Abstract
Recurrent chromosomal abnormalities, especially chromosomal translocations, are strongly associated with certain subtypes of leukemia, lymphoma and solid tumors. The appearance of particular translocations or associated genomic alterations can be important indicators of disease prognosis, and in some cases, certain translocations may indicate appropriate therapy protocols. To date, most of our knowledge about chromosomal translocations has derived from characterization of the highly selected recurrent translocations found in certain cancers. Until recently, mechanisms that promote or suppress chromosomal translocations, in particular, those responsible for their initiation, have not been addressed. For translocations to occur, two distinct chromosomal loci must be broken, brought together (synapsed) and joined. Here, we discuss recent findings on processes and pathways that influence the initiation of chromosomal translocations, including the generation fo DNA double strand breaks (DSBs) by general factors or in the context of the Lymphocyte-specific V(D)J and IgH class-switch recombination processes. We also discuss the role of spatial proximity of DSBs in the interphase nucleus with respect to how DSBs on different chromosomes are justaposed for joining. In addition, we discuss the DNA DSB response and its role in recognizing and tethering chromosomal DSBs to prevent translocations, as well as potential roles of the classical and alternative DSB end-joining pathways in suppressing or promoting translocations. Finally, we discuss the potential roles of long range regulatory elements, such as the 3’IgH enhancer complex, in promoting the expression of certain translocations that are frequent in lymphomas and, thereby, contributing to their frequent appearance in tumors.
1. CHROMOSOMAL TRANSLOCATIONS IN CANCER
1.1. Overview
In 1914, Theodor Boveri postulated the somatic mutation theory of cancer (Boveri, 1914). Microscopic analysis of chromosomal segregation in dividing cells led him to hypothesize that changes in the chromosome constitution of a single cell are the driving force of tumorigenesis. Boveri suggested that a cancer originates from a single cell that has acquired an abnormal chromosomal constitution, and that this abnormality, when passed on to all the descendants overcomes a normal inhibitory mechanism, thereby causing rapid cell proliferation. Today, it is widely accepted that cancer is a genetic disease, caused by genomic aberrations, including point mutations or chromosomal translocations or amplifications that lead to activation of oncogenes or mutations or chromosomal deletions that inactivate tumor suppressor genes (reviewed in Fröhling and Döhner, 2008; Stratton et al., 2009).
Chromosomal translocations are abnormalities caused by visibly aberrant rearrangements of large pieces of chromosomal DNA. Translocations are one of several subtypes of chromosomal rearrangements traditionally distinguished as deletions, amplifications, inversions, duplication/insertions, and translocations. In this context, there are two general types of translocations. Reciprocal translocations appear as a simple swap between two nonhomologous chromosome arms such that the chimeric products are monocentric in metaphase. Nonreciprocal translocations on the other hand can lead to large deletions or duplications of chromosomal segments. Nonreciprocal translocations may or may not originate mechanistically by the same general mechanisms as reciprocal translocations. In either case, the pattern with which the nonhomologous chromosomes swap yield chimeric chromosomes that generate an imbalance; for example, dicentric chromosomes that can lead to deletions or amplifications as the cells progress through the cell cycle. Thus, the definition of reciprocal versus nonreciprocal translocation refers to the outcome of a rearrangement as visualized in cytogenetic analysis and not necessarily to the underlying mechanistic distinctions. The commonality between the initiating molecular mechanistic paths by which visible karyotypic changes from a normal cell are introduced into a premalignant clone, whether reciprocal or nonreciprocal translocations, deletions, amplifications, inversions, and insertions is not well defined at present (Zhang et al., 2009). Further resolution of the mechanistic factors that lead to these two general types of translocations in mammalian cells, for example, when translocation-initiating DNA double-strand breaks (DSBs) are introduced during the cell cycle and when during the cycle and how they are repaired (e.g., Zhu et al., 2002), awaits the development of new techniques to follow the initial generation of translocations before they are further molded by cellular and oncogenic selection.
1.2. Oncogenic translocations in cancer frequently have DSB intermediates
Chromosomal translocations are fundamental pathogenetic events in cancer, both with respect to tumor onset and tumor progression. Recurrent oncogenic translocations are common features of hematopoietic malignancies such as leukemia and lymphomas (Küppers and Dalla-Favera, 2001). Likewise, genomic instability in the form of translocations and related deletions and amplifications occurs in the context of solid tumor progression. Solid tumors, such as certain brain tumors, as well as prostate and lung cancers, also can contain recurrent translocations (Mitelman et al., 2007). Indeed, a major goal of the large cancer genome project is to better define genomic alterations in tumor cells, including translocations, deletions, inversions, and amplifications. DSBs are common intermediates in such genomic aberrations. DSBs can be generated by normal metabolic processes, by genotoxic agents including agents commonly used to treat cancer, and by the programmed processes of V(D)J recombination and immunoglobulin (Ig) heavy (H) chain (IgH) class switch recombination (CSR) in T and/or B lymphocytes. When DSBs occur, highly conserved DNA repair pathways efficiently rejoin broken ends to preserve the genome integrity. Nonetheless, repair sometimes fails, and the resulting unresolved DSBs can lead to chromosomal translocations and other genomic aberrations.
1.3. Chromosomal translocations in hematological malignancies
Chromosomal translocations are most well characterized in hematological malignancies. Several online databases are maintained with frequent updates where all chromosomal abnormalities in the cancer genetic literature are tracked in a searchable form (http://cgap.nci.nih.gov/Chromosomes/Mitelman, http://atlasgeneticsoncology.org/, http://www.sanger.ac.uk/genetics/CGP/Census/; Futreal et al., 2004). At present, more than 300 recurrent chromosomal translocations have been annotated. While some translocations may be late events that occur during the progression of a tumor, there is now clear evidence translocations associated with certain cancers actually contribute to transformation at an early stage of tumorigenesis (Stratton et al., 2009). Hematological malignancies, which account for about 10% of human cancers (Küppers and Dalla-Favera, 2001), frequently harbor characteristic recurrent chromosomal translocations that can activate oncogenes by several different mechanisms. One mechanism is the generation of chimeric proteins that result from chromosomal translocations that fuse the coding sequences of two different genes lying on the two translocation partner chromosomes. Another mechanism is the linkage of a potential oncogene to a regulatory element in a different locus; for example, the enhancer elements found in Ig loci, which lead to the misregulated expression of a translocated, but otherwise intact, structural gene. Indeed, more than 50% of leukemias and lymphomas carry reciprocal chromosomal translocations that recur among different patients and that are highly associated with particular cancer subtypes (Zhang and Rowley, 2006). Examples of recurrent translocations that lead to either novel fusion proteins or deregulated oncogene expression are discussed below.
A translocation called the Philadelphia chromosome by cytogeneticists was the first example of a consistent aberrant chromosomal translocation in a human cancer (Nowell and Hungerford, 1960). The Philadelphia chromosome was initially visualized as an unusually short chromosome that was recurrent in chronic myelogenous leukemia (CML) cells, and subsequently characterized in more detail as a t(9;22) (q34;q11) translocation (Rowley, 1973). In the 1980s, the crossover site on the Philadelphia chromosome was cloned and found to encode a composite gene in which all but the more 5′-exons of the cellular ABL gene (a cellular homologue of the v-Abl Abelson murine leukemia virus oncogene) on chromosome 9 had become joined to a gene termed BCR (for breakpoint cluster region; Rowley, 1973) on chromosome 22. The BCR–ABL fusion protein retained the tyrosine kinase activity of the parent ABL gene and was shown to be oncogenic (Bartram et al., 1983; de Klein et al., 1982; Groffen et al., 1984; Heisterkamp et al., 1983). The BCR-encoded portion of the protein was found to mediate permanent oligomerization of the chimeric protein, which enforced a constitutive and altered kinase activity (Goldman and Melo, 2003).
Having pinpointed the misregulated tyrosine kinase as a driving force in CML, a targeted approach in which tyrosine kinase inhibitors were screened for efficacy and specificity led to the discovery of imatinib mesylate (Gleevec), marking a stunning advance in the treatment of CML and other diseases exhibiting the Philadelphia chromosome (Druker et al., 2006). Oncogenic translocations, as illustrated by the Philadelphia chromosome and Gleevec example, can represent an Achilles’ heel of a tumor, revealing effective targets for drug therapy. Where the unregulated proliferation in cancer cells is largely due to abnormal oncogene expression via formation of a fusion protein or deregulated transcription, it is possible that inactivation of a single oncogene can result in the elimination of most, if not all, tumor cells (Felsher, 2008).
The other classic mechanism by which a reciprocal chromosomal exchange can be tumorigenic is by leading to changes in the level and/or specificity of the expression of a potential cellular oncogene. There are many examples of this form of deregulated protein expression, particular in B and T lymphoid malignancies in which potential oncogenes are placed under the control of tissue-specific gene regulatory elements that lie, respectively, within Ig or T cell receptor (TCR) loci. The classical example of a recurrent translocation that leads to such deregulated proto-oncogene expression is the t(8;14)-related translocations observed in Burkitt’s lymphoma (Janz, 2006). In the context of this translocation, the c-Myc (MYC1) gene, normally on chromosome 8 is fused to an Ig locus, most commonly the IgH locus near the telomere of the long arm of chromosome 14, but also to the Igκ or Igλ light chain loci on the long arm of chromosome 2 or long arm of chromosome 22, respectively (Janz, 2006). The c-Myc proto-oncogene was first identified as the human homolog of the oncogene found in an avian myelocytomatosis retrovirus and it encodes the C-MYC transcription factor. Dramatically increased expression of C-MYC, promotes transition of the cell cycle from G1 to S phase, which favors tumor-associated clonal bursts and further tumorigenesis (Küppers, 2005).
Other classes of human tumors similarly have translocations that deregulate expression of oncogenes such as Bcl-1 or Bcl-6 by fusing them with IgH or IgL loci (Küppers and Dalla-Favera, 2001). Such translocations deregulate c-Myc (or other cellular oncogenes) by putting them under the control of Ig regulatory elements. Both B cell lymphomas and plasmacytomas in mouse tumor models also can have similar recurrent translocations between IgH or IgL loci and the c-Myc locus, which deregulate c-Myc expression (Janz, 2006). While it was originally thought that the intronic enhancer within the IgH locus was a key element in mediating such deregulated expression in translocations involving IgH, recent studies of mouse B cell lymphoma models have shown that the IgH 3′-regulatory region (IgH3′RR), downstream of the IgH locus, plays a key role through its ability to activate oncogenes over long distances (Gostissa et al., 2009a; see Section 3).
Amplification of the c-Myc gene have also been found in association with V(D)J recombination-initiated, nonreciprocal translocations to the IgH locus (referred to a complex translocations with amplifications or “complicons”) in mouse progenitor B cell lymphomas (Difilippantonio et al., 2002; Zhu et al., 2002) and have been occasionally been observed in human B cell lymphomas and multiple myelomas (Janz, 2006; Martín-Subero et al., 2005). In addition, recent studies have implicated recurrent gene amplifications in the context of complicons initiated by V(D)J recombination-associated DSBs in the TCRδ locus in mouse thymic lymphomas that arise in the context of deficiency for the ataxia telangiectasia mutated (ATM) gene (Zha et al., 2010).
1.4. Chromosomal translocations in epithelial tumors
Recurrent chromosomal translocations, excluding amplifications, have not been as frequently described in the context of epithelial carcinomas as in hematological malignancies. This might in some part reflect technical limitations in the generation of metaphase chromosome spreads from epithelial tumors. Moreover, another significant feature of epithelial neoplasms is that they often evolve complex karyotypes during tumor progression so that the multiple cytogenetic changes become difficult to interpret, and clonal markers are not identifiable (Mitelman et al., 2007). Nonetheless there are examples of carcinomas in which some fraction of cases exhibit characteristic gene fusions brought about by recurrent translocations (reviewed in Brenner and Chinnaiyan, 2009).
About ~50% of papillary thyroid carcinomas (PTC) have translocations which connect the RET gene on chromosome 10 to a 5′ fusion partner that both mislocalizes and activates it. The specific identity of the protein can vary; the basic requirement appears to be that it confers dimerization capability (Nikiforov, 2008; Zitzelsberger et al., 2010). Likewise, about 50% of prostate cancers overexpress ETS transcription factors due to a translocation that puts genes such as ERG or ETV1, ETV4 or ETV5 under the control of the 5′-untranslated region of an androgen responsive gene, TMPRSS2. This mechanism of oncogene activation via translocation is reminiscent of the mechanism of misregulated expression of c-Myc or other oncogenes when translocated to an Ig locus in lymphoid tumors. However, while the TMPRSS2–ETS family fusions are common in premalignant prostate lesions, they are insufficient for the initiation of tumors in mouse models (Carver et al., 2009a). More recent work suggests that overexpressed ERG must cooperate with other mutations that inactivate a tumor repressor in promoting the progression from neoplasia to carcinoma (Brenner and Chinnaiyan, 2009; Carver et al., 2009b).
Testing NIH 3T3 cells with a retrovirally delivered cDNA library prepared from a lung cancer specimen led to the description of an inversion within the long arm of chromosome 2 that joins the ALK (anaplastic lymphoma kinase) and EML4 (echinoderm microtubule-associated protein-like 4) loci (Soda et al., 2007). The EML4–ALK fusion protein was shown to have tyrosine kinase activity and the transcript was detected in a significant fraction of patients. However, since then, the detection of EML4–ALK protein in non-small-cell lung cancers (NSCLCs) exhibiting fusion transcripts has been variable and the possibility that this particular rearrangement defines a therapeutic target remains to be determined (Martelli et al., 2009; Soda et al., 2008). An independent phosphotyrosin proteomics-based approach also detected ROS kinase (highly expressed in NSCLCs) fused to the transmembrane portions of either the SLC34A2 or CD74 genes in lung tumors (Rikova et al., 2007).
