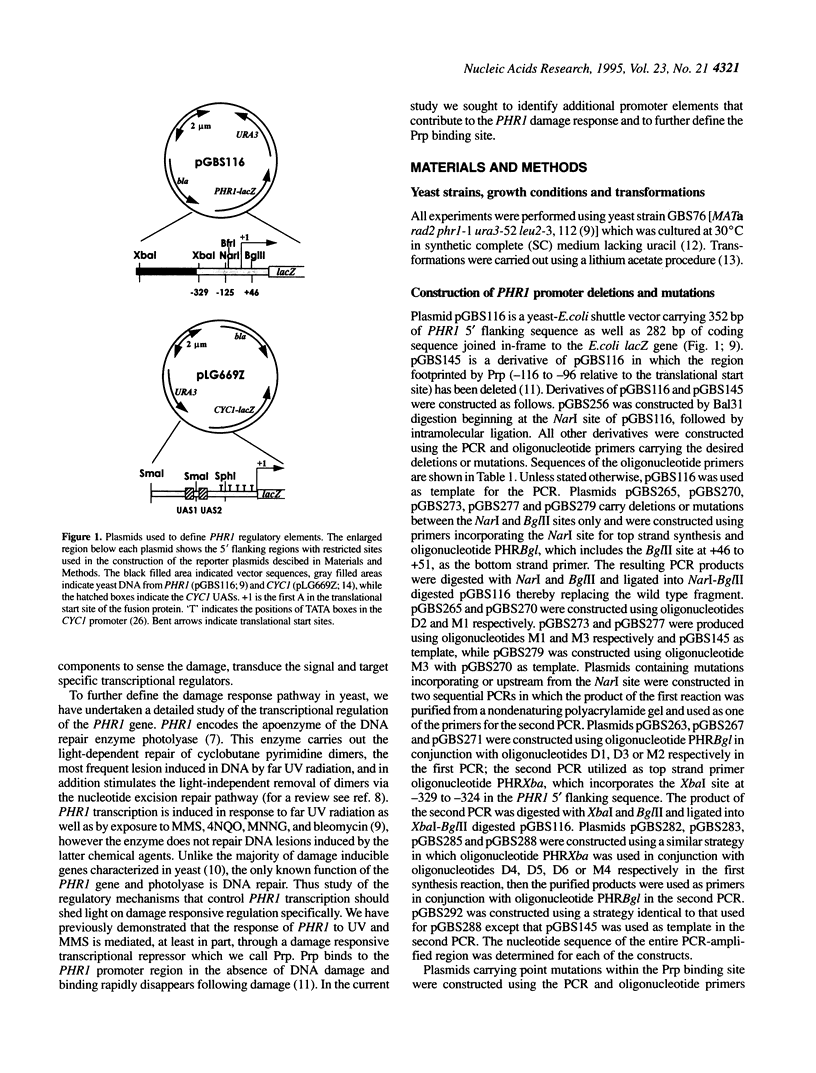
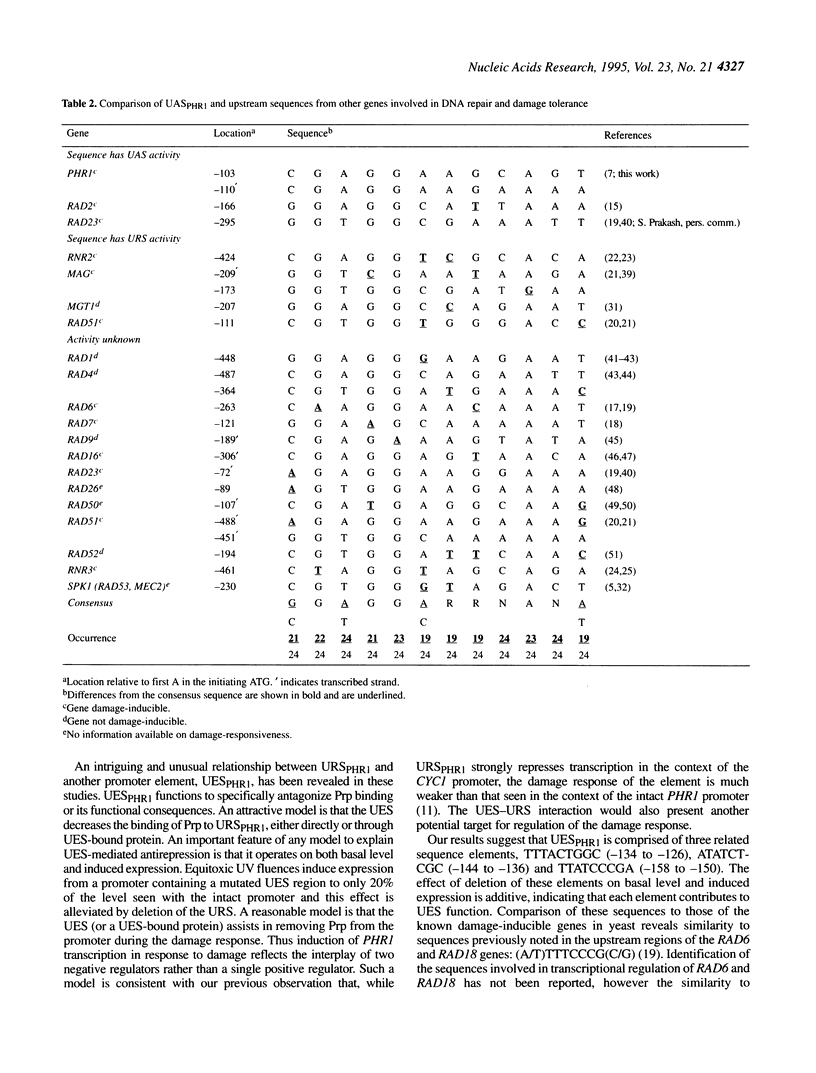
The PHR1 gene of Saccharomyces cerevisiae encodes the apoenzyme for the DNA repair enzyme photolyase. PHR1 transcription is induced in response to 254 nm radiation and a variety of chemical damaging agents. We report here the identification of promoter elements required for PHR1 expression. Transcription is regulated primarily through three sequence elements clustered within a 120 bp region immediately upstream of the translational start site. A 20 bp interrupted palindrome comprises UASPHR1 and is responsible for 80-90% of basal and induced expression. UASPHR1 alone can activate transcription of a CYC1 minimal promoter but does not confer damage responsiveness. In the intact PHR1 promoter UAS function is dependent upon an upstream essential sequence (UES). URSPHR1 contains a binding site for the damage-responsive repressor Prp; consistent with this role, deletion or specific mutations of the URS increase basal level expression and decrease the induction ratio. Deletion of URSPHR1 also eliminates the requirement for UESPHR1 for promoter activation, indicating that the UES attenuates Prp-mediated repression. Sequences within UASPHR1 are similar to regulatory sequences found upstream of both damage responsive and nonresponsive genes involved in DNA repair and metabolism.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alani E., Subbiah S., Kleckner N. The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics. 1989 May;122(1):47–57. doi: 10.1093/genetics/122.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. A., Kremenstova E., Luo L. Complete transposition requires four active monomers in the mu transposase tetramer. Genes Dev. 1994 Oct 15;8(20):2416–2428. doi: 10.1101/gad.8.20.2416. [DOI] [PubMed] [Google Scholar]
- Bang D. D., Timmermans V., Verhage R., Zeeman A. M., van de Putte P., Brouwer J. Regulation of the Saccharomyces cerevisiae DNA repair gene RAD16. Nucleic Acids Res. 1995 May 25;23(10):1679–1685. doi: 10.1093/nar/23.10.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang D. D., Verhage R., Goosen N., Brouwer J., van de Putte P. Molecular cloning of RAD16, a gene involved in differential repair in Saccharomyces cerevisiae. Nucleic Acids Res. 1992 Aug 11;20(15):3925–3931. doi: 10.1093/nar/20.15.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Micklem G., Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987 Dec 4;51(5):709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Kornberg R. D. A yeast ARS-binding protein activates transcription synergistically in combination with other weak activating factors. Mol Cell Biol. 1990 Mar;10(3):887–897. doi: 10.1128/mcb.10.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. Identification of the DNA damage-responsive element of RNR2 and evidence that four distinct cellular factors bind it. Mol Cell Biol. 1989 Dec;9(12):5373–5386. doi: 10.1128/mcb.9.12.5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S. J., Davis R. W. