Abstract
Lymphatic filariasis is caused by the parasitic nematodes Brugia malayi and Wuchereria bancrofti and asparaginyl-tRNA synthetase (AsnRS) is considered an excellent antifilarial target. The discovery of three new tirandamycins (TAMs), TAM E (1), F (2), and G (3), along with TAM A (4) and B (5), from Streptomyces sp. 17944 was reported. Remarkably, 5 selectively inhibits the B. malayi AsnRS and efficiently kills the adult B. malayi parasite, representing a new lead scaffold to discover and develop antifilarial drugs.
Lymphatic filariasis (LF), also known as elephantiasis, is caused by the parasitic nematodes Brugia malayi and Wuchereria bancrofti and represents a worldwide health crisis with over 200 million people infected and another 20% of the global population at risk for infection.1,2 In 1997, the World Health Organization (WHO) passed a resolution, prioritizing “the elimination of LF as a public health problem” and more recently announced the discovery of effective macrofilaricides, drugs that can kill adult female worms, a top priority.3 This action was spurred by concerns related to resistance to current anti-helminth agents in addition to the long-standing desire to impair transmission among hosts. Programs aimed at interrupting disease transmission have been based on mass distribution of ivermectin and albendazole.3–6 However, resistance to both agents is well known and both agents suffer from serious limitations – neither agent is effective at killing adult worms (macrofilariae) and serious side effects are associated with both drugs.7–9 A clear need exists to identify new anti-parasitic agents with new, alternative modes of action.9
Aminoacyl tRNA synthetases (AARSs) were the first filarial targets embraced by the WHO10 and are generally regarded as excellent therapeutic targets because they: (i) perform important primary and secondary transformations within eukaryotes including filarial and other human and veterinary parasites, (ii) are essential to microbial viability, and (iii) demonstrate primary and secondary structural heterogeneity while sharing a common catalytic site topology critical to recognition by inhibitors that block the synthesis or release of the aminoacyl adenylate intermediate.
Among the AARSs, asparaginyl-tRNA synthetase (AsnRS) is, in particular, an excellent filarial target because it (i) is expressed in both sexes, adults, and larvae of B. malay2,11 and (ii) is well-characterized biochemically and structurally in several species, including B. malayi, and (iii) high-throughput screening (HTS) platform with recombinant B. malayi AsnRS for inhibitors has been developed.12 Most recently, the complete atomic structure of the B. malayi AsnRS, has been determined and the structural basis of asparagine and false substrate recognition elucidated.13
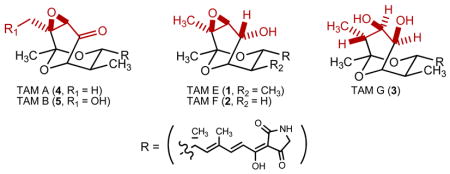
Despite the rise to prominence of combinatorial chemistry, natural products remain a valuable source of new drug leads and have demonstrated almost limitless potential in showcasing new molecular scaffolds with clinically relevant biological activities.14 As part of a drug discovery program targeting the B. malayi AsnRS, we recently completed a HTS campaign of ~73,000 microextracts, from a collection of 36,720 microbial strains, for activity against recombinant B. malayi AsnRS. Of the extracts screened, 199, representing 177 strains, induced at least 70% inhibition. We now report bioassay-guided dereplication of one of these active strains, Streptomyces sp. 17944, discovering three new tirandamycins (TAMs), TAM E (1), F (2), and G (3) along with two known TAMs, TAM A (4) and TAM B (5) (Figure 1A). Remarkably, 5 selectively inhibits the B. malayi AsnRS and efficiently kills the adult B. malayi parasite. To our knowledge TAMs have not been recognized previously for use in the treatment or prevention of LF. Consequently, 5 represents a new lead scaffold for the discovery and development of antifilarial drugs.
Figure 1.
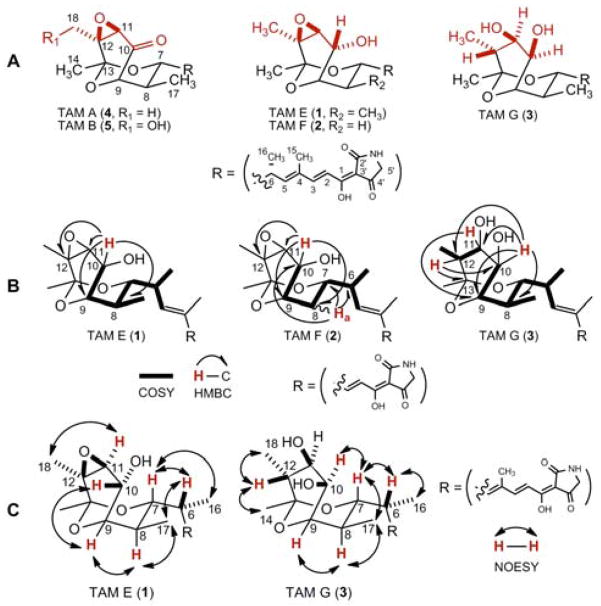
Isolation and structural determination of TAMs from S. sp. 17944: (A) structures of the three new TAMs, TAM E (1), F (2), and G (3), and the two known TAMs, TAM A (4) and B (5); (B) key COSY and HMBC correlations for 1–3; and (C) key NOESY correlations for 1 and 3.