1.5. Chromosomal translocations and cancer development
1.5.1. Roles of translocations in cancer
It has become clear that some translocations involving certain genomic loci are highly associated with specific tumor types (Mitelman et al., 2007). In cases where the approach of targeting the chimeric protein produced by chromosomal translocation leads to a successful therapy (Hughes et al., 2006), there is little room for doubt that tumorigenesis depends upon the acquisition and expression of the translocation product. In less certain situations, in vivo and in vitro methods have been used to investigate the role of particular translocations. For example, expression of the 210-kDa BCR–ABL protein (the product of the Philadelphia chromosome) in retro-virally transduced bone marrow cells is sufficient to cause a CML-like disease in mice (Pear et al., 1998; Zhang and Ren, 1998). Likewise, transgenic overexpression of the c-Myc oncogene, deregulated by linkage to an IgH or IgL enhancer element at a level comparable to the deregulated expression resulting from Ig/Myc translocations, clearly demonstrated that the deregulated oncogene expression that results from such a chromosomal rearrangement can lead to the development of malignancy (reviewed in Morgenbesser and DePinho, 1994). However, even in the case of the MYC transgenic models, the tumors that arise are clonal, indicating that additional genetic alterations are involved.
The BCL-2 protein has antiapoptotic activities and thus can promote the survival of aberrant cells otherwise targeted for death via apoptosis (Youle and Strasser, 2008). The t(14;18) translocation, which fuses the IgH and BCL-2 loci, is a common genetic aberration found in certain human lymphoid malignancies. About 90% of follicular lymphomas and 20–30% of diffuse large B cell lymphomas have translocations that lead to deregulated expression of the translocated bcl-2 gene (Küppers, 2005). Notably, over 50% of healthy individuals have small numbers of B cells with t(14;18) translocation that never progress to cancer (Schüler et al., 2003). The reason for this is that even though high levels of the BCL-2 protein protect cells from early death by apoptosis, deregulation of BCL-2 alone is not sufficient for the development of B cell lymphomas. Thus, there is a requirement for additional genetic events. In this regard, mice carrying an IgH–bcl-2 fusion transgene exhibited an abnormal expansion of resting B cells and lymphoid hyperplasia (McDonnell et al., 1989). Yet, only after about 16 months, roughly 10% of these transgenic mice developed high-grade diffuse large B cell lymphomas. Such lymphomas that do develop in these older IgH–bcl-2 transgenic mice frequently exhibit a rearranged c-Myc gene (McDonnell and Korsmeyer, 1991). Thus, studies with this mouse model provide a clear illustration that progression from benign follicular hyperplasia to malignant lymphoma requires multiple genetic changes.
1.5.2. Passenger versus driver translocations
Major efforts and substantial resources are now being devoted to projects aimed at characterizing cancer genomes, with the goal of better elucidating sets of mutations in cancer cells (Stratton et al., 2009). However, such studies only examine the final outcome of the multistep transformation process, and the complex set of genomic alterations obtained can be difficult to fully interpret. Thus, while the cancer genome sequencing projects are data-rich, many of the genetic alterations in given tumors may not play a critical role in tumor development (Stratton et al., 2009). The concept of “driver” versus “passenger” translocations usefully distinguishes between mutations that are causally implicated in the initiation of oncogenesis and those that may occur, even recurrently, without being integral to the process. Indeed, it now appears that the complexity of genomic alterations in cancer is much higher than expected. In particular, making the distinction between real oncogenic “driver” mutations or translocations from “passenger” events can be difficult.
Cancers, as illustrated by the c-Myc translocations in the lymphomas arising in IgH–bcl-2 transgenic mice, may have required more than one driver to take hold, and likely the number of drivers will vary between cancer types. Moreover, mouse B cell lymphoma modeling studies have suggested that mechanistic factors, in the absence of selection, can drive the appearance of recurrent translocations in tumor progenitor cells at sufficiently high frequency that they appear as recurrent “passenger” translocations in the tumors (Wang et al., 2009). In this context, some recurrent translocations, like those in advanced stage prostate cancer, could theoretically be promoted by mechanistic factors, such as proximity induced by androgen-dependent transcription in early stage tumors (Lin et al., 2009; Mani et al., 2009). Also, mechanistic factors have been implicated in promoting the appearance of the translocation of one oncogene versus another one with equivalent transforming activity in mouse tumor models (Gostissa et al., 2009b; see below). In meeting the goal of identifying all driver mutations within cancer genomes, it will be necessary to more fully understand the mechanistic factors that can give rise to adventitious passenger translocations or promote particular types of recurrent translocations. The influence of mechanistic factors on translocations will be discussed in more depth below.
2. DNA RECOMBINATION IN B AND T CELL DEVELOPMENT
2.1. Overview
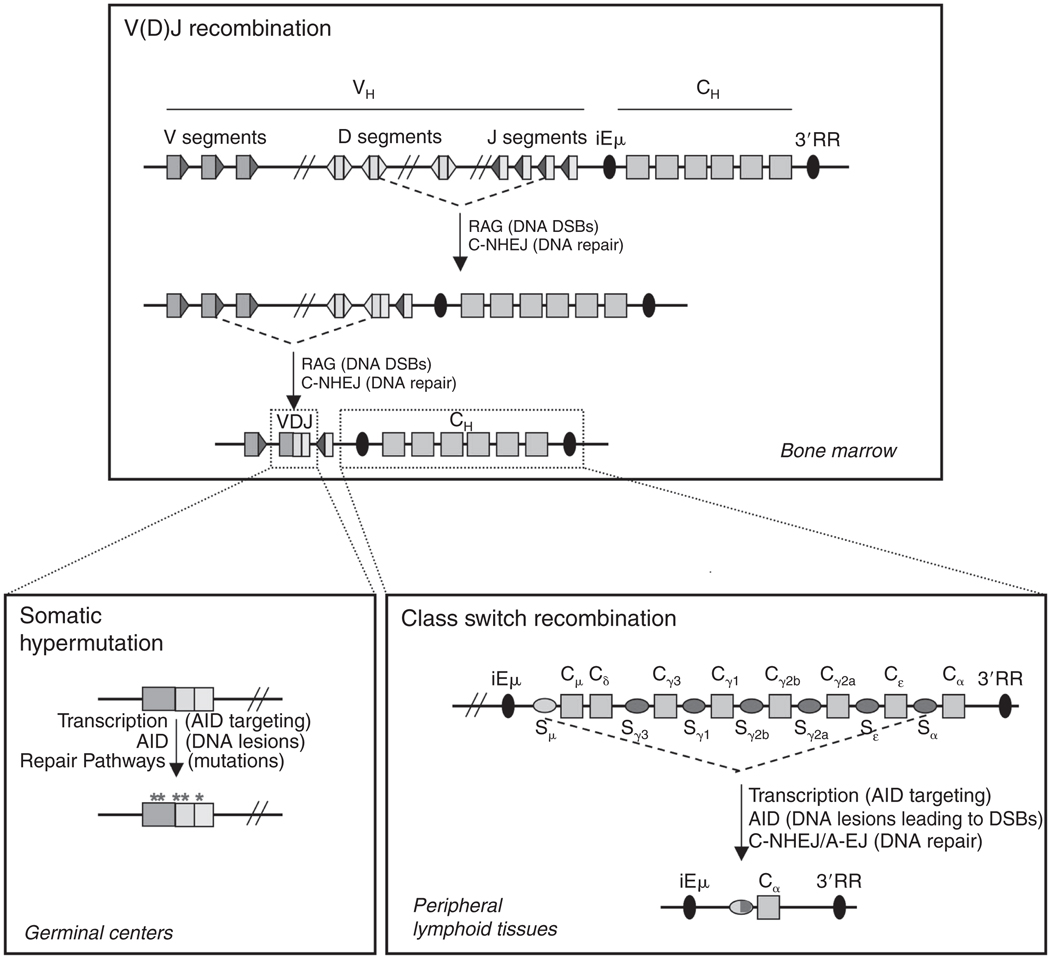
The V(D)J recombination events employed to assemble antigen receptor variable region exons in developing B and T lineage cells and the IgH CSR events that exchange IgH constant region exons in mature B cells are all initiated by DNA DSBs (Fig. 4.1). These lymphocyte-specific programmed DNA DSBs are normally repaired by general cellular DNA repair pathways that join broken DNA ends in the absence of large regions of homology and which are termed end-joining pathways. As will be outlined in more depth below, abnormalities in these lymphocyte-specific DNA breakage and joining pathways, whether it be at the introduction or joining of the programmed DSBs (or both), represent a major source of potential initiating events for chromosomal translocations that lead to lymphoid malignancies.
FIGURE 4.1.
DNA rearrangement events in the IgH locus during B cell development. (See Text for details.)
2.2. V(D) J recombination
Ig and TCR receptor variable region exons are assembled from V, (D), and J gene segments. Ig heavy chain and TCRβ and TCRδ variable exons are assembled from V, D, and J segments; while IgL and TCRα and TCRγ variable region exons are assembled from just V and J segments (Dudley et al., 2005). While the overall organization and content of individual Ig and TCR loci varies, each has numerous V, D, and J segments spread over large chromosomal distances. For example, the mouse IgH locus has 4 J segments, 11 D segments, and hundreds of V segments organized as clusters that are spread over several megabases at the telomeric end of chromosome 12 (Fig. 4.1). The process of “V(D)J recombination” assembles the germ line V, D, and J segments into Ig and TCR variable region exons, respectively, in developing B and T lymphocytes. The TCRα and TCRδ locus have an unusual organization in that the TCRδ locus is fully contained within the TCRα locus, such that TCRα V(D)J recombination deletes the TCRδ locus (Dudley et al., 2005). This unusual organization was the source of some confusion regarding the recurrent translocations that appear in ATM-deficient mouse thymic lymphomas. Thus, ATM-deficient thymic lymphomas were for years thought to have recurrent translocations that involved aberrant TCRα locus V(D)J recombination leading to translocations and oncogene activation by the TCRα enhancer; but, in fact, more recent studies have shown that these translocations arise during attempted TCRδ locus recombination and are independent of the TCRα enhancer (Zha et al., 2010).
Recombination activating gene 1/2 (RAG) endonuclease initiates V(D)J recombination by cleaving V, D, and J segments (Oettinger et al., 1990; Schatz and Baltimore, 1988; Schatz et al., 1989), which are then joined by general cellular end-joining pathways to form V(D)J exons (Taccioli et al., 1993). Thus, the “V(D)J recombinase” is comprised of both lymphoid-specific and generally expressed components. The RAG target site, called a recombination signal sequence (RSS), consists of seven base pair palindromic heptamer (canonical heptamer is CACAGTG) separated by a spacer of 12 or 23 base pairs from an AT-rich nonamer (Hesse et al., 1989; Max et al., 1979; Sakano et al., 1979). The RAG proteins follow a “12/23” rule, in that two gene segments will be cleaved only if one segment is abutted by a 12-bp spacer RSS, and the other by an RSS with a 23-bp spacer (Tonegawa, 1983). The RSS motifs can vary in sequence, within limits, from one gene segment to the next and still support RAG cleavage. This is significant because there are many sequences in a mammalian genome that fortuitously resemble RSSs and which may be targeted for cleavage in developing B and T cells (Ferguson and Alt, 2001; Kim et al., 2000), or even in more mature B cells (Wang et al., 2009), that express RAG. As will be discussed later (Section 3), the V(D)J recombination process entails a certain degree of risk, and evidence of this risk is seen as oncogenic translocations in some lymphoid and myeloid tumors.
