Abstract
Foxa is a forkhead transcription factor that is expressed in the endoderm lineage across metazoans. Orthologs of foxa are expressed in cells that intercalate, polarize and form tight junctions in the digestive tracts of the mouse, the sea urchin and the nematode and in the chordate notochord. The loss of foxa expression eliminates these morphogenetic processes. The remarkable similarity in foxa phenotypes in these diverse organisms raises the following questions – why is the developmental role of Foxa so highly conserved? Is foxa transcriptional regulation as conserved as its developmental role? Comparison of the regulation of foxa orthologs in sea urchin and in C. elegans shows that foxa transcriptional regulation has diverged significantly between these two organisms, particularly in the cells that contribute to the C. elegans pharynx formation. We suggest that the similarity of foxa phenotype is due to its role in an ancestral gene regulatory network that controlled intercalation followed by mesenchymal to epithelial transition. foxa transcriptional regulation had evolved to support the developmental program in each species so foxa would play its role controlling morphogenesis at the necessary embryonic address.
Keywords: gene regulatory networks, cis-regulatory analysis, evolution, mesenchymal to epithelial transition
Introduction
Regulatory genes play a major role in the evolution of diverse body plans as they control the differential expression of regulatory and structural genes (Davidson, 2006; Ben-Tabou de-Leon and Davidson, 2007; Ben-Tabou de-Leon and Davidson, 2009). There are examples for evolutionary rearrangement of the architecture of developmental gene regulatory networks (GRNs) that result in change in the organism morphological features (Hinman and Davidson, 2007; Gao and Davidson, 2008; Fraser et al., 2009; Lemons et al., 2010) and examples for conservation of morphological features that relate to preservation of ancestral developmental GRN (see e.g., (Hinman et al., 2003; Fraser et al., 2009; McCauley et al., 2010)).
A common feature of developmental GRNs is that the specific combinations of transcription factors define the specification state of a cell, and not individual transcription factors (Davidson, 2006). Yet, there are certain regulatory genes that seem to be involved in a similar developmental task in a wide range of organisms. Examples for such regulatory genes are the transcription factor Pax6 that is involved in eye development across metazoans (Pichaud and Desplan, 2002; Kozmik, 2005) and the transcription factor Tin/Nkx2.5 that is involved in heart development across bilaterians (Harvey, 1996; Holland et al., 2003; Davidson, 2006). Apparently, these genes were key regulators of ancestral GRNs that initiated the development of these organs. However, it is not clear why the developmental role of these particular genes is so well conserved and whether the transcriptional regulation of these genes is as conserved as their developmental roles.
In order to shed light on these fundamental questions we consider here a particular example, the forkhead transcription factor, Foxa. We review the conserved developmental role of Foxa and the transcriptional regulation of foxa orthologs in different organisms. Foxa is critical for similar morphogenetic processes in the digestive tracts of the mouse (Burtscher and Lickert, 2009), sea urchin (Oliveri et al., 2006) and nematode (Horner et al., 1998) embryos, and in the notochord formation in chordates (Friedman and Kaestner, 2006; Kumano et al., 2006; Kim et al., 2007; Passamaneck et al., 2009). A recent study reveals the cis-regulatory code that controls foxa expression in the sea urchin embryo (Ben-Tabou de Leon and Davidson, 2010). Detailed functional analyses of the regulation of foxa ortholog were done in the nematode, C. elegans, and some regulatory information is available for other species. We review this information and try to understand why foxa developmental role is so highly conserved and whether its transcriptional regulation as conserved as its developmental role.