The S. sp. 17944 strain was cultivated in the ISP2 medium, and natural product isolation was guided by bioassay for inhibitory activity against recombinant B. malayi AsnRS (see Supporting Information). Thus, solid phase extract of 7.2 L of the resultant fermentation, prepared using 3% Amberlite XAD-16 resin and previously described procedures,15 was subjected to silica gel and Sephadex LH-20 chromatography, followed by further purification with reversed-phase HPLC, to afford 1–5 (see Supporting Information). Analysis of high resolution ESI-MS (HRESIMS) data and 1H and 13C NMR spectra (Table S1) suggested 4 and 5 to be TAM A and B, respectively, whose identities were unambiguously confirmed by extensive 1D and 2D NMR (gCOSY, gHSQC, and gHMBC) analyses, as well as, comparison to previously reported spectroscopic data (Figure 1A).16
The molecular formula of 1 was determined to be C22H29NO7 by HRESIMS, affording an [M + Na]+ ion at m/z 442.18355 (calculated [M + Na]+ ion at m/z 442.18362) and indicating that 1 differs from 4 (C22H27NO7) by the presence of two additional protons. The 1H and 13C NMR spectral data (Tables 1 and S1) supported a close structural relationship between 1 and 4, demonstrated by the consistent appearance of the olefinic and methyl signals of the dienoyl acyl chain and the geminal protons of the tetramic acid methylene carbon. Correlations observed in gDQCOSY, gHSQC, and gHMBC experiments further confirmed 4 and 1 have identical carbon backbone except for the oxygenation patterns of the bicyclic ketal moiety. Close examination of the NMR data for 1 and 4 (Tables 1 and S1) revealed that disappearence of the C-10 carbonyl carbon of 4 (δC 202.8) with the concomitant appearance of the oxygenated methine carbon (δC 67.3, C-10) and an additional proton signal (δH 4.41, H-10), together with the MS data, suggesting that 4 was likely a C-10 reduced congener of 1 (Figure 1A). This conclusion was further confirmed by gHMBC correlations of H-10 (δH 4.41) with C-8 (δC 36.6), C-9 (δC 71.8), C-11 (δC 64.2), and C-12 (δC 56.4) and the COSY correlation between H-10 and H-9 (Figure 1B). Since the absolute stereochemistry of 4 was known,17 1, with the exception of C-10, was assigned the same stereochemistry as 4 on the basis of its biosynthetic origin. Finally, the C-10 stereochemistry of 1 was deduced on the basis of the NOESY spectrum. Key correlations between H-10/H-9, H-11/H-18, and H-9/H-8 (Figure 1C) are consistent with the assignment of an R configuration to C-10 of 1, which we named TAM E (Figure 1A).
Table 1.
Summary of 1H (500 MHz) and 13C (125 MHz) Spectroscopic Data for 1, 2, and 3 (in CDCl3).