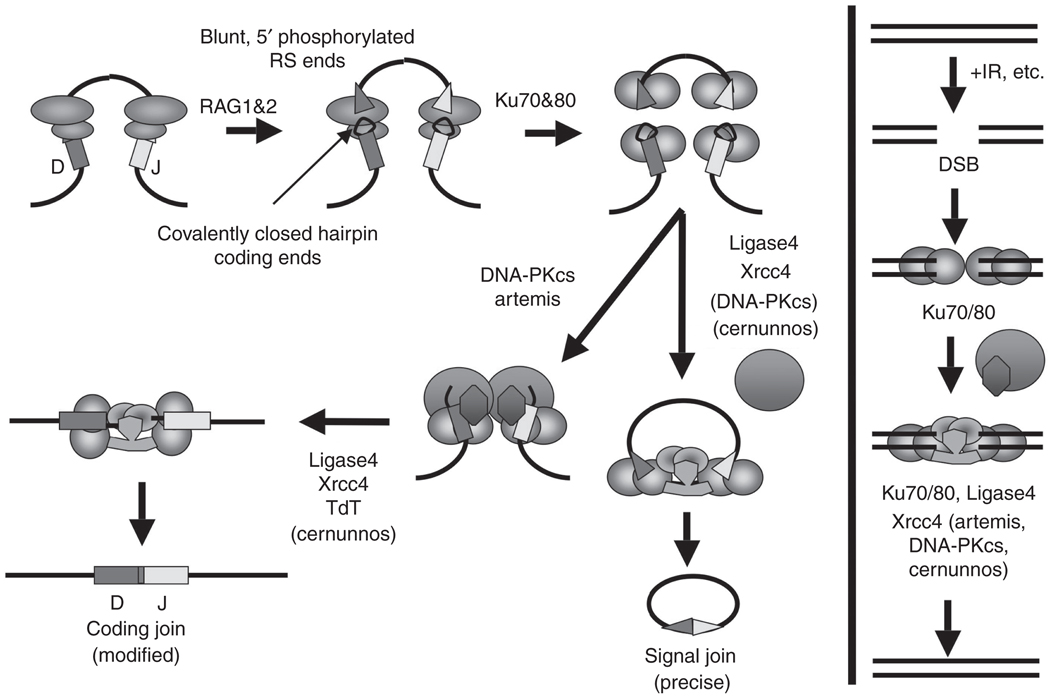
V(D)J recombination can be divided into several steps (Dudley et al., 2005) (Fig. 4.2). Once an appropriate 12/23 pair of RSSs are engaged by RAG, one strand at the site just between each RSS and its adjacent gene segment is hydrolyzed. The 3′-OH at the break is then used as the nucleophile in the attack of the second strand just opposite. This process creates a pair of DSBs, at which each RSS has a blunt 3′-hydroxylated DSB end and each potential V, D, and J coding gene segment has a hairpin terminus. Notably, the RAG proteins carry out a transposition-related reaction and appear to have evolved from a primordial transposase; these potentially harmful transposition activities of the RAG proteins are normally suppressed by regions of the RAG proteins not required for normal catalytic activity (Mundy et al., 2002; Sekiguchi et al., 2001). In the second step of the V(D)J recombination reaction, the two broken RSS ends and two broken coding ends are, respectively, joined by the generally expressed classical nonhomologous DNA end-joining pathway (C-NHEJ; see below for more detailed description). The RSS ends are directly ligated without further processing; but the coding end hairpins must be opened and usually further processed before joining. All but one of the known proteins (Tdt) (Alt and Baltimore, 1982; Gilfillan et al., 1993; Komori et al., 1993) involved in trimming and joining the coding ends are general DNA repair factors in the C-NHEJ pathway (see below).
FIGURE 4.2.
Classic nonhomologous end joining in V(D)J recombination and general double-strand break repair. (Adapted from Dudley et al., 2005). (See text for details.)
RAG expression is restricted primarily to developing B and T lymphocytes in the bone marrow and thymus, respectively (Monroe et al., 1999; Yannoutsos et al., 2001; Yu et al., 1999); although some recent studies suggest that it may also occur in certain mature B lymphocytes (Wang et al., 2009). RAG expression is also restricted to the G1 phase of the cell cycle (Desiderio et al., 1996). Gene targeted mutations showed that RAG-1 and RAG-2 are necessary for initiation of V(D)J recombination in vivo (Mombaerts et al., 1992; Shinkai et al., 1992); thus, RAG-1 or -2 deficient mice are blocked in T and B cell differentiation and exhibit a severe combined immunodeficiency (SCID). In humans, hypomorphic, “leaky” mutations of the RAG genes are the most common cause of Omenn Syndrome (OS), a combined immunodeficiency associated with severe organ damage due to infiltrating activated, anergic, and oligoclonal T lymphocytes (Villa et al., 1999).
The RAG endonuclease appears to have additional functions beyond RSS recognition and cleavage that might serve to direct its activity and, thus, impact on its potential to initiate translocations. A PHD finger in the C-terminal of RAG-2 has been identified recently by several groups (Liu et al., 2007; Matthews et al., 2007; Ramón-Maiques et al., 2007). This motif specifically recognizes trimethylated histone H3 tail (H3K4me3), which leads to RAG-2 binding to sites of this histone modification genome wide and not just within Ig loci (Ji et al., 2010). In addition, recent biochemical studies suggested H3K4me3 not only tethers the RAG enzyme complex to a region of DNA, but also induces a substantial increase in the catalytic turnover number (k(cat)) of the RAG complex (Shimazaki et al., 2009). Of great relevance to potential roles in translocations, several studies have indicated that a postcleavage RAG1/2 complex is involved in later stages of V(D)J recombination and that RAG may be required to direct the repair of V(D)J recombination DSBs by general cellular DSB response and repair machinery (Huye et al., 2002; Qiu et al., 2001; Yarnell et al., 2001). Most notably, the C-terminus of RAG-2 is important in ensuring that RAG-generated DSB intermediates are delivered to the C-NHEJ machinery (Corneo et al., 2007; Cui and Meek, 2007; Lee et al., 2004). If the RAG-2 protein contains a frameshift replaces the 166 carboxy-terminal amino acids with 27 novel amino acids, it forms a mutant RAG complex that will be able to catalyze V(D)J recombination in NHEJ-deficient cells via an alternative end-joining (A-EJ) pathway (Corneo et al., 2007). Notably, A-EJ appears potentially more prone to catalyzing translocations than C-NHEJ (Simsek and Jasin, 2010; Yan et al., 2007; Zhu et al., 2002; see below).
During B cell differentiation in the bone marrow (or fetal liver), RAG (and consequently the V(D)J recombination reaction) first assembles IgH variable region gene exons in progenitor-B cells. Following the successful assembly and expression (as a µ IgH chain) of an IgH V(D)J, RAG is then retargeted to the IgL (Igκ and Igλ) loci in precursor B cells. Following assembly and expression of an IgL chain that associates with the preexisting IgH chain, the resulting B lymphocytes express the complete Ig molecule on their surface and migrate to the periphery where they can be activated to undergo IgH CSR, a separate form of IgH locus rearrangement (see below). During T cell development in the thymus, developing T cells first undergo V(D)J recombination at the TCRβ, TCRδ, and TCRγ loci in so-called double negative progenitor T cells (Kreslavsky et al., 2010). Successful rearrangement of TCRδ and TCRγ genes leads to the development of T cells that express a γ/δ TCR; whereas successful rearrangement and expression of TCRβ genes leads to the subsequent rearrangement of TCRα genes and the ultimate development of α/β TCRs and α/β T cells (Kreslavsky et al., 2010). Following assembly of the gene segments leading to TCR expression, T cells do not undergo any further genomic alterations.
Aberrant joining of RAG-initiated DSBs in both Ig and TCR loci generally has been linked to the generation of oncogenic translocations associated with immature human and mouse B and T lymphomas and leukemias. The identity of the locus that participates in the translocations also can give information about the developmental stage in which the translocations occurred. For example, the recurrent TCRδ locus translocations found in ATM-deficient thymic lymphomas indicates that the translocations were initiated in thymocytes at the early double negative developmental stage (Zha et al., 2010). However, oncogenic translocations that are apparently RAG-initiated are also found in certain human mature B cell lymphomas and might either result from the persistence of unrepaired RAG-initiated DSBs through development in the absence of DSB checkpoints (e.g., in association with ATM deficiency; Callén et al., 2007) or from the persistence of translocations that occurred early in development but which were not activated until later when the IgH3′RR became active (Gostissa et al., 2009a; Janz, 2006; see below). Alternatively, they might also be initiated by RAG expression in certain “mature” lymphocyte populations in the periphery (e.g., Wang et al., 2009) (see below).
2.3. IgH class switch recombination (CSR) and somatic hypermutation (SHM)
Surface IgM expressing B lymphocytes migrate to peripheral lymphoid tissues (e.g., spleen) where they can be activated to undergo CSR, which changes the expressed IgH constant region exons from Cμ to one of a set of downstream constant region exons (generically called “CH genes,” e.g., Cγ1) (Chaudhuri et al., 2007). In the mouse, there are seven CH genes imbedded in the 200 kb region downstream of the Cμ gene (Fig. 4.1). Each of the germline CH genes (except Cδ) is flanked just upstream by long (up to 12 kb), repetitive switch (S) regions. CSR involves the introduction of DSBs into the “donor” S region upstream of Cμ (Sμ) and into a downstream acceptor S region, which are then end-joined to complete CSR. As a consequence, CSR introduces a large deletion of all the sequences between the donor and acceptor junction, including the Cμ gene; thereby, placing a different CH gene downstream of the expressed V(D)J and allowing the B cell to express Ig with a different IgH constant region and effector function (Chaudhuri et al., 2007; Stavnezer et al., 2008).
CSR is initiated by the activation-induced cytidine deaminase (AID) enzyme (Muramatsu et al., 2000) which introduces lesions into S regions that are processed through the co-opted activities of normal cellular DNA lesion repair pathways to generate DSBs (di Noia and Neuberger, 2007). AID is a single-strand DNA-specific cytidine deaminase that is targeted to duplex S regions via transcription. Each S region is preceded by its own transcriptional promoter that is responsive to certain cytokines and activators allowing CSR. This organization allows CSR to be transcriptionally directed to a particular acceptor S region by outside signals from T cells and other immune cells. (Chaudhuri et al., 2007). AID is induced in antigen-activated B cells and is essential not only for introducing DSBs during CSR but also for introducing SHMs into variable region exons in germinal center (GC) B cells (Muramatsu et al., 2007). Exactly how the DSB versus mutational activities of AID are modulated in CSR and SHM is not known; but it likely has some relationship to differences in the S region and variable region exon target sequences.
Not surprisingly, AID activity during the CSR process has been implicated in generating chromosomal translocations (Franco et al., 2006a; Ramiro et al., 2006). Additional evidence has also implicated AID activity during SHM in chromosomal translocation initiation (Dorsett et al., 2007; Pasqualucci et al., 2008). Indeed, the DSB-initiating activity of AID on S regions is so robust that activated B cells in culture can generate sufficient IgH locus DSBs, such that well over 50% can be induced to undergo CSR in culture over a several day period (Boboila et al., 2010a; Cheng et al., 2009; Zhang et al., 2010). How the activities of AID are directly primarily to the IgH S regions in B cells activated for CSR remain unknown. During the process of SHM in antigen-activated GC B cells, AID introduces point mutations, and more rarely, small insertions or deletions, in the variable region exons of the IgH and IgL genes. The mutation frequency is substantial, at about 10−3 to 10−4 per basepair per generation, allowing the selection of GC B cells with improved antigen specificity and affinity (Li et al., 2004). AID targets so-called RGYW and WRCY motifs, of which AGCT is a consensus sequence (Rogozin and Kolchanov, 1992); these sequence motifs are abundant both within variable region exons and even more so in S regions (Chaudhuri et al., 2007). However, these motifs alone, even with specific transcription, do not appear sufficient to explain the great specificity of AID for Ig loci (Wagner et al., 1995). Despite the great specificity of AID for Ig loci, the enzyme has been found to have numerous other targets, albeit at highly reduced efficiency (Liu et al., 2008; Odegard and Schatz, 2006; Robbiani et al., 2009). Indeed, AID can induce the DSBs in both the IgH and c-Myc loci that mediate IgH to c-Myc translocations in activated mouse B cells in culture (Robbiani et al., 2009).
2.4. DNA double strand break (DSB) repair during V(D) J recombination and IgH CSR
There are several known DSB repair pathways in mammalian cells. Following DNA replication, DSBs can be repaired by homologous recombination (HR) using the homologous chromosome as a template for accurate repair (reviewed by Bernstein and Rothstein, 2009; Li and Heyer, 2008; Strathern et al., 1995). HR, as its name implies, requires long stretches of homology to initiate repair. This pathway is not thought to function in G1 phase cells where V(D)J and CSR are initiated. Single-strand annealing (SSA) is also a homology-dependent mode of joining and is closely related to HR (Ivanov et al., 1996; Liang et al., 1998; Lin et al., 1984). SSA mainly occurs at DSBs between two close direct repeats (distances smaller than 25 kb). DNA ends at DSBs are generally processed to 3′-single-stranded tails. Thus, in SSA, direct repeat sequences next to each side of a DSB can anneal afterward and mediate religation, whereas extruded, noncomplementary regions between both homologous sequences are cleaved away. Accordingly, SSA introduces deletions into the repaired product.
In the G1 cell-cycle phase, DSBs appear to be repaired mainly by classical nonhomologous end-joining (C-NHEJ) which is a pathway that joins ends independent of homology (termed “blunt” or “direct” joining) or ends with very short (usually 1–2 bp) overlap (termed microhomology (MH)-mediated joining). In contrast to HR and SSA, C-NHEJ is unlikely to recreate the same sequence that existed prior to damage, unless the ends happen to be cohesive; but it can rejoin ends with minimal loss of sequence (Clikeman et al., 2001; Guirouilh-Barbat et al., 2004; Lin et al., 1999). C-NHEJ operates throughout the cell cycle (Mills et al., 2004; Takata et al., 1998), although perhaps less robustly in S phase (Chen et al., 2005; Lee et al., 1997). In the absence of C-NHEJ factors, ends can also be joined by “alternative” end-joining, a still poorly characterized end-joining pathway or pathways. Because of their relevance to V(D)J and CSR, we will describe in more detail below what is known about C-NHEJ and A-EJ pathways.
2.5. Classical nonhomologous end-joining (C-NHEJ)
The C-NHEJ pathway was initially revealed by the finding that three noncomplementing IR-sensitive Chinese hamster ovary cell mutants were deficient for ability to join RAG-initiated DSBs (Taccioli et al., 1993), ultimately leading to the identification of the Ku80, Xrcc4, and DNA-PKcs genes as C-NHEJ components (Lieber et al., 2004; Rooney et al., 2004b). Subsequently, additional components (e.g., Ku70 which forms a DNA end binding complex with Ku80 and DNA ligase 4 which forms a ligation complex with Xrcc4) were identified based on interactions with known C-NHEJ components and all were shown to function in DNA DSB repair by knockout studies in mice (reviewed by Franco et al., 2006b; Lieber et al., 2004; Rooney et al., 2004b; Zha et al., 2009). Another C-NHEJ factor, Artemis, was identified based on its mutation in certain radio-sensitive SCID patients (Moshous et al., 2001). Most recently, yet another potential C-NHEJ factor, termed Cernunnos or XLF, was also identified based on its mutation in radio-sensitive, immune-deficient patients (Ahnesorg et al., 2006; Buck et al., 2006). It has been shown that XLF/Cernunnos stimulates the ligation reaction of noncompatible DNA ends in vitro (Ahnesorg et al., 2006) and is required for gap filling of partially cohesive ends by polymerase λ or µ (Akopiant et al., 2009). However, unlike all of the other C-NHEJ factors mentioned above, XLF is not required for V(D)J recombination in mice so its precise role in C-NHEJ remains to be fully determined (Li et al., 2008).