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 1990 May;4(5):740–751. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R. A., Schiestl R. H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992 Mar 25;20(6):1425–1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Prakash S. Cloning and nucleotide sequence analysis of the Saccharomyces cerevisiae RAD4 gene required for excision repair of UV-damaged DNA. Gene. 1988 Dec 30;74(2):535–541. doi: 10.1016/0378-1119(88)90186-2. [DOI] [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Guarente L., Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd H. K., Roberts J. W. Upstream regulatory sequences of the yeast RNR2 gene include a repression sequence and an activation site that binds the RAP1 protein. Mol Cell Biol. 1989 Dec;9(12):5359–5372. doi: 10.1128/mcb.9.12.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. S., Prakash L. Transcript levels of the Saccharomyces cerevisiae DNA repair gene RAD18 increase in UV irradiated cells and during meiosis but not during the mitotic cell cycle. Nucleic Acids Res. 1991 Feb 25;19(4):893–898. doi: 10.1093/nar/19.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher C. A., Goutte C., Johnson A. D. The yeast cell-type-specific repressor alpha 2 acts cooperatively with a non-cell-type-specific protein. Cell. 1988 Jun 17;53(6):927–936. doi: 10.1016/s0092-8674(88)90449-7. [DOI] [PubMed] [Google Scholar]
- Li W. Z., Sherman F. Two types of TATA elements for the CYC1 gene of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991 Feb;11(2):666–676. doi: 10.1128/mcb.11.2.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K., Prakash S. Nucleotide sequence, transcript mapping, and regulation of the RAD2 gene of Saccharomyces cerevisiae. J Bacteriol. 1986 Jun;166(3):914–923. doi: 10.1128/jb.166.3.914-923.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal M. L., Higgins D. R., Prakash S. Expression of the RAD1 and RAD3 genes of Saccharomyces cerevisiae is not affected by DNA damage or during the cell division cycle. Mol Gen Genet. 1985;199(1):59–63. doi: 10.1007/BF00327510. [DOI] [PubMed] [Google Scholar]
- Navas T. A., Zhou Z., Elledge S. J. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995 Jan 13;80(1):29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- Passmore S., Elble R., Tye B. K. A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Genes Dev. 1989 Jul;3(7):921–935. doi: 10.1101/gad.3.7.921. [DOI] [PubMed] [Google Scholar]