Foxa conserved developmental role in regulating mesenchymal to epithelial transition
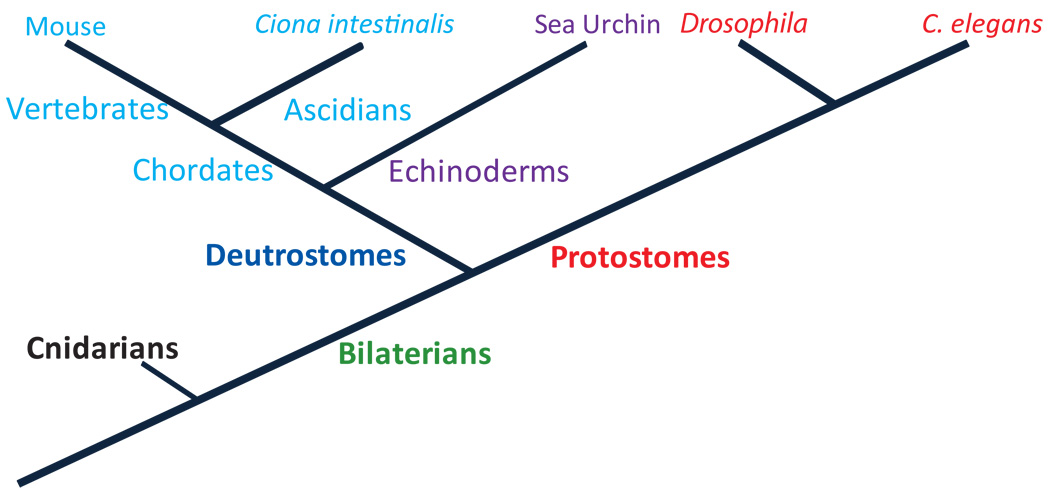
The expression of foxa orthologs in the endoderm lineage is highly conserved across metazoans (Mango et al., 1994; Horner et al., 1998; Koinuma et al., 2000; Fritzenwanker et al., 2004; Suri et al., 2004; Friedman and Kaestner, 2006; Oliveri et al., 2006; Kimura-Yoshida et al., 2007; Boyle and Seaver, 2008; Burtscher and Lickert, 2009). In this section we review the role of Foxa in early development in the species where it was extensively studied, i.e., sea urchin, ascidians, mouse and nematode (see phylogenetic tree, Fig. 1).
Figure 1.
Simplified phylogenetic tree of the organisms discussed in this review in the context of Foxa conserved developmental role and the transcriptional regulation of foxa orthologes.
In the sea urchin embryo, gut formation involves two major processes: local shifts in position of cells that form the archenteron wall and polarized motility of the cells as they rearrange and form tight junctions (Hardin, 1989; Barnet et al., Submitted). In other words, the gut is formed by the local movement of cells that climb on top of each other (intercalation) and then elongate and generate tight junctions to form epithelial sheet. These processes clearly involve the regulation of structural genes and cell adhesion molecules. When foxa is downregulated by the injection of morpholino antisense oligonucleotides these processes do not occur, and there is a failure of gut formation (Oliveri et al., 2006). In addition, the embryo produces excess numbers of pigment cells that are mesenchymal mesoderm derivatives (Oliveri et al., 2006). The suppression of mesodermal fate in the endoderm is mediated, at least in part, by Foxa repression of the gene that encodes the transcription factor GCM (Oliveri et al., 2006), a key regulator of mesodermal fate in the sea urchin embryo (Ransick and Davidson, 2006). Foxa has two other known targets, it activates the transcription of the gene that encodes the ligand, Hedgehog, and it represses its own gene expression (Oliveri et al., 2006). However, these regulatory interactions cannot explain the severe morphological phenotype of foxa downregulation. Most likely, foxa regulates structural genes and cell adhesion molecules that are necessary for intercalation and elongation. This is similar to its role in the definite endoderm (Burtscher and Lickert, 2009) and the notochord (Ang and Rossant, 1994) of the mouse and the pharynx of C. elegans (Gaudet and Mango, 2002; Mango, 2009), as explained below.
Mammals have three orthologs of foxa (Friedman and Kaestner, 2006). foxa ortholog, foxa2, plays a critical role in the specification and morphogenesis of the definitive endoderm (Burtscher and Lickert, 2009) and of the notochord (Ang and Rossant, 1994; Friedman and Kaestner, 2006). The foxa2 gene is the first of foxa orthologs to be activated during mouse embryogenesis and its expression is detected in the anterior primitive streak and the node (Friedman and Kaestner, 2006). foxa2 is expressed in mesoderm and definitive endoderm cells migrating from the node and the expression is maintained in the notochord and throughout the definitive endoderm (Sasaki and Hogan, 1993; Friedman and Kaestner, 2006; Burtscher and Lickert, 2009). A recent study has demonstrated explicitly the role of foxa2 in the definitive endoderm morphogenesis, as follows (Burtscher and Lickert, 2009). At the onset of gastrulation, cells from the definitive endoderm migrate individually and intercalate into the visceral endoderm. Once the cells are within the visceral endoderm they elongate and form tight cell junctions and epithelial sheet. In foxa2 mutant mice these processes do not take place. Most cells do not intercalate and if they do penetrate to the visceral ectoderm they do not polarize nor elongate, and tight junctions are not formed (Burtscher and Lickert, 2009). In these cells the adherent junction proteins E-cadherins and ZO-1 fail to aggregate in the cell junctions. The expression of claudin4, an important cell-adhesion molecule, vanishes in foxa2 mutants (Burtscher and Lickert, 2009). Claudins bind specifically to ZO-1 so foxa2 transcriptional regulation of claudin4 might explain the failure of ZO-1 aggregation. Another direct target of foxa2 in the definitive endoderm is the gene that encodes the transcription factor Gata4, a key endoderm regulator (Rojas et al., 2010). All in all, these studies reveal the important role of foxa2 in controlling intercalation, elongation and formation of epithelial tissue in the definitive endoderm of the mouse by the regulation of regulatory and structural genes that are critical for these processes.