C-NHEJ is initiated by binding of the Ku70/Ku80 heterodimer to the broken ends (Fig. 4.2). Ku70/Ku80 forms a ring-like structure that can accommodate double-stranded DNA while serving to protect the ends and recruit other components of the C-NHEJ machinery (Mahajan et al., 2002; Mari et al., 2006; Mickelsen et al., 1999; Walker et al., 2001). The DNA-PK catalytic subunit (DNA-PKcs) bound to DNA with Ku70 and K80 form the fully functional DNA-PK holoenzyme (Falck et al., 2005; Gell and Jackson, 1999; Singleton et al., 1999). Like ATM, DNA-PK belongs to the PIKK (Phosphatidylinositol-3-phosphate-related kinases) family of protein kinases, and once assembled with Ku at a DNA end, DNA-PKcs is activated. One of its main functions in C-NHEJ is to promote synapsing of two broken DNA ends, while exposing them to enzymes that can accomplish necessary modifications prior to ligation (DeFazio et al., 2002; Spagnolo et al., 2006; Yaneva et al., 1997). Recently, in vitro studies have indicated that Ku70/80 on its own is a 5𠈲-dRP/AP lyase that can act upon an abasic site occurring at double-strand end (Roberts et al., 2010). Other end-modifying functions would include 5𠈲–3𠈲 exonucleolytic trimming by Artemis (Dahm, 2007; Niewolik et al., 2006). Gap filling apparently is accomplished by polymerase µ or λ (Uchiyama et al., 2009). Finally, ligation of the processed DNA ends is mediated by a C-NHEJ factor complex consisting of ligase 4 and its cofactor Xrcc4 (Grawunder et al., 1997).
Ku70, Ku80, Xrcc4, and ligase 4 are conserved NHEJ factors from yeast to mammalian cells and are absolutely required for both the coding and RSS joining reactions during V(D)J recombination; these four factors are considered “core” C-NHEJ factors (Rooney et al., 2004b; Zha et al., 2009). In contrast, the nonevolutionarily conserved factors DNA-PKcs and Artemis, which have major roles in joining ends that require processing, are relatively dispensable for V(D)J RSS joining, but absolutely required to open V(D)J coding end hairpins prior to joining (Lieber et al., 2004; Rooney et al., 2004a). Deficiencies for any of these C-NHEJ factors lead to severe combined immune deficiency due to the inability of progenitor lymphocytes to join V(D)J coding ends (Mills et al., 2003; Rooney et al., 2004b). Indeed, deficiency for Xrcc4 or ligase 4 leads to late embryonic lethality associated with the widespread death of newly differentiated neurons due to a p53-dependent response to unrepaired DSBs, indicating the broad importance of this pathway in repairing DNA damage (Gao et al., 1998). In addition, deficiency for any core C-NHEJ factor, as well as for DNA-PKcs or Artemis, leads to frequent chromosomal translocations in a wide variety of cell types, clearly indicating that C-NHEJ factors are required to suppress translocations and further indicating that, in the absence of C-NHEJ, other repair pathways can robustly generate translocations (Difilippantonio et al., 2000; Ferguson et al., 2000; Gao et al., 2000). Finally, the role of C-NHEJ pathway in suppressing translocations might also reflect the propensity of this pathway to join DSBs within a chromosome, as opposed to joining to DSBs on other chromosomes to generate translocations (Ferguson et al., 2000; Zarrin et al., 2007). How C-NHEJ might be preferentially restricted to intrachromosomal joins is unknown but one hypothesis is that such a restriction might be imposed if C-NHEJ was somehow restricted to an intrachromosomal DSB response (Boboila et al., 2010b), which is also required to prevent DSBs from separating into chromosomal breaks and generating translocations (Franco et al., 2006a,b; see below).
2.6. Alternative end-joining (A-EJ)
Evidence for an alternative end-joining pathway or pathways came from studies that showed C-NHEJ-deficient cell could still fuse DNA ends via end-joining (Boulton and Jackson, 1996; Kabotyanski et al., 1998; Wang et al., 2003; Zha et al., 2009). Indeed, alternative modes of end-joining can be differentiated from C-NHEJ both by biochemical or genetic requirements in various systems (Iliakis et al., 2004; Wang et al., 2006; Yan et al., 2007). Factors including NBS1, Mre11, CtlP, DNA Ligase 3, Parp1, and XRCC1 have been suggested to be components of the A-EJ pathways (Audebert et al., 2004; Deriano et al., 2009; Dinkelmann et al., 2009; Rass et al., 2009; Wang et al., 2005, 2006; Xie et al., 2009), but there is still no agreement as to whether A-EJ pathway is a single or multiple pathways. Indeed, it seems likely that there is more than one form of A-EJ and, to date, the best definition of A-EJ would be any form of end-joining that occurs in the absence of C-NHEJ factors.
End-joining activities that occur in the absence of C-NHEJ have been demonstrated in experiments where naked linear DNA with various end structures is transfected into C-NHEJ deficient cells. Notably, cells deficient in DNA-PKcs (Chang et al., 1993; Harrington et al., 1992), Ku (Kabotyanski et al., 1998), Xrcc4 (Kabotyanski et al., 1998), or DNA ligase 4 (Verkaik et al., 2002) were able to carryout linear DNA recircularization via end joining. Likewise, DSBs introduced at chromatinized genomic locations were also repaired in NHEJ-deficient cells (Guirouilh-Barbat et al., 2004; Rass et al., 2009; Xie et al., 2009). Further, it has been found that CSR can be catalyzed at levels approaching 50% of those in WT cells in the absence of any of the core C-NHEJ factors (Boboila et al., 2010a; Han and Yu, 2008; Yan et al., 2007). One possibility is that in the absence of one C-NHEJ component, C-NHEJ proceeds but at reduced levels perhaps by some other factor filling in for the missing component (e.g., DNA Ligase 1 or Ligase 3, substituting for Ligase 4). However, it was recently found that CSR can still occur at similar levels in B cells deficient in both Ku70 and DNA ligase 4, the DSB recognition and joining components of C-NHEJ. These latter findings provide the strongest evidence to date that A-EJ is a totally distinct pathway (or pathways) from C-NHEJ. Notably, A-EJ does not function at all in V(D)J recombination, which is completely reliant on C-NHEJ for junction formation. This restriction is mediated by RAG which shepherds the V(D)J recombination reaction specifically into C-NHEJ and excludes other repair pathways (Corneo et al., 2007).
During both V(D)J recombination and CSR, C-NHEJ generates direct junctions and junctions with short MH at relatively similar levels (Boboila et al., 2010a; Komori et al., 1993; Yan et al., 2007). Notably, in several of the systems outlined above, A-EJ was associated with the increased usage of MH-mediated end-joining. Thus, in Xrcc4 or Ligase 4-deficient cells, CSR junctions were essentially all MH-mediated and the MHs were on average longer than those associated with C-NHEJ. However, the frequent usage of MH by A-EJ may be manifested predominantly in situations where the substrate, such as S regions, readily provides long MH. In other cases, for example joining of chromosomally integrated I-SceI substrates, a substantial proportion of joins in Ku or Ligase 4-deficient cells were direct (Guirouilh-Barbat et al., 2004). Again, as outlined above, the most useful current definition of A-EJ is end-joining not reliant on large stretches of homology (e.g., as required by SSA) that occurs in the absence of C-NHEJ components.
The relevance for C-NHEJ in suppressing oncogenic translocations in mice is well established as C-NHEJ-deficient mice (whether they lack Ku80, Xrcc4, Ligase 4, DNA-PKcs, or Artemis) that are also deficient for p53 routinely develop pro-B cell lymphomas harboring oncogenic chromosomal translocations involving the IgH and c-Myc, or in the case of Artemis deficiency, N-myc genes (Difilippantonio et al., 2000; Frank et al., 2000; Gao et al., 2000; Rooney et al., 2004a; Zhu et al., 2002). Notably, Xrcc4 or Ligase 4-deficient progenitor-B cells, when also deficient for the p53 checkpoint protein, give rise to lymphomas with RAG-dependent IgH translocations in the vicinity of c-Myc, all of which are catalyzed by A-EJ and have MH-mediated translocation junctions (Zhu et al., 2002). This finding led to the first suggestion that A-EJ may be a translocation-prone pathway (Zhu et al., 2002). Similarly, in C-NHEJ-deficient activated B cells, A-EJ frequently joins IgH locus breaks to other chromosomes to generate translocations, including IgH to c-Myc translocations that also appear as oncogenic translocations in p53-deficient B cells (Wang et al., 2009; Yan et al., 2007). Likewise, p53-deficient mice that lack Xrcc4 in the central nervous system routinely develop medulloblastomas with recurrent translocations (Yan et al., 2006).
A recent report indicated that Ligase 4 and Xrcc4 repress the formation of translocations (Simsek and Jasin, 2010), and found no difference in the fine structure of the translocation junctions formed in their model system with or without Ligase 4 or Xrcc4. This is consistent with the idea that A-EJ is the major pathway mediating chromosomal translocations in the absence of C-NHEJ (Boboila et al., 2010b; Wang et al., 2008; Yan et al., 2007). Analyses of chromosomal translocation junctions also led to the suggestion that A-EJ might predispose to translocations in humans (e.g., Zhang and Rowley, 2006). The reason for frequent A-EJ-catalyzed translocations in the absence of C-NHEJ is not well understood. It could be that this pathway is simply not as biased to forming intrachromosomal joins as is C-NHEJ (Boboila et al., 2010b). Alternatively, it may be that the apparently high frequency of translocations catalyzed by A-EJ simply reflects the fact that more DSBs are not rapidly repaired in the absence of C-NHEJ and, therefore, persist as substrates for A-EJ (Yan et al., 2007).
2.7. DSB response factors
Upon acquisition of a DSB in the genome, the DSB response is activated and DSB response factors are recruited to the broken DNA ends where they activate signal pathways that lead to cell-cycle arrest, initiate repair of the broken DNA ends, and induce apoptosis or senescence if the repair of DSBs cannot be achieved (Sancar et al., 2004). Of particular relevance to the V(D)J and CSR processes, ATM is activated upon formation of DNA DSBs in the G1 phase of the cell cycle (Abraham, 2001; Canman et al., 1998). During the DSB response, ATM phosphorylates and activates p53, which monitors DSBs in the context of G1 and G2/M checkpoints and signals cell-cycle arrest or apoptosis (Vogelstein et al., 2000). ATM also phosphorylates additional substrates that are key in cell-cycle checkpoint responses, in recruiting or regulating repair factors, and in immobilizing broken DNA ends prior to repair; these substrates include histone H2AX, MDC1, and 53BP1 (Bassing and Alt, 2004).
The histone variant H2AX is one of the most conserved H2A variants across species (Kinner et al., 2008). Upon formation of a DSB, H2AX molecules in nucleosomes flanking DSBs are phosphorylated on serine 139 to form γ-H2AX (Rogakou et al., 1998). The phosphorylation event is initially localized to the site of the DSB but then spreads to a large region (up to 100 kb) around the DSB (Rogakou et al., 1998, 1999; Savic et al., 2009). γ-H2AX carries out its DSB response functions by signaling DSBs and forming a platform for the recruitment of multiple proteins of the DSB response pathway to form large γ-H2AX foci (Kinner et al., 2008). Although evidence suggests that MDC1 is a main interaction partner for γ-H2AX in higher eukaryotes, other interaction partners (e.g., NBS1, 53BP1) have been described (Kobayashi et al., 2002; Stewart et al., 2003; Ward et al., 2003), and are recruited to γ-H2AX foci. Phosphorylated serine 139 on H2AX plays a role as an anchor residue for protein–protein interactions, apart from which it may also serve to promote open chromatin structures due to its negative charge. It has been observed that local chromatin density decreases after generation of DSBs (Downs et al., 2000).
DSB response factor deficiencies lead to unrepaired chromosomal breaks and translocations. For example, ATM, H2AX, and 53BP1 are needed to prevent IgH locus DSBs generated by AID from progressing to chromosomal breaks and translocations, including oncogenic IgH/c-Myc translocations (Franco et al., 2006a; Ramiro et al., 2006). In addition, H2AX deficiency combined with p53 deficiency leads to T and B cell lymphomas that harbor clonal translocations; in B cells, often recurrent IgH to c-Myc translocations (Bassing et al., 2003; Celeste et al., 2003). These findings led to the proposal that DSB response foci tether chromosomal DSBs to promote C-NHEJ joining within a chromosome and prevent translocations (Bassing and Alt, 2004; Bassing et al., 2003; Boboila et al., 2010b; Franco et al., 2006b; Zarrin et al., 2007). Although deficiency in any one of the described DSB response factors increases genomic instability and reduces CSR, the extent varies for the different factors. The highest levels of genomic instability and chromosomal translocations are associated with ATM deficiency and the most severe CSR defect is observed in 53BP1-deficient cells, the latter possibly suggesting CSR-specific functions of 53BP1 beyond those served by the DSB response per se (Franco et al., 2006a; Ramiro et al., 2006). ATM and, to a lesser extent, 53BP1 are also required for fully normal joining of chromosomal DSBs during V(D)J recombination (Bredemeyer et al., 2006; Difilippantonio et al., 2008).