| position | 1(J in Hz) | 2(J in Hz) | 3(J in Hz) | |||
|---|---|---|---|---|---|---|
| δH mult | δC | δH mult | δC | δH mult | δC | |
| 1 | 175.7, s | 175.7, s | 175.5, s | |||
| 2 | 7.15, d (16.0) | 116.5, d | 7.16, d (15.5) | 116.6, d | 7.14, d (16.0) | 116.1, d |
| 3 | 7.60, d (15.5) | 150.6, d | 7.58, d (16.0) | 150.5, d | 7.59, d (15.5) | 150.5, d |
| 4 | 134.7, s | 135.3, s | 134.4, s | |||
| 5 | 6.29, d (9.5) | 145.4, d | 6.07, d (9.5) | 147.0, d | 6.24, d (9.5) | 146.0, d |
| 6 | 2.89, m | 35.0, d | 2.64, m | 39.2, d | 2.87, m | 34.3, d |
| 7 | 3.92, d (10.5) | 76.2, d | 3.93, m | 71.1, d | 3.58, d (11.0) | 79.2, d |
| 8 | 2.04, m | 36.6, d | 1.90, m 1.66, m |
26.1, t | 1.98, m | 35.3, d |
| 9 | 3.98, m | 71.8, d | 4.15, m | 68.3, d | 4.01, m | 73.2, d |
| 10 | 4.41, d (7.0) | 67.3, d | 4.30, d (6.5) | 63.7, d | 3.96, m | 77.1, d |
| 11 | 3.19, s | 64.2, d | 3.15, s | 63.9, d | 3.95, m | 75.8, d |
| 12 | 56.4, s | 56.3, s | 1.78, m | 47.1, d | ||
| 13 | 96.5, s | 96.4, s | 99.3, s | |||
| 14 | 1.42, s | 22.3, q | 1.37, s | 22.5, q | 1.36, s | 26.9, q |
| 15 | 1.93, s | 12.5, q | 1.92, s | 12.9, q | 1.91, s | 12.3, q |
| 16 | 1.14, d (7.0) | 17.3, q | 1.08, d (7.0) | 16.3, q | 1.13, d (6.0) | 17.1, q |
| 17 | 0.96, d (7.5) | 13.2, q | - | - | 0.96, d (7.5) | 12.8, q |
| 18 | 1.40, s | 16.6, q | 1.35, s | 16.7, q | 1.14, d (6.5) | 12.8, q |
| 2′ | 176.7, s | 176.8, s | 176.5, s | |||
| 3′ | 100.1, s | 100.2, s | 99.8, s | |||
| 4′ | 192.8, s | 192.8, s | 192.4, s | |||
| 5′ | 3.82, s | 51.8, t | 3.82, s | 51.8, t | 3.82, s | 51.5, t |
The molecular formula of 2 was determined to be C21H27NO7 by HRESIMS, yielding an [M + Na]+ ion at m/z 428.16998 (calculated [M + Na]+ ion at m/z 428.16797), which differs from 1 by the absence of a CH2 unit. The structure of 2 was established by careful comparison of the NMR data between 1 and 2 (Table 1). The absence of a doublet methyl signal and a methine signal with the concomitant presence of one new methylene signal suggested 2 as a desmethyl congener of 1. This conclusion was further supported by gHMBC correlation of Ha-8 (δH 1.66) with C-6 (δC 39.2), C-7 (δC 71.1), and C-10 (δC 63.7) and COSY correlations among Ha-8 (δH 1.66), H-7 (δH 3.93), and H-9 (δH 4.15) (Figure 1B). Thus, 2 was finally assigned as 8-desmethyl TAM E, hereafter regarded as TAM F, with the same absolute stereochemistry as 1 on the basis of their common biosynthetic origin (Figure 1A).
The molecular formula of 3 was determined to be C22H31NO7 by HRESIMS, giving rise to an [M + Na]+ ion at m/z 444.19900 (calculated [M + Na]+ ion at m/z 444.19927), which differs from 1 by the presence of two additional protons. Careful analysis of the 1H and 13C NMR data for 1 and 3 indicated that the characteristic 11, 12-epoxide resonances in 1 (δC 64.2 for C-11 and δC 56.4 for C-12) were replaced by a downfield shifted oxygenated methine signal (δC 75.8 for C-11) and an upfield shifted methine signal (δC 47.1 for C-12) in 3 (Table 1). Together with the MS data, 3 was suggested to be the epoxide ring–reduced congener of 1, which was further supported by gHMBC correlations of H-11 (δH 3.95) with C-9 (δC 73.2) and H-12 (δH 1.78) with C-10 (δC 77.1) and C-13 (δC 99.3), as well as, COSY correlations among H-12 (δH 1.78), H-11 (δH 3.95), and H-18 (δH 1.14) (Figure 1B). The stereochemistry of 3 was assigned as 10R, 11R, and 12R on the basis of a NOSEY experiment, and key observations included correlations between H-10/H-7, H-12/H-18, and H-12/H-14 (Figure 1C). Finally, 3 was named TAM G, with the rest of its stereochemistry to be the same as 4 on the basis of their common biosynthetic origin (Figure 1A).