Later in mouse development, foxa orthologs, foxa1 and foxa2 are required for normal development of endoderm-derived organs such as the pancreas and the lungs (Wan et al., 2005; Friedman and Kaestner, 2006). Interestingly, recent studies show that foxa2 functions as a suppressor of tumor metastasis by inhibition of epithelial-to-mesenchymal transition in human lung and pancreatic cancers (Song et al., 2010; Tang et al., 2010). In human lung cancer cells Foxa2 directly represses the gene that encodes the transcription factor Slug (Tang et al., 2010), a key factor in epithelial-to-mesenchymal transition. This could be a part of the regulatory control that foxa mandates to promote epithelial formation and prevent mesenchymal fate. Apparently, the regulatory role that foxa plays through development is relevant to the normal function of differentiated adult cells that express foxa orthologs.
foxa2 plays a role in the notochord formation across chordates (Ang and Rossant, 1994; Weinstein et al., 1994; Ruiz i Altaba et al., 1995; Shimeld, 1997; Friedman and Kaestner, 2006; Yamanaka et al., 2007). The notochord is a mesoderm derivative that is a hallmark of all chordates (Stemple, 2004). In its final form, the notochord is a rod of large cells positioned between the developing spinal cord and gut (Stemple, 2004). The notochord produces a variety of secreted signaling factors, such as Sonic hedgehog, which induce particular specification states in the notochord surrounding tissues (Stemple, 2004). Similarly to the definitive endoderm in the mouse, the notochord is formed through cells intercalation, elongation and formation of tight junctions (Munro and Odell, 2002; Yamanaka et al., 2007). In foxa2 mutant mouse the notochord is completely missing (Ang and Rossant, 1994; Yamanaka et al., 2007) indicating a critical role of foxa2 in regulating the formation of this organ. In the mouse embryo, Foxa2 drives the expression of the gene that encodes the T-box transcription factor, Brachyury, and together, Foxa2 and Brachyury drive not and sonic hedgehog, key notochord regulatory genes (Jeong and Epstein, 2003; Abdelkhalek et al., 2004;Yamanaka et al., 2007). Multiple studies show that Foxa and Brachyury directly regulate gene expression in the ascidian notochord, which suggests a conserved regulatory role of these two factors in the notochord development (Kumano et al., 2006; Passamaneck et al., 2009). This is another example of a developmental process that involves intercalation, elongation and formation of epithelial tissue that is entirely abolished by the knock-out of foxa ortholog.
pha-4 is the ortholog of foxa in the nematode, C. elegans. pha-4 is expressed in all the cells that generate the pharynx and the gut, that is, in all the daughters of the E cell (endoderm), and all the daughters of MS and AB cells that contribute to the pharynx (mesoderm) (Azzaria et al., 1996; Murray et al., 2008). During pharynx development, cells that originate from different lineages but are fated to form the pharynx, cluster together, ingress and go through mesenchymal to epithelial transition (Mango, 2009). In pha-4 mutants the entire pharynx is deleted while other organs, including the gut, appear to be normal (Mango et al., 1994; Horner et al., 1998; Mango, 2009). The pharyngeal precursors that normally cluster together and ingress during gastrulation remain dispersed in the embryo surface when pha-4 is mutated (Horner et al., 1998). This elimination of clustering and epithelialization is similar to foxa2 phenotypes in other organisms discussed above. Different studies show that Pha-4 regulates many of the regulatory and structural genes that are necessary for the specification and morphogenesis of the pharynx (Gaudet and Mango, 2002; Anokye-Danso et al., 2008; Mango, 2009) as well as represses ectoderm regulatory genes (Kiefer et al., 2007). Overall, Pha-4 is the central regulator of the pharynx development and its expression is necessary for this organ formation, in particular, for cells clustering, migration and mesenchymal to epithelial transition.