53BP1 also has been proposed to play a role in regulating the mobility of DNA ends, in particular at deprotected telomeres (de Lange, 2009), possibly indicating that 53BP1 influences chromatin dynamics and facilitates repair of breaks at distant sites (Difilippantonio et al., 2008). DSB response factors that are involved in tethering of broken DNA ends also include NBS1, MRE11, and Rad50, which together comprise the MRN complex. In addition to its function in tethering broken DNA ends, the MRN complex is involved in many cellular processes such as DNA replication, DNA repair, cell cycle checkpoints, and telomere maintenance (D’Amours and Jackson, 2002; Van den Bosch et al., 2003). Genetic studies have suggested a role for the MRN complex in both C-NHEJ and A-EJ during CSR (Deriano et al., 2009; Dinkelmann et al., 2009; Rass et al., 2009; Xie et al., 2009).
3. MECHANISTIC FACTORS THAT INFLUENCE THE APPEARANCE OF CHROMOSOMAL TRANSLOCATIONS
3.1. Overview
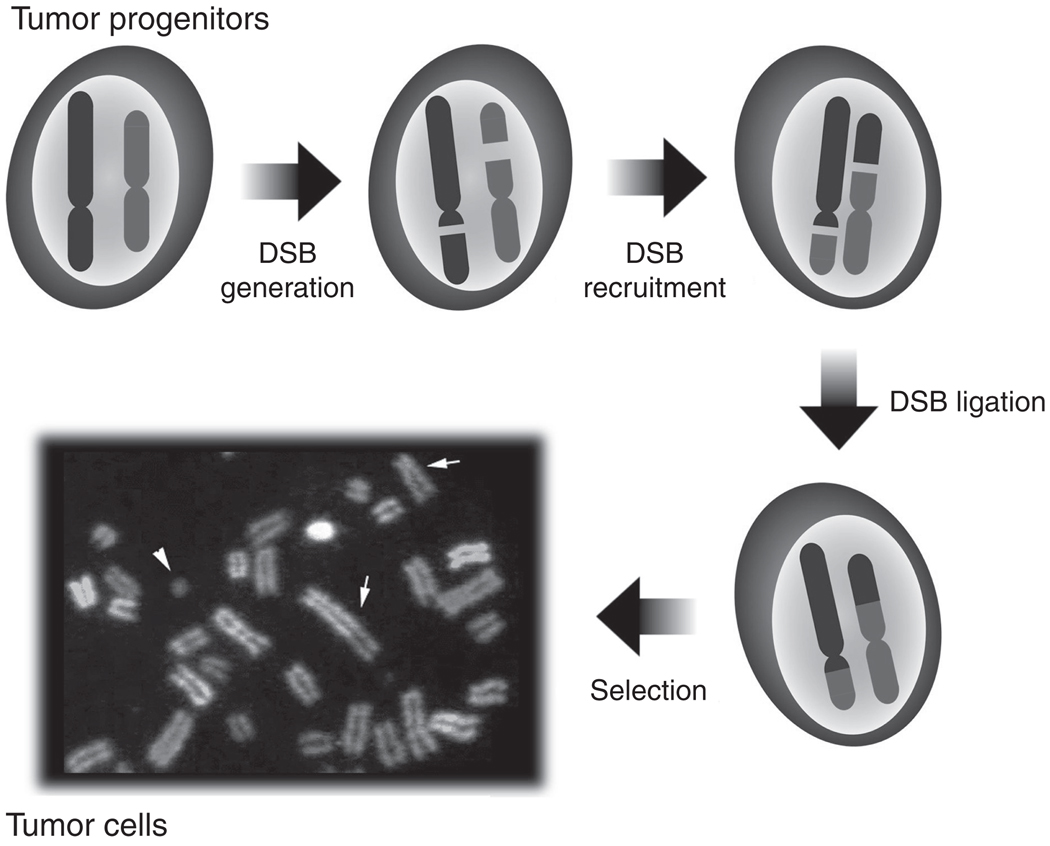
Recurrent translocations in tumors generally are thought to arise spontaneously as low-frequency events that are selected at the cellular level for contribution to oncogenesis (Fig. 4.3). Thus, underlying mechanistic features that influence the generation of well-studied translocations may be masked. While selection for oncogenic activities is clearly a key factor in the appearance of oncogenic translocations, recent studies have shown that mechanistic factors also are basic to the process and can influence appearance of sequences, including cellular oncogenes, in recurrent translocations (Gostissa et al., 2009a,b; Wang et al., 2009). Mechanistic factors that might influence translocations include DSB frequency at translocating loci and factors that influence such DSBs (Robbiani et al., 2008; Wang et al., 2009), factors that contribute to two translocating loci lying in close enough proximity in the interphase nucleus to be joined (Meaburn et al., 2007; Wang et al., 2009), and mechanisms that circumvent functions of the cellular DSB response and repair pathways that promote joining of DSBs within a chromosome and suppress joining of DSBs between chromosomes (Ferguson et al., 2000; Franco et al., 2006a,b; Ramiro et al., 2006).
FIGURE 4.3.
Formation of recurrent reciprocal chromosomal translocations in tumors. First, two DSBs occur at two nonhomologous chromosomes. Then the four broken ends, after escape from normal DSB repair, are synapsed and ligated to form reciprocal chromosomal translocations. Finally, recurrent translocations in tumors are generally thought to arise spontaneously as very low-frequency events that are strongly selected at the cellular level via their contributions to oncogenesis.
3.2. Role of mechanistic factors in promoting chromosomal translocations
3.2.1. Chromosomal position
Accumulating evidence suggests that the genome of eukaryotes is not distributed randomly in the interphase nucleus but rather follows a distinct spatial order (Lanctô t et al., 2007; Lieberman-Aiden et al., 2009; Meaburn et al., 2007). Importantly, spatial genome organization is highly tissue- and cell-type specific and is affected by transcriptional activity, cellular proliferation status, and possibly many pathways (Bridger et al., 2000; Cremer et al., 2003; Parada et al., 2004). The defined spatial localization of a chromosome within the nucleus is referred to as chromosome territory (Cremer and Cremer, 2001; Foster and Bridger, 2005; Parada and Misteli, 2002).
Chromosomal position of two, otherwise, relatively equivalent oncogenes, can greatly influence their ability to contribute to oncogenic translocations. Thus, the c-myc and N-myc cellular oncogenes belong to the same family, have similar functional activity in vivo including oncogenic potential, and are expressed in developing lymphocytes and neuronal cells (Kohl et al., 1983; Malynn et al., 2000; Zimmerman et al., 1986). Yet, translocations in B cell lymphomas, such as the pro-B lymphomas that arise from Xrcc4/p53 deficient developing mouse B cells or in human Burkitt’s lymphomas (Küppers and Dalla-Favera, 2001), frequently involve c-myc but not N-myc (e.g., Difilippantonio et al., 2000; Zhu et al., 2002); while N-myc, but not c-myc, is amplified in human neuroblastomas (e.g., Kohl et al., 1983) and mouse neuroblastomas that arise from Xrcc4/p53 deficient developing neuronal cells (Yan et al., 2006). However, when inserted in place of c-myc coding sequence, N-myc coding sequences compete well with the normal c-myc allele to generate recurrent translocations leading to pro-B cell lymphomas (Gostissa et al., 2009b). Thus, these gene replacement studies rule out cellular selection as the only factor that promotes recurrent c-myc but not N-myc translocations in developing B lineage cells and clearly demonstrate that chromosomal location can markedly influence ability of a cellular oncogene to contribute to recurrent translocations. In this context, chromosomal environment might influence the frequency of DSBs around a potential oncogene, influence its spatial proximity to a translocation partner, alter its chromatin struction or transcriptional regulation, or predispose it to a particular form of DNA repair.
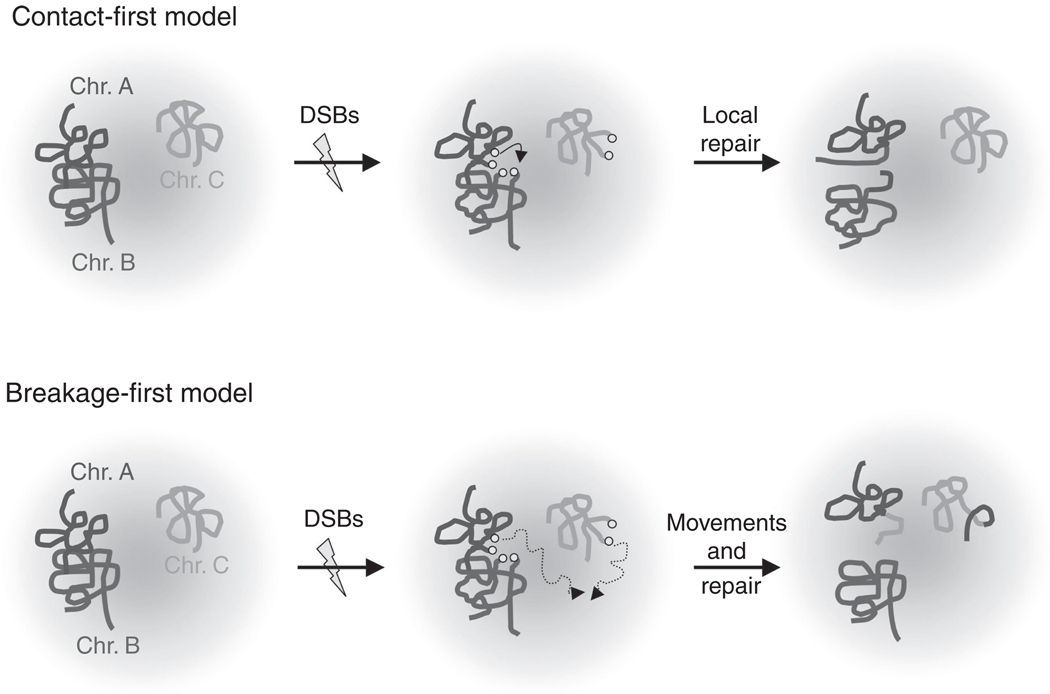
Recent studies have provided a detailed picture of the organization of the genome within the mammalian nucleus that is based on nonrandomly positioned genes and chromosomes (e.g., Lieberman-Aiden et al., 2009). In this context, there are two long-standing models for translocation initiation (Fig. 4.4). The “contact-first” model postulates that translocations are restricted to DSBs that arise in proximally positioned regions of the genome, while the “breakage-first” model suggests DSBs initially located far apart can move together (Meaburn et al., 2007). It should be noted that the breakage- and contact-first models, while extremes, are not necessarily mutually exclusive. In any case, the spatial disposition of chromosomes (i.e., the relative positioning of chromosomal domains or whole chromosomes with respect to each other) likely impacts substantially on patterns of genome rearrangements. Numerous studies have indicated that loci that are proximal to each other in 3D appear to have increased likelihood of engaging in a translocation event (reviewed by Meaburn et al., 2007). Imaging studies have demonstrated that certain test DSBs are relatively immobile in the nucleus (Soutoglou et al., 2007). However, the possibility that some DSBs on distant chromosomes are brought together (synapsed) for repair after their generation, as has been reported for DSBs in yeast cells (Lisby et al., 2003), has not been ruled out and awaits further experimentation.
FIGURE 4.4.
The “contact-first” and “breakage-first” models for chromosomal translocation. (See text for details).
3.2.2. DSBs
Despite the key role of DSBs in translocations and genomic instability, a unified understanding of the elements that predispose to DSBs in many tumors is still lacking. In this regard, it is also worth reemphasizing increased levels of particular DSBs could come either by their increased generation or by their decreased repair.
As outlined above, in lymphomas, many oncogenic translocations arise from joining of DSBs in antigen receptor loci to DSBs located in or around a potential oncogene. In developing B and T lymphocytes, RAG can generate IgH DSBs that lead to oncogenic translocations by joining to either RAG-generated or non-RAG-generated DSBs elsewhere in the genome (Fig. 4.5) (e.g., Difilippantonio et al., 2002; Gladdy et al., 2003; Zhu et al., 2002). In mature B cells, AID plays a key role in generating DSBs including IgH breaks that lead to high levels of translocations in DNA repair and DSB response deficient backgrounds (Franco et al., 2006a; Ramiro et al., 2006; Wang et al., 2009) and IgH DSBs that underlie oncogenic IgH/c-myc translocations in mouse lymphoma models (Ramiro et al., 2004). As mentioned above, RAG-initiated DSBs are handled differentially from most DSBs, as RAG proteins channel their joining exclusively into C-NHEJ (Corneo et al., 2007). CSR breaks, on the other hand, are handled much like general cellular DSBs in that they require the DSB response for normal joining over the 100–200 kb distance involved in CSR and are less dependent on C-NHEJ. Thus, CSR DSBs might be considered a model for “intrachromosomal translocations.” Indeed, AID-initiated IgH locus DSBs can be efficiently joined to yeast I-SceI endonuclease-generated DSBs to cause IgH deletions leading to class switching (Zarrin et al., 2007) or to chromosomal translocations (Robbiani et al., 2008; Wang et al., 2009).
FIGURE 4.5.