- Perozzi G., Prakash S. RAD7 gene of Saccharomyces cerevisiae: transcripts, nucleotide sequence analysis, and functional relationship between the RAD7 and RAD23 gene products. Mol Cell Biol. 1986 May;6(5):1497–1507. doi: 10.1128/mcb.6.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C., Cayrol C., Lesca C., Kaiser P., Thompson C., Defais M. Characterization of dinY, a new Escherichia coli DNA repair gene whose products are damage inducible even in a lexA(Def) background. J Bacteriol. 1993 Feb;175(3):642–646. doi: 10.1128/jb.175.3.642-646.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond W. E., Kleckner N. Expression of the Saccharomyces cerevisiae RAD50 gene during meiosis: steady-state transcript levels rise and fall while steady-state protein levels remain constant. Mol Gen Genet. 1993 Apr;238(3):390–400. doi: 10.1007/BF00291998. [DOI] [PubMed] [Google Scholar]
- Reynolds P., Weber S., Prakash L. RAD6 gene of Saccharomyces cerevisiae encodes a protein containing a tract of 13 consecutive aspartates. Proc Natl Acad Sci U S A. 1985 Jan;82(1):168–172. doi: 10.1073/pnas.82.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G. W., Nicolet C. M., Kalainov D., Friedberg E. C. A yeast excision-repair gene is inducible by DNA damaging agents. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1842–1846. doi: 10.1073/pnas.83.6.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar G. B. DNA photolyases: physical properties, action mechanism, and roles in dark repair. Mutat Res. 1990 Sep-Nov;236(2-3):147–160. doi: 10.1016/0921-8777(90)90002-m. [DOI] [PubMed] [Google Scholar]
- Sancar G. B. Sequence of the Saccharomyces cerevisiae PHR1 gene and homology of the PHR1 photolyase to E. coli photolyase. Nucleic Acids Res. 1985 Nov 25;13(22):8231–8246. doi: 10.1093/nar/13.22.8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R. H., Reynolds P., Prakash S., Prakash L. Cloning and sequence analysis of the Saccharomyces cerevisiae RAD9 gene and further evidence that its product is required for cell cycle arrest induced by DNA damage. Mol Cell Biol. 1989 May;9(5):1882–1896. doi: 10.1128/mcb.9.5.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian J., Kraus B., Sancar G. B. Expression of the yeast PHR1 gene is induced by DNA-damaging agents. Mol Cell Biol. 1990 Sep;10(9):4630–4637. doi: 10.1128/mcb.10.9.4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian J., Sancar G. B. A damage-responsive DNA binding protein regulates transcription of the yeast DNA repair gene PHR1. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11251–11255. doi: 10.1073/pnas.88.24.11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A., Ogawa H., Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992 May 1;69(3):457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Siede W., Friedberg E. C. Regulation of the yeast RAD2 gene: DNA damage-dependent induction correlates with protein binding to regulatory sequences and their deletion influences survival. Mol Gen Genet. 1992 Mar;232(2):247–256. doi: 10.1007/BF00280003. [DOI] [PubMed] [Google Scholar]
- Siede W., Robinson G. W., Kalainov D., Malley T., Friedberg E. C. Regulation of the RAD2 gene of Saccharomyces cerevisiae. Mol Microbiol. 1989 Dec;3(12):1697–1707. doi: 10.1111/j.1365-2958.1989.tb00155.x. [DOI] [PubMed] [Google Scholar]