It is important to note that the regulatory role of Foxa is not limited to intercalation and epithelialization nor can it be the only regulator of these processes. For example, Pha-4 has different regulatory roles later in C. elegans development and aging (Panowski et al., 2007; Chen and Riddle, 2008). On the other hand, the gut formation in C. elegans is unaffected in pha-4 mutant so other factors are regulating epithelialization there. foxa2 and other foxa orthologs in the mouse are important to various regulatory processes (see e.g., (Kimura-Yoshida et al., 2007; Gao et al., 2008; Lin et al., 2009)). Yet, the similarity of the phenotypes observed when foxa orthologs are knocked down in diverse organisms such as the mouse, the nematode and the sea urchin is quite striking. This conserved role of foxa in embryogenesis raises the following question: is foxa regulation as conserved as its role in early development, or did it change to accommodate evolutionary changes that involve foxa function? The next sections describe foxa regulation in different organisms, and shed light on this question.
foxa transcriptional regulation in the sea urchin embryo
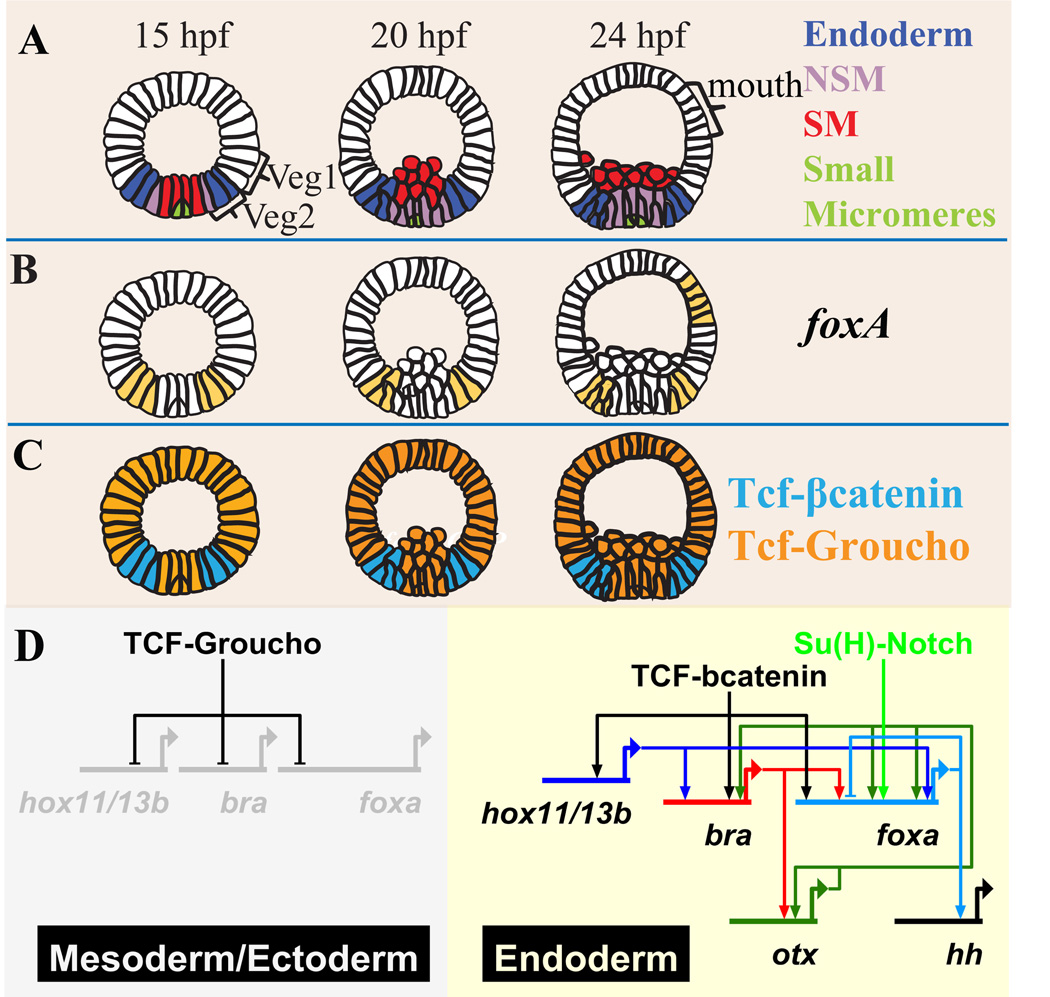
A detailed cis-regulatory analysis was recently conducted to decipher the genomic code that controls foxa expression early in sea urchin development (Ben-Tabou de Leon and Davidson, 2010). Here we review the main results of this work. In the sea urchin embryo foxa is expressed in the presumptive mesoderm and endoderm. At mid-blastula stage, (about 15 hours post fertilization (hpf)) foxa is expressed in both non-skeletogenic mesoderm (NSM) and endoderm precursors (Fig. 2A, B) (Peter and Davidson, 2010). At mesenchyme blastula stage (18–20 hpf), foxa expression is shut down in the NSM progenitors and transcription continues from then on only in the endoderm (Fig. 2A, B) (Peter and Davidson, 2010). Later, foxa is also expressed in a patch of cells in the oral ectoderm where the mouth will form (Fig. 2B). One of the important aspects of foxa regulation is the transition between the broad expression in both the NSM and endoderm to the specific expression in the endoderm. Understanding this transition can illuminate the mechanisms that control the endoderm versus mesoderm cell fate decision.