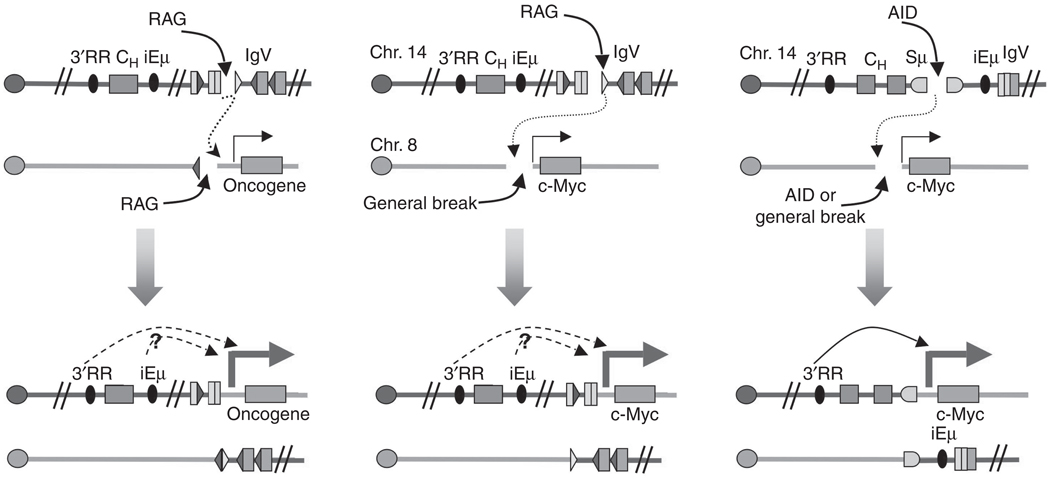
Chromosomal translocations involving aberrant V(D)J recombination and CSR. Left: aberrant V(D)J recombination between RSS in the IgH locus and cryptic RSS in a different chromosome. Middle: Formation of reciprocal translocations between RAG-generated and non-RAG-generated DSBs. In both these cases, the iEμ and IgH3′RR are both linked to c-myc and can potentially contribute to its over-expression (dotted arrows). Right: Formation of reciprocal translocations between AID-generated breaks at the IgH and c-Myc loci; only the IgH3′RR is present in this configuration and it has been shown to enhance c-myc expression (solid arrow). (see text for further details.)
DSBs in translocation partners other than antigen receptor loci might be caused by intrinsic factors, such as oxidative metabolism and replication stress, or extrinsic factors such as ionizing radiation or chemotherapeutic agents (Hoeijmakers, 2009). Another source of localized DSBs in most cells types is chromosome fragile sites (Durkin and Glover, 2007). However, the mechanisms that predispose to fragile site formation have not been fully elucidated. In addition, unusual DNA sequence and structures have also been implicated in generating DSBs and translocations (Zhao et al., 2010). On the other hand, AID can generate “off-target” DSBs in c-Myc that are joined to AID-initiated IgH breaks to form IgH/c-Myc translocations like those found in Burkitt’s lymphoma (Robbiani et al., 2008) (Fig. 4.5). In addition, recent studies suggested a potential collaboration between AID and RAG in initiating DSBs at nonclassical RAG target sites in developing B cells, thereby predisposing to oncogenic translocations found in human acute lymphoblastic leukemias (Tsai et al., 2008). Notably, transgenic overexpression of AID in B cells leads to rather widespread genome instability (Robbiani et al., 2009). Identifications of such AID “off-targets” should provide important insights into the targeting of chromosomal translocations in cancers derived from activated B cells.
Surprisingly, AID has been suggested to be involved in the generation of chromosomal translocations and mutations in nonlymphoid tumors (Lin et al., 2009; Okazaki et al., 2007). In this context, AID has been reported to be expressed in embryonic stem cells, where it is proposed to have a role in demethylation of 5-methylcytosine residues and, thereby, a role in epigenetic regulation (Bhutani et al., 2010). AID has also been implicated in gastric cancers and in recurrent translocations in prostate cancers (Lin et al., 2009; Matsumoto et al., 2007). Thus, further understanding of the role of AID in destabilizing the genome of cells outside the immune system is needed.
3.2.3. Chromosomal features that promote selection
Chromosomal translocations can be subject to various negative and positive selection events in dividing cells. As discussed above, “driver” translocations are causally implicated in the initiation of oncogenesis via positive selection for a conferred growth advantage of the cancer progenitor cell in the microenvironment of the tissue in which it arises. In this regard, most initial chromosome translocation events are indeed quite rare. For example, IgH/c-Myc translocations generally appear to occur in less than 1 in 106 activated mouse B cells (Ramiro et al., 2006; Wang et al., 2009), consistent with strong oncogenic selection playing a major role in their recurrent appearance in mature B cell lymphomas.
Chromosomal features of particular loci beyond their propensity to break or their spatial proximity to translocation partner loci can play a role in the appearance of recurrent oncogenic translocations. Thus, transgenic mice bearing IgH intronic enhancer (iEμ) or IgH3′RR sequences fused to c-Myc, respectively, are predisposed to the development of early and mature B lineage lymphomas, demonstrating that both elements can confer oncogenic c-Myc expression (Adams et al., 1985; Schmidt et al., 1988; Truffinet et al., 2007). However, in most mature B cell lymphomas that arise as a result of errors in IgH CSR (such as certain Burkitt’s lymphomas), IgH/Myc translocations delete iEμ and place c-Myc up to 200 kb upstream of the IgH3′RR (Janz, 2006; Küppers and Dalla-Favera, 2001). However, recent studies have shown that the IgH3′RR, which was previously shown to regulate transcription of CH gene promoters in the context of CSR over long distances (Cogné et al., 1994), can also activate expression of the c-myc oncogene over 200 kb distances and, as such, is required for development of tumors in a mouse model for peripheral B cell lymphomas. (Gostissa et al., 2009a) (Fig. 4.5). Thus, the IgH3′RR is a locus-specific element that plays a critical role in driving the expression of oncogenic IgH/c-Myc translocation in mature B cell lymphomas. Indeed, it is possible that the iEμ element does not play a major role in activating translocated oncogene expression even in IgH locus translocations in which it is retained (Janz, 2006; Gostissa et al., 2009a). Finally, other recent studies have shown that deletion of a 2.2 kb promoter region of c-myc gene severely impairs its transcription and drastically reduces the frequency of IgH/Myc translocations in peripheral B cells (Robbiani et al., 2008). It seems likely that transcriptional regulatory elements in other translocation target loci may similarly play a role in promoting the occurrence of oncogenic translocations.
3.2.4. DNA repair
The potential roles of DNA DSB response and repair pathways in the suppression of translocations have been discussed above. In this context, it is notable that the vast majority of translocation junctions mapped in tumors shows typical characteristics of C-NHEJ or A-EJ (e.g., junctions with no or limited microhomology as well as deletions, insertions, and end modifications) (Gillert et al., 1999; Zhang and Rowley, 2006). These observations are in accordance with studies in model systems revealing extensive use of C-NHEJ or A-EJ in translocation formation (Weinstock et al., 2006a,b). Of note, in model systems, NHEJ-mediated translocation junctions exhibit more extensive use of deletions, insertions, and micro-homologies than junctions of standard NHEJ-repaired DNA DSBs (Weinstock et al., 2006a,b). Tumor patient-derived junctions of reciprocal translocations show comparable patterns (Stephens et al., 2009; Weinstock et al., 2006a), thus, suggesting the existence of potential mechanistic differences.
3.3. Recurrent “passenger” translocations promoted by multiple mechanistic factors
Xrcc4-deficient B cells very frequently translocate RAG-initiated DSBs in the Igλ locus on chromosome 16 to AID-initiated IgH breaks on chromosome 12, likely because the two loci are both frequently broken in the same cells and because they frequently lie in close spatial proximity, and potentially because they are joined by A-EJ. Notably, colocalization of Igλ and IgH in these cells, as measured by 3D interphase FISH, was relatively focal with respect to the position of Igλ on chromosome 16, implicating aspects of particular chromosomal regions, as opposed to broader chromosomal territories, as important in determining spatial proximity and translocation frequency (Wang et al., 2009). Strikingly, these mechanistic factors led to such a high frequency of Igλ to IgH locus translocations in progenitors of Xrcc4/p53 double-deficient B cell lymphomas, that they appear as recurrent translocations in the tumors apparently without conferring any known selective advantage. This finding highlights the potential role of mechanistic factors in helping to promote translocations.
4. PERSPECTIVE
Thus far, most studies of translocation mechanisms have been focused on and limited to a few specific translocations, namely those observed as recurrent translocations in tumors. Although studies from tumor models have provided enormously useful insights on the mechanistic factors involved in chromosomal translocations, new experimental systems will be necessary to advance such studies. Thus, in tumor studies, strong in vivo oncogenic selection for translocation products could have masked the identification of factors influencing early translocation steps. A more representative system to study translocation-initiating events should minimize cellular selection. Recently, work on primary B cells, A-MuLV transformed pro-B cell lines, ES cell lines, and prostate cancer cell lines (Ramiro et al., 2006; Wang et al., 2009; Mahowald et al., 2009; Weinstock et al., 2008; Lin et al., 2009) has provided some direction into the requirements for establishing experimental systems to study the initiation phase of the translocations. In addition, cancer genome studies have provided further insights and suggested that there are more rearrangements in tumors than previously expected (Stephens et al., 2009; Stratton et al., 2009). Also, studies on primary B cells activated for CSR suggested that 1–5% of the metaphases harbor IgH-related chromosomal translocations in DSB response and DSB repair deficient backgrounds (Franco et al., 2006a,b; Wang et al., 2009; Yan et al., 2007). Most of these IgH locus translocations involve unidentified partners, raising the potentialy tractable question of determining the unselected pattern of IgH locus translocations from a genome-wide viewpoint. To further our understanding of the role of various mechanistic factors in promoting translocations, future studies should focus on the development of methods to study these events genome-wide in various cell types and tumor progenitors, an effort that should benefit greatly from the rapid evolution of the genomic tools, especially high-throughput sequencing techniques.