- Singer V. L., Wobbe C. R., Struhl K. A wide variety of DNA sequences can functionally replace a yeast TATA element for transcriptional activation. Genes Dev. 1990 Apr;4(4):636–645. doi: 10.1101/gad.4.4.636. [DOI] [PubMed] [Google Scholar]
- Walker G. C. Inducible DNA repair systems. Annu Rev Biochem. 1985;54:425–457. doi: 10.1146/annurev.bi.54.070185.002233. [DOI] [PubMed] [Google Scholar]
- Watkins J. F., Sung P., Prakash L., Prakash S. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol Cell Biol. 1993 Dec;13(12):7757–7765. doi: 10.1128/mcb.13.12.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobbe C. R., Struhl K. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol Cell Biol. 1990 Aug;10(8):3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Derfler B., Chen J., Samson L. Primary sequence and biological functions of a Saccharomyces cerevisiae O6-methylguanine/O4-methylthymine DNA repair methyltransferase gene. EMBO J. 1991 Aug;10(8):2179–2186. doi: 10.1002/j.1460-2075.1991.tb07753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Samson L. The Saccharomyces cerevisiae MGT1 DNA repair methyltransferase gene: its promoter and entire coding sequence, regulation and in vivo biological functions. Nucleic Acids Res. 1992 Jul 25;20(14):3599–3606. doi: 10.1093/nar/20.14.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Singh K. K., Chen B., Samson L. A common element involved in transcriptional regulation of two DNA alkylation repair genes (MAG and MGT1) of Saccharomyces cerevisiae. Mol Cell Biol. 1993 Dec;13(12):7213–7221. doi: 10.1128/mcb.13.12.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagle K., McEntee K. The DNA damage-inducible gene DIN1 of Saccharomyces cerevisiae encodes a regulatory subunit of ribonucleotide reductase and is identical to RNR3. Mol Cell Biol. 1990 Oct;10(10):5553–5557. doi: 10.1128/mcb.10.10.5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang E., Friedberg E. C. Molecular cloning and nucleotide sequence analysis of the Saccharomyces cerevisiae RAD1 gene. Mol Cell Biol. 1984 Oct;4(10):2161–2169. doi: 10.1128/mcb.4.10.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P., Fay D. S., Burton J., Xiao H., Pinkham J. L., Stern D. F. SPK1 is an essential S-phase-specific gene of Saccharomyces cerevisiae that encodes a nuclear serine/threonine/tyrosine kinase. Mol Cell Biol. 1993 Sep;13(9):5829–5842. doi: 10.1128/mcb.13.9.5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Elledge S. J. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell. 1993 Dec 17;75(6):1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]
- van Gool A. J., Verhage R., Swagemakers S. M., van de Putte P., Brouwer J., Troelstra C., Bootsma D., Hoeijmakers J. H. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 1994 Nov 15;13(22):5361–5369. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