Figure 2.
foxa expression and cis-regulation in the sea urchin embryo (Ben-Tabou de Leon and Davidson, 2010). A. Lineage fate map showing lateral view of the sea urchin embryo at 15, 20 and 24 hpf. The SM lineage is marked in red, the NSM lineage is marked in purple, veg2 endoderm lineage is marked in blue and veg1 and the ectoderm lineages are marked in white. The most vegetal descendents of veg1 contribute to the endoderm. B. foxa spatial expression at 15, 20 and 24 hpf. C. Diagrams of Tcf activity mode at 15, 20 and 24hpf. The cells where Tcf binds to the co-repressor Groucho to form a repressive complex are marked in orange. The cells where Tcf binds to βcatenin to form a permissive complex are marked in cyan. D. Diagram of the gene regulatory network that drives foxa expression in the sea urchin embryo. Tcf-Groucho represses foxa and other endodermal genes in the ectoderm and progressively in the mesoderm. The multiple additive activators that contribute to foxa expression are Hox11/13b, Su(H)-Notch, Otx and Brachyury. Foxa represses its own gene expression.
cis-regulatory analysis of foxa reveals that the spatial expression of foxa is restricted by Tcf-Groucho/β-catenin toggle switch (Fig. 2C, D) (Weitzel et al., 2004; Wikramanayake et al., 2004; Range et al., 2005; Ben-Tabou de Leon and Davidson, 2010). Tcf is a transcription factor that can act either as a repressor or as an activator, depending on its cofactors (Range et al., 2005; Ben-Tabou de-Leon and Davidson, 2007). When Tcf binds to β-catenin they form a complex permissive for transcription, otherwise Tcf and the co-repressor Groucho form dominant repressor complex. Therefore Tcf targets are expressed only in cells where β-catenin is nuclear localized and repressed elsewhere. Maternal anisotropies in the sea urchin egg leads to β-catenin nuclearization in the vegetal cells of the embryo early in development (Logan et al., 1999; Weitzel et al., 2004). By mid-blastula stage nuclear β-catenin has cleared from the skeletogenic mesoderm (SM) nuclei and is localized in the veg2 lineage nuclei, i.e., in cells that give rise to NSM plus endoderm (Fig. 2C, (Logan et al., 1999)). At this time foxa expression is likewise restricted to these cells (Fig. 2 B,C, (Peter and Davidson, 2010)). At mesenchyme blastula stage β-catenin clears from the NSM nuclei as well, and remains visible only in the nuclei of veg2 and veg1 endoderm (Fig. 2C) (Logan et al., 1999). This leads to silencing of foxa expression in the NSM due to Tcf-Groucho repression there. Expression of foxa is henceforth restricted to the veg2 endoderm (Fig. 2B, C). Thus the Tcf-Groucho/β-catenin system initially enables broad foxa expression throughout the veg2 endomesoderm and later restricts it to the endodermal domain of this lineage.
foxa is activated by multiple additive inputs (Fig. 2D) (Ben-Tabou de Leon and Davidson, 2010). One of the early activators of foxa is the transcription factor Hox11/13b that is co-expressed with foxa at blastula stage (Peter and Davidson, 2010). Later in development hox11/13b expression turns off in veg2 and the gene becomes active in veg1, where foxa is not expressed. At blastula stage foxa is also a target of the Delta-Notch signaling that occurs in veg2 and its NSM descendents due to reception of the Delta ligand produced by the adjacent SM cells (Fig. 2D) (Ben-Tabou de Leon and Davidson, 2010). At mesenchyme blastula stage the transcription factors Otx and Brachyury boost foxa expression and Foxa represses its own gene expression (Fig. 2D) (Ben-Tabou de Leon and Davidson, 2010). Different Otx isoforms are expressed everywhere in the embryo throughout development. Therefore the Otx input probably acts to boost foxa level, providing no spatial information. brachyury is co-expressed with foxa starting at blastula stage (Peter and Davidson, 2010). At mesenchyme blastula stage brachyury expression begins to fade in veg2 and becomes active in the veg1 ring of cells. Thus after 24hpf the expression domains of these genes have a small overlap.