ACKNOWLEDGMENTS
We thank members of the Alt laboratory for helpful comments. This work was supported by grants from NIH/NCI 5PO1 CA109901-05 and 5PO1 CA92625-09. Y. Z. was a fellow of the Cancer Research Institute. M. G. was a fellow of the Leukemia and Lymphoma Society of America. C. B. was supported by a training grant from the Cancer Research Institute. F. W. A. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous endjoining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Akopiant K, Zhou R-Z, Mohapatra S, Valerie K, Lees-Miller SP, Lee K-J, Chen DJ, Revy P, de Villartay J-P, Povirk LF. Requirement for XLF/Cernunnos in alignment-based gap filling by DNA polymerases l and m for nonhomolgous end joining in human whole-cell extracts. Nucleic Acids Res. 2009;37:4055–4062. doi: 10.1093/nar/gkp283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt FW, Baltimore D. Joining of immunoglobulin heavy chain gene segments: Implications from a chromosome with evidence of three D-JH fusions. Proc. Natl. Acad. Sci. USA. 1982;79:4118–4122. doi: 10.1073/pnas.79.13.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- Bartram CR, de Klein A, Hagemeijer A, van Agthoven T, Geurts van Kessel A, Bootsma D, Grosveld G, Ferguson-Smith MA, Davies T, Stone M, et al. Translocation of c-ab1 oncogene correlates with the presence of a Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1983;306:277–280. doi: 10.1038/306277a0. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Swat W, Alt FW. The mechanisms and regulation chromosomal V(D)J recombination. Cell. 2002;109:45–55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. Histone H2AX: A dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Alt FW. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004;3:149–153. doi: 10.4161/cc.3.2.689. [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Rothstein R. At loose ends: Resecting a double-strand break. Cell. 2009;137:807–810. doi: 10.1016/j.cell.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boboila C, Yan C, Wesemann DR, Jankovic M, Wang JH, Manis J, Nussenzweig A, Nussenzweig M, Alt FW, Nussenzweig MC, et al. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J. Exp. Med. 2010a;207:417–427. doi: 10.1084/jem.20092449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boboila C, Jankovic M, Yan CT, Wang JH, Wesemann DR, Zhang T, Fazeli A, Feldman L, Nussenzweig A, Nussenzweig M, Alt FW. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc. Natl. Acad. Sci. USA. 2010b;107:3034–3039. doi: 10.1073/pnas.0915067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Boveri T. Zur Frage der Entstehung maligner Tumoren. Gustav-Fischer-Verlag: Jena; 1914. p. 164. [English translation by Boveri, M. (1929). Arch. Intern. Med. 44, 910] [Google Scholar]
- Bredemeyer AL, Sharma GG, Huang C, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, Sleckman BP. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- Brenner JC, Chinnaiyan AM. Translocations in epithelial cancers. Biochim. Biophys. Acta. 2009;1796:201–215. doi: 10.1016/j.bbcan.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger JM, Boyle S, Kill IR, Bickmore WA. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr. Biol. 2000;10:149–152. doi: 10.1016/s0960-9822(00)00312-2. [DOI] [PubMed] [Google Scholar]
- Buck D, Malivert L, de Chasseval R, Barraud A, Fondanèche MC, Sanal O, Plebani A, Stéphan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, et al. Cernunnos, a novel 111 nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Callén E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, Celeste A, Pellegrini M, McBride K, Wangsa D, Bredemeyer AL, Sleckman BP, Ried T, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Carver BS, Tran J, Chen Z, Carracedo-Perez A, Alimonti A, Nardella C, Gopalan A, Scardino PT, Cordon-Cardo C, Gerald W, Pandolfi PP. ETS rearrangements and prostate cancer initiation. Nature. 2009a;457:E1. doi: 10.1038/nature07738. discussion E2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, Cordon-Cardo C, Gerald W, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat. Genet. 2009b;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Biedermann KA, Mezzina M, Brown JM. Characterization of the DNA double strand break repair defect in scid mice. Cancer Res. 1993;53:1244–1248. [PubMed] [Google Scholar]
- Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, Manis J, Alt FW. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- Chen BP, Chan DW, Kobayashi J, et al. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J. Biol. Chem. 2005;280:14709–14715. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- Cheng HL, Vuong BQ, Basu U, Franklin A, Schwer B, Astarita J, Phan RT, Datta A, Manis J, Alt FW, Chaudhuri J. Integrity of the AID serine-38 phosphorylation site is critical for class switch recombination and somatic hypermutation in mice. Proc. Natl. Acad Sci. USA. 2009;106:2717–2722. doi: 10.1073/pnas.0812304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clikeman JA, Khalsa GJ, Barton SL, Nickoloff JA. Homologous recombinational repair of double-strand breaks in yeast is enhanced by MAT heterozygosity through yKu-dependent and -independent mechanisms. Genetics. 2001;157:579–589. doi: 10.1093/genetics/157.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogné M, Lansford R, Bottaro A, Zhang J, Gorman J, Young F, Cheng HL, Alt FW. A class switch control region at the 3’ end of the immunoglobulin heavy chain locus. Cell. 1994;77:737–747. doi: 10.1016/0092-8674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, Shah S, Brandt VL, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Cremer M, Küpper K, Wagler B, Wizelman L, von Hase J, Weiland Y, Kreja L, Diebold J, Speicher MR, Cremer T. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J. Cell Biol. 2003;162:809–820. doi: 10.1083/jcb.200304096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Meek K. Linking double-stranded DNA breaks to the recombination activating gene complex directs repair to the nonhomologous end-joining pathway. Proc. Natl. Acad. Sci. USA. 2007;104:17046–17051. doi: 10.1073/pnas.0610928104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm K. Functions and regulation of human artemis in double strand break repair. J. Cell. Biochem. 2007;100:1346–1351. doi: 10.1002/jcb.21226. [DOI] [PubMed] [Google Scholar]
- D’Amours D, Jackson SP. The Mre11 complex: At the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Klein A, van Kessel AG, Grosveld G. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982;300:765–767. doi: 10.1038/300765a0. [DOI] [PubMed] [Google Scholar]
- de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol. Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiderio S, Lin WC, Li Z. The cell cycle and V(D)J recombination. Curr. Top. Microbiol. Immunol. 1996;217:45–59. doi: 10.1007/978-3-642-50140-1_4. [DOI] [PubMed] [Google Scholar]
- Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio M, Petersen S, Chen HT, Johnson R, Jasin M, Kanaar R, Ried T, Nussenzweig A. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J. Exp. Med. 2002;196:469–480. doi: 10.1084/jem.20020851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, Sleckman BP, Nussenzweig A. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkelmann M, Spehalski E, Stoneham T, Buis J, Wu Y, Sekiguchi JM, Ferguson DO. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat. Struct. Mol. Biol. 2009;16:808–813. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Dorsett Y, Robbiani DF, Jankovic M, Reina-San-Martin B, Eisenreich TR, Nussenzweig MC. A role for AID in chromosome translocations between c-myc and the IgH variable region. J. Exp. Med. 2007;204:2225–2232. doi: 10.1084/jem.20070884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Guilhot F, O’Brien SG. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: Similarities and differences. Adv. Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- Durkin SG, Glover TW. Chromosome fragile sites. Annu. Rev. Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKCS to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Felsher DW. Tumor dormancy and oncogene addiction. APMIS. 2008;116:629–637. doi: 10.1111/j.1600-0463.2008.01037.x. [DOI] [PubMed] [Google Scholar]
- Ferguson DO, Alt FW. DNA double strand break repair and chromosomal translocation: Lessons from animal models. Oncogene. 2001;20(40):5572–5579. doi: 10.1038/sj.onc.1204767. [DOI] [PubMed] [Google Scholar]
- Ferguson DO, Sekiguchi JM, Chang S, Frank KM, Gao Y, DePinho RA, Alt FW. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl. Acad. Sci. USA. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster HA, Bridger JM. The genome and the nucleus: A marriage made by evolution. Genome organisation and nuclear architecture. Chromosoma. 2005;114:212–229. doi: 10.1007/s00412-005-0016-6. [DOI] [PubMed] [Google Scholar]
- Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA, Yan C, Tepsuporn S, Morales JC, Adams MM, Lou Z, Bassing CH, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 2006a;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Franco S, Alt FW, Manis JP. Pathways that suppress programmed DNA breaks from progressing to chromosomal breaks and translocations. DNA Repair (Amst.) 2006b;5:1030–1041. doi: 10.1016/j.dnarep.2006.05.024. [DOI] [PubMed] [Google Scholar]
- Frank KM, Sharpless NE, Gao Y, Sekiguchi JM, Ferguson DO, Zhu C, Manis JP, Horner J, DePinho RA, Alt FW. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- Fröhling S, Döhner H. Chromosomal abnormalities in cancer. N. Engl. J. Med. 2008;359:722–734. doi: 10.1056/NEJMra0803109. [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, Bronson RT, Malynn BA, Bryans M, Zhu C, Chaudhuri J, Davidson L, Ferrini R, Stamato T, Orkin SH, Greenberg ME, Alt FW. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, Chaudhuri J, Horner J, DePinho RA, Alt FW. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- Gell D, Jackson SP. Mapping of protein–protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 1999;27:3494–3502. doi: 10.1093/nar/27.17.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: Mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- Gillert E, Leis T, Repp R, Reichel M, Hösch A, Breitenlohner I, Angermüller S, Borkhardt A, Harbott J, Lampert F, Griesinger F, Greil J, et al. DNA damage repair mechanism is involved in the origin of chromosomal translocations. Oncogene. 1999;18:4663–4671. doi: 10.1038/sj.onc.1202842. [DOI] [PubMed] [Google Scholar]
- Gladdy RA, Taylor MD, Williams CJ, Grandal I, Karaskova J, Squire JA, Rutka JT, Guidos CJ, Danska JS. The RAG-1/2 endonuclease causes genomic instability and controls CNS complications of lymphoblastic leukemia in p53/Prkdc-deficient mice. Cancer Cell. 2003;3:37–50. doi: 10.1016/s1535-6108(02)00236-2. [DOI] [PubMed] [Google Scholar]
- Goldman JM, Melo JV. Chronic myeloid leukemia—Advances in biology and new approaches to treatment. N. Engl. J. Med. 2003;349:1451–1464. doi: 10.1056/NEJMra020777. [DOI] [PubMed] [Google Scholar]
- Gostissa M, Yan CT, Bianco JM, Cogné M, Pinaud E, Alt FW. Long-range oncogenic activation of Igh-c-myc translocations by the Igh 3’ regulatory region. Nature. 2009a;462:803–807. doi: 10.1038/nature08633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Ranganath S, Bianco JM, Alt FW. Chromosomal location targets different MYC family gene members for oncogenic translocations. Proc. Natl. Acad. Sci. USA. 2009b;106:2265–2270. doi: 10.1073/pnas.0812763106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36:93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Han L, Yu K. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV-deficient B cells. J. Exp. Med. 2008;205:2745–2753. doi: 10.1084/jem.20081623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington J, Hsieh CL, Gerton J, Bosma G, Lieber MR. Analysis of the defect in DNA end joining in the murine scid mutation. Mol. Cell Biol. 1992;12:4758–4768. doi: 10.1128/mcb.12.10.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisterkamp N, Stephenson JR, Groffen J. Localization of the c-ab1 oncogene adjacent to a translocation break point in chronicmyelocytic leukaemia. Nature. 1983;306:239–242. doi: 10.1038/306239a0. [DOI] [PubMed] [Google Scholar]
- Hesse JE, Lieber MR, Mizuuchi K, Gellert M. V(D)J recombination: A functional definition of the joining signals. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. DNA damage, aging, and cancer. N. Engl. J. Med. 2009;361:1475. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Hughes T, Deininger M, Hochhaus A, Branford S, Radich J, Kaeda J, Baccarani M, Cortes J, Cross NC, Druker BJ, Gabert J, Grimwade D, et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: Review and recommendations for harmonizing current methodology for detecting BCR–ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108:28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huye LE, Purugganan MM, Jiang MM, Roth DB. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol. Cell Biol. 2002;22:3460–3473. doi: 10.1128/MCB.22.10.3460-3473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliakis G, Wang H, Perrault AR, Boecker W, Rosidi B, Windhofer F, Wu W, Guan J, Terzoudi G, Pantelias G. Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet. Genome Res. 2004;104:14–20. doi: 10.1159/000077461. [DOI] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz S. Myc translocations in B cell and plasma cell neoplasms. DNA Repair. 2006;5:1213–1224. doi: 10.1016/j.dnarep.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DR, Park SJ, Oettinger MA. V(D)J recombination: Site-specific cleavage and repair. Mol. Cells. 2000;10:367–374. [PubMed] [Google Scholar]
- Kinner A, Wu W, Staudt C, Iliakis G. γ-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, Kobayashi T, Tamai K, Tanimoto K, Komatsu K. NBS1 localizes to g-H2AX foci through interaction with the FHA/BRCT domain. Curr. Biol. 2002;12:1846–1851. doi: 10.1016/s0960-9822(02)01259-9. [DOI] [PubMed] [Google Scholar]
- Kohl NE, Kanda N, Schreck RR, Bruns G, Latt SA, Gilbert F, Alt FW. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell. 1983;35:359–367. doi: 10.1016/0092-8674(83)90169-1. [DOI] [PubMed] [Google Scholar]
- Komori T, Okada A, Stewart V, Alt FW. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- Kreslavsky T, Gleimer M, von Boehmer H. Alphabeta versus gammadelta lineage choice at the first TCR-controlled checkpoint. Curr. Opin Immunol. 2010;22:185–192. doi: 10.1016/j.coi.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat. Rev. Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- Küppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Lee SE, Mitchell RA, Cheng A, Hendrickson EA. Evidence for DNA-PK-dependent and DNA-PK-independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol. Cell Biol. 1997;17:1425–1433. doi: 10.1128/mcb.17.3.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Woo CJ, Iglesias-Ussel MD, Ronai D, Scharff MD. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 2004;18:1–11. doi: 10.1101/gad.1161904. [DOI] [PubMed] [Google Scholar]
- Li G, Alt FW, Cheng HL, Brush JW, Goff PH, Murphy MM, Franco S, Zhang Y, Zha S. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Mol. Cell. 2008;31:631–640. doi: 10.1016/j.molcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Han M, Romanienko PJ, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl. Acad. Sci. USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst.) 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FL, Sperle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: Role for DNA ends in the recombination process. Mol. Cell. Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Lukacsovich T, Waldman AS. Multiple pathways for repair of double-strand breaks in mammalian chromosomes. Mol. Cell. Biol. 1999;19:8353–8360. doi: 10.1128/mcb.19.12.8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, Rosenfeld MG. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 2003;5:572–577. doi: 10.1038/ncb997. [DOI] [PubMed] [Google Scholar]
- Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: Role for pol mu in end-joining double-strand break repair. Mol. Cell. Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald GK, Baron JM, Mahowald MA, Kulkarni S, Bredemeyer AL, Bassing CH, Sleckman BP. Aberrantly resolved RAG-mediated DNA breaks in Atm-deficient lymphocytes target chromosomal breakpoints in cis. Proc. Natl. Acad. Sci. USA. 2009;106:18339–18344. doi: 10.1073/pnas.0902545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malynn BA, de Alboran IM, O‘Hagan RC, Bronson R, Davidson L, DePinho RA, Alt FW. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14:1390–1399. [PMC free article] [PubMed] [Google Scholar]
- Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari PO, Florea BI, Persengiev SP, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli MP, Sozzi G, Hernandez L, Pettirossi V, Navarro A, Conte D, Gasparini P, Perrone F, Modena P, Pastorino U, Carbone A, Fabbri A, et al. EML4–ALK rearrangement in non-small cell lung cancer and non-tumor lung tissues. Am. J. Pathol. 2009;174:661–670. doi: 10.2353/ajpath.2009.080755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Subero I, Odero MD, Hernandez R, Cigudosa JC, Agirre X, Saez B, Sanz-García E, Ardanaz MT, Novo FJ, Gascoyne RD, Calasanz MJ, Siebert R. Amplification of IGH/MYC fusion in clinically aggressive IGH/BCL2-positive germinal center B-cell lymphomas. Genes Chromosomes Cancer. 2005;43:414–423. doi: 10.1002/gcc.20187. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Marusawa H, Kinoshita K, Endo Y, Kou T, Morisawa T, Azuma T, Okazaki IM, Honjo T, Chiba T. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat. Med. 2007;13:470–476. doi: 10.1038/nm1566. [DOI] [PubMed] [Google Scholar]
- Matthews AG, Oettinger MA. Regulation of RAG transposition. Adv. Exp. Med. Biol. 2009;650:16–31. doi: 10.1007/978-1-4419-0296-2_2. [DOI] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Max EE, Seidman JG, Leder P. Sequences of five potential recombination sites encoded close to an immunoglobulin kappa constant region gene. Proc. Natl. Acad. Sci. USA. 1979;76:3450–3454. doi: 10.1073/pnas.76.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell TJ, Korsmeyer SJ. Progression from lymphoid hyperplasia to high-grade malignant lymphoma in mice transgenic for the t(14;18) Nature. 1991;349:254–256. doi: 10.1038/349254a0. [DOI] [PubMed] [Google Scholar]
- McDonnell TJ, Deane N, Platt FM, Nunez G, Jaeger U, McKearn JP, Korsmeyer SJ. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell. 1989;57:79–88. doi: 10.1016/0092-8674(89)90174-8. [DOI] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T, Soutoglou E. Spatial genome organization in the formation of chromosomal translocations. Semin. Cancer Biol. 2007;17:80–90. doi: 10.1016/j.semcancer.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelsen S, Snyder C, Trujillo K, Bogue M, Roth DB, Meek K. Modulation of terminal deoxynucleotidyltransferase activity by the DNA-dependent protein kinase. J. Immunol. 1999;163:834–843. [PubMed] [Google Scholar]
- Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- Mills KD, Ferguson DO, Essers J, Eckersdorff M, Kanaar R, Alt FW. Rad54 and DNA Ligase 4 cooperate to maintain mammalian chromatid stability. Genes Dev. 2004;18:1283–1292. doi: 10.1101/gad.1204304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- Monroe RJ, Seidl KJ, Gaertner F, Han S, Chen F, Sekiguchi J, Wang J, Ferrini R, Davidson L, Kelsoe G, Alt FW. RAG2:GFP knockin mice reveal novel aspects of RAG2 expression in primary and peripheral lymphoid tissues. Immunity. 1999;11:201–212. doi: 10.1016/s1074-7613(00)80095-3. [DOI] [PubMed] [Google Scholar]
- Morgenbesser SD, DePinho RA. Use of transgenic mice to study myc family gene function in normal mammalian development and in cancer. Semin. Cancer Biol. 1994;5:21–36. [PubMed] [Google Scholar]
- Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- Mundy CL, Patenge N, Matthews AG, Oettinger MA. Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell Biol. 2002;22:69–77. doi: 10.1128/MCB.22.1.69-77.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Nagaoka H, Shinkura R, Begum NA, Honjo T. Discovery of activation-induced cytidine deaminase, the engraver of antibody memory. Adv. Immunol. 2007;94:1–36. doi: 10.1016/S0065-2776(06)94001-2. [DOI] [PubMed] [Google Scholar]
- Niewolik D, Pannicke U, Lu H, et al. DNA-PKCS dependence of Artemis endonucleolytic activity, differences between hairpins and 5′ or 3′ overhangs. J. Biol. Chem. 2006;28:33900–33909. doi: 10.1074/jbc.M606023200. [DOI] [PubMed] [Google Scholar]
- Nikiforov YE. Thyroid carcinoma: Molecular pathways and therapeutic targets. Mod. Pathol. 2008;21:37–43. doi: 10.1038/modpathol.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC, Hungerford DA. Chromosome studies on normal and leukemic human leukocytes. J. Natl. Cancer Inst. 1960;25:85–109. [PubMed] [Google Scholar]
- Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat. Rev. Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- Okazaki IM, Kotani A, Honjo T. Role of AID in tumorigenesis. Adv. Immunol. 2007;94:245–273. doi: 10.1016/S0065-2776(06)94008-5. [DOI] [PubMed] [Google Scholar]
- Parada L, Misteli T. Chromosome positioning in the interphase nucleus. Trends Cell Biol. 2002;12(9):425–432. doi: 10.1016/s0962-8924(02)02351-6. [DOI] [PubMed] [Google Scholar]
- Parada L, Sotiriou S, Misteli T. Spatial genome organization. Exp. Cell Res. 2004;269:64–70. doi: 10.1016/j.yexcr.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC, III, Nussenzweig MC, Dalla-Favera R. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, Baltimore D. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- Qiu JX, Kale SB, Yarnell SH, Roth DB. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol. Cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, Nussenzweig A, Nussenzweig MC. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc. Natl. Acad. Sci. USA. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B, Difilippantonio S, Hanitsch L, Masilamani RF, Nussenzweig A, Nussenzweig MC. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, Nussenzweig MC. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride KM, Klein IA, Stone G, Eisenreich TR, Ried T, Nussenzweig A, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol. Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, Hasty P, Ramsden DA. Ku is a 5’-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin IB, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- Rooney S, Sekiguchi J, Whitlow S, Eckersdorff M, Manis JP, Lee C, Ferguson DO, Alt FW. Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells. Proc. Natl. Acad. Sci. USA. 2004a;101:2410–2415. doi: 10.1073/pnas.0308757101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol. Rev. 2004b;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- Sakano H, Hüppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Savic V, Yin B, Maas NL, Bredemeyer AL, Carpenter AC, Helmink BA, Yang-Iott KS, Sleckman BP, Bassing CH. Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol. Cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz DG, Baltimore D. Stable expression of immunoglobulin gene V(D)J recombinase activity by gene transfer into 3T3 fibroblasts. Cell. 1988;53:107–115. doi: 10.1016/0092-8674(88)90492-8. [DOI] [PubMed] [Google Scholar]
- Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- Schmidt EV, Pattengale PK, Weir L, Leder P. Transgenic mice bearing the human c-myc gene activated by an immunoglobulin enhancer: A pre-B-cell lymphoma model. Proc. Natl. Acad. Sci. USA. 1988;85:6047–6051. doi: 10.1073/pnas.85.16.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüler F, Hirt C, Dölken G. Chromosomal translocation t(14;18) in healthy individuals. Semin. Cancer Biol. 2003;13:203–209. doi: 10.1016/s1044-579x(03)00016-6. [DOI] [PubMed] [Google Scholar]
- Sekiguchi JA, Whitlow S, Alt FW. Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol. Cell. 2001;8:1383–1390. doi: 10.1016/s1097-2765(01)00423-3. [DOI] [PubMed] [Google Scholar]
- Shimazaki N, Tsai AG, Lieber MR. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: Implications for translocations. Mol. Cell. 2009;34:535–544. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, Alt FW. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat. Struct. Mol. Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton BK, Torres-Arzayus MI, Rottinghaus ST, Taccioli GE, Jeggo PA. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol. Cell. Biol. 1999;19:3267–3277. doi: 10.1128/mcb.19.5.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- Soda M, Takada S, Takeuchi K, Choi YL, Enomoto M, Ueno T, Haruta H, Hamada T, Yamashita Y, Ishikawa Y, Sugiyama Y, Mano H. A mouse model for EML4–ALK-positive lung cancer. Proc. Natl. Acad. Sci. USA. 2008;105:19893–19897. doi: 10.1073/pnas.0805381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA–PKCS/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, Greenman CD, Jia M, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, Alt FW. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, et al. Homologous recombination and non-homologous end-joining pathways of DNA doublestrand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Truffinet V, Pinaud E, Cogné N, Petit B, Guglielmi L, Cogné M, Denizot Y. The 3’ IgH locus control region is sufficient to deregulate a c-myc transgene and promote mature B cell malignancies with a predominant Burkitt-like phenotype. J. Immunol. 2007;179:6033–6042. doi: 10.4049/jimmunol.179.9.6033. [DOI] [PubMed] [Google Scholar]
- Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama Y, Takeuchi R, Kodera H, Sakaguchi K. Distribution and roles of X-family DNA polymerases in eukaryotes. Biochimie. 2009;91:165–170. doi: 10.1016/j.biochi.2008.07.005. [DOI] [PubMed] [Google Scholar]
- van den Bosch M, Bree RT, Lowndes NF. The MRN complex: Coordinating and mediating the response to broken chromosomes. EMBO Rep. 2003;4:844–849. doi: 10.1038/sj.embor.embor925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkaik NS, Esveldt-van Lange RE, van Heemst D, Brüggenwirth HT, Hoeijmakers JH, Zdzienicka MZ, van Gent DC. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur. J. Immunol. 2002;32:701–709. doi: 10.1002/1521-4141(200203)32:3<701::AID-IMMU701>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Villa A, Santagata S, Bozzi F, Imberti L, Notarangelo LD. Omenn syndrome: A disorder of Rag1 and Rag2 genes. J. Clin. Immunol. 1999;19:87–97. doi: 10.1023/a:1020550432126. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Wagner SD, Milstein C, Neuberger MS. Codon bias targets mutation. Nature. 1995;376:732. doi: 10.1038/376732a0. [DOI] [PubMed] [Google Scholar]
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Wang H, Perrault AR, Takeda Y, Qin W, Wang H, Iliakis G. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 2003;31:5377–5388. doi: 10.1093/nar/gkg728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Alt FW, Gostissa M, Datta A, Murphy M, Alimzhanov MB, Coakley KM, Rajewsky K, Manis JP, Yan CT. Oncogenic transformation in the absence of Xrcc4 targets peripheral B cells that have undergone editing and switching. J. Exp. Med. 2008;205:3079–3090. doi: 10.1084/jem.20082271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E, Difilippantonio S, Wesemann DR, Zarrin AA, Rajewsky K, Nussenzweig A, Alt FW. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Minn K, Jorda KG, Chen J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 2003;278:19579–19582. doi: 10.1074/jbc.C300117200. [DOI] [PubMed] [Google Scholar]
- Weinstock DM, Richardson CA, Elliott B, Jasin M. Modeling oncogenic translocations: Distinct roles of double strand break repair pathways in translocation formation in mammalian cells. DNA Repair. 2006a;5:1065–1074. doi: 10.1016/j.dnarep.2006.05.028. [DOI] [PubMed] [Google Scholar]
- Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: Differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006b;107:777–780. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Brunet E, Jasin M. Induction of chromosomal translocations in mouse and human cells using site-specific endonucleases. J. Natl. Cancer Inst. Monogr. 2008;39:20–24. doi: 10.1093/jncimonographs/lgn009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CT, Kaushal D, Murphy M, Zhang Y, Datta A, Chen C, Monroe B, Mostoslavsky G, Coakley K, Gao Y, Mills KD, Fazeli AP, et al. XRCC4 suppresses medulloblastomas with recurrent translocations in p53-deficient mice. Proc. Natl. Acad. Sci. USA. 2006;103:7378–7383. doi: 10.1073/pnas.0601938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- Yaneva M, Kowalewski T, Lieber MR. Interaction of DNA-dependent protein kinase with DNA and with Ku: Biochemical and atomic-force microscopy studies. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannoutsos N, Wilson P, Yu W, Chen HT, Nussenzweig A, Petrie H, Nussenzweig MC. The role of recombination activating gene (RAG) reinduction in thymocyte development in vivo. J. Exp. Med. 2001;194:471–480. doi: 10.1084/jem.194.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnell SH, Landree MA, Qiu JX, Kale SB, Roth DB. Joining-deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro. Mol. Cell. 2001;7:65–75. doi: 10.1016/s1097-2765(01)00155-1. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Strasser A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- Yu W, Nagaoka H, Misulovin Z, Meffre E, Suh H, Jankovic M, Yannoutsos N, Casellas R, Besmer E, Papavasiliou F, Qin X, Nussenzweig MC. RAG expression in B cells in secondary lymphoid tissues. Cold Spring Harb. Symp. Quant. Biol. 1999;64:207–210. doi: 10.1101/sqb.1999.64.207. [DOI] [PubMed] [Google Scholar]
- Zarrin AA, Del Vecchio C, Tseng E, Gleason M, Zarin P, Tian M, Alt FW. Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science. 2007;315:377–381. doi: 10.1126/science.1136386. [DOI] [PubMed] [Google Scholar]
- Zha S, Boboila C, Alt FW. Mre11: Roles in DNA repair beyond homologous recombination. Nat. Struct. Mol. Biol. 2009;16:798–800. doi: 10.1038/nsmb0809-798. [DOI] [PubMed] [Google Scholar]
- Zha S, Bassing CH, Sanda T, Brush JW, Patel H, Goff PH, Murphy MM, Tepsuporn S, Gatti RA, Look AT, Alt FW. ATM-deficient thymic lymphoma is associated with aberrant Tcrδ rearrangement and gene amplification. J. Exp. Med. 2010;207:1369–1380. doi: 10.1084/jem.20100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ren R. Bcr-Abl efficiently induces a myeloproliferative disease and production of excess interleukin-3 and granulocyte-macrophage colony-stimulating factor in mice: a novel model for chronic myelogenous leukemia. Blood. 1998;92:3829–3840. [PubMed] [Google Scholar]
- Zhang Y, Rowley JD. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair. 2006;5:1282–1297. doi: 10.1016/j.dnarep.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Zhang F, Carvalho CM, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Franklin A, Boboila C, McQuay A, Gallagher MP, Manis JP, Khamlichi AA, Alt FW. Downstream class switching leads to IgE antibody production by B lymphocytes lacking IgM switch regions. Proc. Natl. Acad. Sci. USA. 2010;107:3040–3045. doi: 10.1073/pnas.0915072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Bacolla A, Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability and evolution. Cell Mol. Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- Zimmerman KA, Yancopoulos GD, Collum RG, Smith RK, Kohl NE, Denis KA, Nau MM, Witte ON, Toran-Allerand D, Gee CE, Minna JD, Alt FW. Differential expression of myc family genes during murine development. Nature. 1986;319:780–783. doi: 10.1038/319780a0. [DOI] [PubMed] [Google Scholar]
- Zitzelsberger H, Bauer V, Thomas G, Unger K. Molecular rearrangements in papillary thyroid carcinomas. Clin. Chim. Acta. 2010;411:301–308. doi: 10.1016/j.cca.2009.11.028. [DOI] [PubMed] [Google Scholar]