Having each of the TAMs purified and their structures established, we re-evaluated their inhibitory activity against the B. malayi AsnRS, taking advantage of our recently reported non-radioactive assay that uses malachite green to measure rates and yields of phosphate generated by the recombinant AsnRS.12 This “pre-transfer editing assay” exploits the novel asparagine substrate mimic L-aspartate β-hydroxamate to drive the enzymatic activity of AsnRS. A molecular explanation why this asparagine mimic functions so efficiently in the assay has been provided by our recently solved B. malayi AsnRS structure.13 Assays of pure 1–5 revealed that 5 inhibited B. malayi AsnRS with an IC50 of 30 μM; the other four TAMs showed IC50 > 200 μM. Remarkably, under the identical assay conditions, 5 exhibited minimally 10-fold selectivity for the B. malayi AsnRS over the human AsnRS (Figure 2A), suggesting 5 as a promising lead for antifilarial drug discovery.
Figure 2.
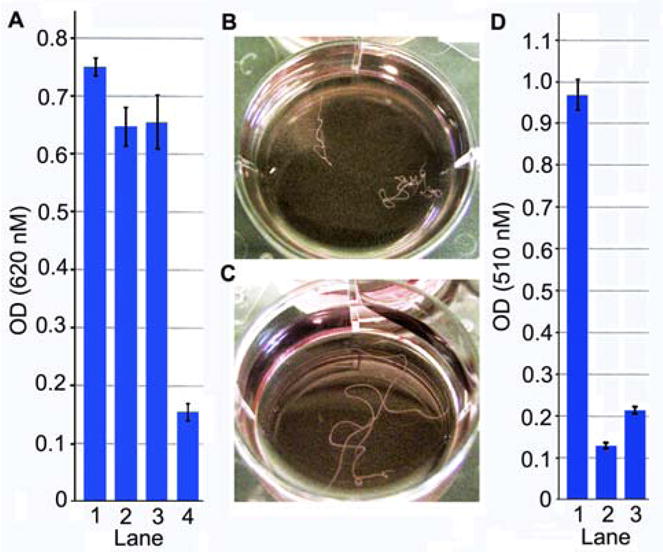
Summary of anti-filarial activity of 5. (A) In vitro analysis of AsnRS inhibition by 5 against human and filarial AsnRS using pre-transfer editing assay. Lane/Reaction contents: (1) human AsnRS; (2) human AsnRS, 150 μM 5; (3) B. malayi AsnRS; (4) B. malayi AsnRS, 150 μM 5. Assays employed limiting (50%) maximal enzyme activity. (B) Live worms are coiled and motile whereas dead worms (C) are elongated and nonmotile. (D) Live worms reduce MTT which leads to absorption at 510 nm. Lane/Reaction contents: (1) B. malayi, DMSO; (2) B. malayi, 100 μM albendazole as positive control; (3) B. malayi, 100 μM 5.
Finally, we showed that 5 can efficiently kill the adult B. malayi worm in vitro. Live adult female B. malayi worms were maintained in 6-well plates, and 5 (100 μM) was added to select wells compared with albendazole, a known LF drug, serving as a positive control.8,9 B. malayi are very amenable to visual health assessment as healthy worms are vigorously motile and coiled, whereas dead worms assume an elongated morphology devoid of movement (Figures 2B and 2C). Worm death can also be differentiated from simple paralysis, using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT); live worms reduce MTT to formazan, a spectroscopically unique species whose absorption at 510 nm provides a real-time measure of filarial viability.18 As shown in Figure 2D, 5 kills the adult worms with a yield similar to that observed with albendazole. The efficiency with which 5 (100 μM) kills adult worms is remarkable – whereas 100 μM albendazole requires >10 days to effectively kill worms, 5 induces dramatic filarial death after less than 24 h. The IC50 for 5 to kill the adult worms calculated using the MTT assay was found to be 1 μM. This finding, in combination with the ability of 5 to selectively inhibit the filarial AsnRS, thereby targeting B. malayi, constitutes the basis for continued chemical and biological efforts to understand the mode of action of 5 and the application of both combinatorial biosynthesis and medicinal chemistry methods to the 5 scaffold for structure-and-activity relationship study. The fact that the TAM biosynthetic gene cluster has been recently cloned and partially characterized19 further heightens enthusiasm for the application of combinatorial biosynthesis strategies to overproduce 5 and to enhance structural diversity. Although other activities20 known for the TAMs may complicate future drug development effort, 5 represents an outstanding natural product lead that could be exploited to combat the global health crisis of LF.
Supplementary Material
Acknowledgments
We thank the Filariasis Research Reagent Resource, Division of Microbiology and Infectious Diseases, NIAID, NIH for providing adult B. malayi and the Analytical Instrumentation Center of the School of Pharmacy, UW-Madison for support in obtaining MS and NMR data. This work was supported in party by NIH grants U01 A1053877 (MK) and GM086184 (BS).