Summing up foxa cis-regulation at early development of the sea urchin embryo, foxa is regulated by the activators Hox11/13b, Su(H), Brachyury and Otx, that act additively and partially overlap in time and embryonic space with foxa expression. Foxa is an auto-repressor. foxa expression is restricted spatially by Tcf-Groucho repression that prevents expression in the ectoderm and progressively in the mesoderm. In regulatory logic terms, foxa cis-regulatory modules execute additive OR logic on all its positive inputs, and NOT logic on Tcf-Groucho.
The transcriptional regulation of foxa ortholog, pha-4, in C. elegans
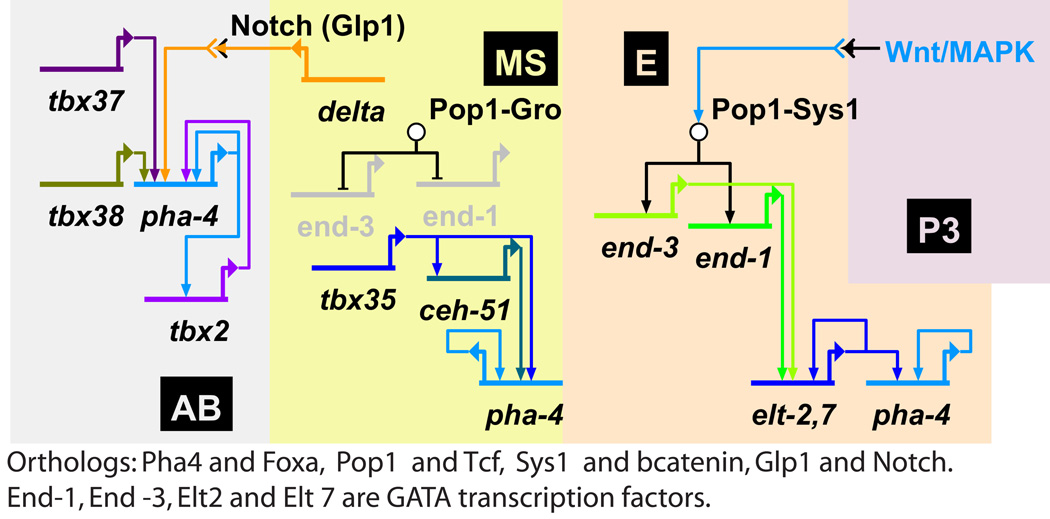
As described above, pha-4, the C. elegans ortholog of foxa, is expressed in the gut and in the pharynx of the nematode (Horner et al., 1998). The direct regulation of pha-4 in C. elegans was not studied by a cis-regulatory analysis. Yet, a comprehensive study of mutants and knock-down phenotypes enabled the construction of a model of the endomesoderm GRN, that includes pha-4 predicted connections (Owraghi et al., 2010). Based on this work, we present a partial GRN diagram that highlights the predications for pha-4 inputs in different cell lineages, Fig. 3 (Owraghi et al., 2010).The gut is formed from all the daughters of the E cell. Reception of Wnt/MapK signaling in the E cell activates the orthologs of the Tcf-βcatenin switch (Pop1-Sys1) and as a result the genes encoding the GATA factors End-1 and End-3 are turned on in this cell (Owraghi et al., 2010). These genes are repressed by Tcf- Groucho in the MS cell that does not receive the Wnt/MapK signaling. Apparently, pha-4 does not have a direct link to Tcf since it is expressed in both the E and the MS cells. In the descendents of the E cell, End-1 and End-3 activate the genes encoding the GATA factors Elt-2 and Elt-7, which then form a positive feedback regulatory loop and activate the expression of the pha-4 gene (Fig. 3).
Figure 3.
The predicted transcriptional regulation of foxa ortholog, pha-4, in the nematode C. elegans (based on (Owraghi et al., 2010)). Different lineages are indicated by different background colors. Pha-4 is expressed in all the daughters of the E cell and in the daughters of the AB and MS cells that gives rise to the pharynx. Pop1 is ortholog of Tcf, Sys1 is ortholog of βcatenin, End-1, End-3, Elt-2 and Elt-7 are GATA transcription factors. In each lineage pha-4 is activated by different set of transcription factors. In all the lineages Pha-4 positively regulates its own gene expression.
Regulatory roles of Tcf-βcatenin and GATA factors in the endoderm specification were observed in other organisms. Gata4/5/6 family of transcription factors plays a central role in the endoderm specification in vertebrates (Soudais et al., 1995; Bossard and Zaret, 1998; Capo-Chichi et al., 2005; Zorn and Wells, 2007) and Drosophila (Murakami et al., 2005). As stated above, in the sea urchin embryo the Tcf-βcatenin switch restricts the spatial expression of foxa and other endodermal genes (Fig. 2D) (Ben-Tabou de Leon and Davidson, 2010). In the embryo of the chordate, Ciona intestinalis, βcatenin is important to the endoderm specification (Imai et al., 2000). Deletion of βcatenin in the mouse definitive endoderm changes cell fate from endoderm to precardiac mesoderm and lead to the formation of multiple hearts (Lickert et al., 2002). The widely shared role of GATA factors and Tcf-Groucho/βcatenin in the endoderm specification suggests that these factors are part of an ancestral endoderm GRN. The GRN that controls the E cell lineage seem to be a derived form of this ancestral GRN. However, the GRN that controls the pharynx formation is significantly different.
In the daughters of the MS cell that contributes to the pharynx, pha-4 is downstream of the T-box transcription factor Tbx35 and the Nk-2 class homeodomain transcription factor, Ceh-51 (Fig. 3) (Owraghi et al., 2010). These two factors are expressed in the MS lineage and are essential to the production of MS-derived tissues (Broitman-Maduro et al., 2009). The MS cells secrete the ligand Delta and its reception activates pha-4 expression in the daughters of the AB cell that contribute to the pharynx (Fig. 3) (Owraghi et al., 2010). In these cells the T-box transcription factors, Tbx37 and Tbx38, contributes to the expression of pha-4 (Fig. 3). In AB daughter cells, Pha-4 activates the gene that encodes the T-box transcription factor, Tbx-2 (Fig. 3) (Smith and Mango, 2007). Tbx-2 feeds back and activates pha-4 expression so the two genes form a positive feedback loop essential for the maintenance of pha-4 expression (Smith and Mango, 2007). Interestingly, the regulatory interaction between foxa orthologs to T-box transcription factors is common to the AB and MS cells in C. elegans, the chordate notochord and the endoderm of the sea urchin. The T-box transcription factor, Brachyury, interacts with Foxa orthologs in the chordate notochord and in the sea urchin endoderm (Jeong and Epstein, 2003; Abdelkhalek et al., 2004; Hotta et al., 2008; Tamplin et al., 2008; Ben-Tabou de Leon and Davidson, 2010). The reason for this similarity could be the ancestral regulatory role of Tbox transcription factors, particularly Brachyury (Technau, 2001; Marcellini et al., 2003; Hotta et al., 2008) in controlling morphogenetic processes in cooperation with Foxa.
Pha-4 positively regulates its own gene expression in all the cells where it is expressed, but it is not clear whether this regulatory interaction is direct or indirect. The regulatory control of pha-4 in the MS and AB cells seem to be C. elegans specific, except from the Delta-Notch input. As stated above, the reception of the SM Delta signal contributes to foxa expression in the NSM lineage in the sea urchin (Fig. 2D) (Ben-Tabou de Leon and Davidson, 2010). It is hard to conclude about the evolution of Delta-foxa regulatory connection based on echinoderm and nematodes only. Without further evidence for Delta activation of foxa orthologs in other organisms, this link seems to be the result of convergent evolution and not a part of an ancestral GRN.
All in all, pha-4 is activated by different activators in each of the three lineages that form the gut and the pharynx in C. elegans. It appears that pha-4 does not have a direct link to Tcf so its expression outside of the E cell lineage is not restricted by Tcf-Groucho repression. Apparently, pha-4 gained new activators that drive its expression in the cells that form the pharynx. The absence of a restricting repressor and the gain of new activators allow the expression of pha-4 in mesodermal lineages where pha-4 drives its distinctive developmental program.
Discussion
The embryo morphologies of the sea urchin, the mouse and nematode, C. elegans, are significantly different. Yet, in all these organisms, orthologs of foxa are expressed in cells that intercalate, polarize and form tight junctions. The loss of foxa expression eliminates these morphogenetic processes. The transcriptional regulation of foxa in C. elegans is quite different from its regulation in the sea urchin embryo (compare Fig. 3 to Fig. 2D). foxa activating inputs are not only different between the two organisms, but in the case of C. elegans, unique in every lineage where pha-4 is expressed (Fig. 3). Apparently, the upstream regulation of foxa is rather flexible while the downstream phenotypes are highly conserved. Davidson and Erwin recently suggested that basal ancestral GRNs were shallow and consisted of few regulatory genes that controlled batteries of structural genes (Davidson and Erwin, 2009). They propose that evolution had added regulatory links into the GRN of each species, to either advance the network function or to redeploy the ancestral GRN into a new embryonic address. Taking this view and the evidences presented above, Foxa might have been a member of such ancestral GRN that controlled structural genes necessary for cell motility and migration and for mesenchymal to epithelial transition. foxa regulation had changed through evolution according to the developmental program in each species so it plays its role controlling morphogenesis at the relevant embryonic lineage.