Footnotes
Supporting Information Available Experimental procedures, MS and 1H and 13C NMR data of 1–5. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dzenowagis J, editor. WHO. Lymphatic Filariasis: Reasons for Hope. Geneva: World Health Organization; 1997. [Google Scholar]
- 2.Kron M, Marquard K, Hartlein M, Price S, Lederman R. FEBS Lett. 1995;374:122–124. doi: 10.1016/0014-5793(95)01092-s. [DOI] [PubMed] [Google Scholar]
- 3.Ridley RG, Kita K. Expert Opin Drug Discov. 2007;2(Suppl 1):S1. doi: 10.1517/17460441.2.S1.S1. [DOI] [PubMed] [Google Scholar]
- 4.Woods DJ, Lauret C, Geary TG. Expert Opin Drug Discov. 2007;2:S25–S33. doi: 10.1517/17460441.2.S1.S25. [DOI] [PubMed] [Google Scholar]
- 5.Geary TG. Trends Parasitol. 2005;21:530–532. doi: 10.1016/j.pt.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Molyneux DH, Bradley M, Hoerauf A, Kyelem D, Taylor MJ. Trends Parasitol. 2003;19:516–522. doi: 10.1016/j.pt.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Prichard RK. Expert Opin Drug Discov. 2007;2:S41–S52. doi: 10.1517/17460441.2.S1.S41. [DOI] [PubMed] [Google Scholar]
- 8.Fox LM, Furness BW, Haser JK, Desire D, Brissau JM, Milord MD, Lafontant J, Lammie PJ, Beach MJ. Am J Tropical Med & Hygeine. 2005;73:115–121. [PubMed] [Google Scholar]
- 9.(a) Kron MA, Kuhn LA, Sanschagrin PC, Hartlein M, Grotli M, Cusack S. J Parasitol. 2003;89:S226–S235. [Google Scholar]; (b) Hoerauf A. Curr Opin Infect Disease. 2008;21:673–681. doi: 10.1097/QCO.0b013e328315cde7. [DOI] [PubMed] [Google Scholar]; (c) Bockarie MJ, Deb RM. Curr Opin Infect Disease. 2010;23:617–620. doi: 10.1097/QCO.0b013e32833fdee5. [DOI] [PubMed] [Google Scholar]
- 10.Kron MA, Ramirez BL, Ramirez Y. Expert Opin Drug Discov. 2007;2:S1–S8. doi: 10.1517/17460441.2.S1.S75. [DOI] [PubMed] [Google Scholar]
- 11.Nilsen TW, Maroney PA, Goodwin RG, Perrine KG, Denkar JA, Nanduri J, Kazura J. Proc Natl Acad Sci USA. 1988;85:3604–3607. doi: 10.1073/pnas.85.10.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danel F, Caspers P, Nuoffer C, Hartlein M, Kron MA, Page MG. Curr Drug Discov Technol. 2010;8:66–75. doi: 10.2174/157016311794519947. [DOI] [PubMed] [Google Scholar]
- 13.Crepin T, Peterson F, Hartlein M, Jensen D, Wang C, Cusack S, Kron M. J Mol Biol. 2011;405:1056–1069. doi: 10.1016/j.jmb.2010.11.049. [DOI] [PubMed] [Google Scholar]
- 14.Newman DJ, Cragg GM. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 15.Ju J, Seo JW, Her Y, Lim SK, Shen B. Org Lett. 2007;9:5183–5186. doi: 10.1021/ol702249g. [DOI] [PubMed] [Google Scholar]
- 16.(a) Meyer CE. J Antibiot. 1971;24:558–560. doi: 10.7164/antibiotics.24.558. [DOI] [PubMed] [Google Scholar]; (b) Carlson JC, Li S, Burr DA, Sherman DH. J Nat Prod. 2009;72:2076–2079. doi: 10.1021/np9005597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duchamp DJ, Branfman AR, Button AC, Rinehart KL. J Am Chem Soc. 1973;95:4077–4078. doi: 10.1021/ja00793a058. [DOI] [PubMed] [Google Scholar]
- 18.Comley JCW, Rees MJ, Turner CH, Jenkins DC. Int J Parasitol. 1989;19:77–83. doi: 10.1016/0020-7519(89)90024-6. [DOI] [PubMed] [Google Scholar]
- 19.Carlson JC, Fortman JL, Anzai Y, Li S, Burr DA, Sherman DH. ChemBioChem. 2010;11:564–572. doi: 10.1002/cbic.200900658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reusser F. Infect Immun. 1970;2:82–88. doi: 10.1128/iai.2.1.82-88.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]; Reusser F. Antimicrob Agents Chermother. 1976;10:618–622. doi: 10.1128/aac.10.4.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.