An apparent example of regulatory changes that enabled the activation of foxa in a new embryonic territory is the activation of pha-4 in the pharynx of the C. elegans. pha-4 is driven by a specific set of transcription factors in the MS lineage, and a different set of transcription factors specific to the AB lineage. This regulatory code enabled the mesodermal expression of pha-4 and the cooption of the developmental program that it drives. Evolutionary modification of the regulatory code must have led to foxa2 expression in the notochord, a chordate mesoderm derivative (Ang and Rossant, 1994; Weinstein et al., 1994; Yamanaka et al., 2007)
The logic applied on foxa inputs might have contributed to the flexibility of foxa regulation. In the sea urchin embryo foxa expression is regulated by a combination of multiple additive inputs, that is, OR logic (Istrail and Davidson, 2005; Istrail et al., 2007; Ben-Tabou de Leon and Davidson, 2010). In cis-regulatory modules that execute OR logic, loss of a binding site does not eliminate the expression of the downstream gene since other inputs are still driving it. Furthermore, in Or logic, an addition of a functional binding site does not require the presence of other specific binding sites. On the other hand, in cis-regulatory modules that execute AND logic (multiple necessary inputs (Istrail and Davidson, 2005; Istrail et al., 2007)), loss of any binding site causes complete loss of the downstream gene expression. Gain of function in AND logic can only happen when there is a gain of binding sites of all the necessary inputs. Therefore, in principle, OR logic might be more flexible to evolutionary changes of addition or deletion of binding sites. If the logic applied on foxa inputs in other organisms is also OR logic, it might have facilitated the evolutionary flexibility of foxa transcriptional regulation. It would be illuminating to identify the logic applied on foxa inputs in other organisms, and learn about the role of regulatory logic in GRN evolution.
The connection between the evolution of developmental GRN to the evolution of diverse body plans is more and more evident as models of GRNs of diverse organisms become available (Davidson and Erwin, 2009; Erwin and Davidson, 2009). It is now apparent that different parts of GRNs evolve at a different pace and some parts are more conserved then others (Hinman et al., 2003; Hinman and Davidson, 2007; Davidson and Erwin, 2009; Erwin and Davidson, 2009; McCauley et al., 2010). Here we focused on one regulatory gene and tried to understand the reasons for its highly conserved developmental role and whether its regulation is as conserved as its role. We learned that Foxa orthologs in a wide spectrum of organisms are essential to a specific morphogenetic process: cells intercalation followed by elongation and epithelialization. Foxa controls this process mainly in the endoderm, but also in the mesoderm of some organisms. The extreme conservation of Foxa developmental role is most likely due to its ancestral direct control of batteries of structural genes that mandate mesenchymal to epithelial transition and initially enabled the gut formation. Foxa regulation had changed through evolution to accommodate foxa expression at novel embryonic addresses, for example, the C. elegans pharynx and the chordate notochord, both mesodermal derivatives. That is, evolution had added layers of regulatory control in order to either redeploy or improve the function of the conserved morphogenetic program controlled by Foxa.
Acknowledgments
The author thanks Eric Davidson for insightful discussions and critical review of the manuscript that helped shape the paper final form. The author thanks Ute Deichmann, Jongmin Nam, Shlomo Ben-Tabou de-Leon and Veronica Hinman for inspiration. Research was supported by NIH grant GM61005.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelkhalek HB, Beckers A, Schuster-Gossler K, Pavlova MN, Burkhardt H, Lickert H, Rossant J, Reinhardt R, Schalkwyk LC, Muller I, Herrmann BG, Ceolin M, Rivera-Pomar R, Gossler A. The mouse homeobox gene Not is required for caudal notochord development and affected by the truncate mutation. Genes Dev. 2004;18:1725–1736. doi: 10.1101/gad.303504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang SL, Rossant J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. doi: 10.1016/0092-8674(94)90522-3. [DOI] [PubMed] [Google Scholar]
- Anokye-Danso F, Anyanful A, Sakube Y, Kagawa H. Transcription factors GATA/ELT-2 and forkhead/HNF-3/PHA-4 regulate the tropomyosin gene expression in the pharynx and intestine of Caenorhabditis elegans. J Mol Biol. 2008;379:201–211. doi: 10.1016/j.jmb.2007.11.103. [DOI] [PubMed] [Google Scholar]
- Azzaria M, Goszczynski B, Chung MA, Kalb JM, McGhee JD. A fork head/HNF-3 homolog expressed in the pharynx and intestine of the Caenorhabditis elegans embryo. Dev Biol. 1996;178:289–303. doi: 10.1006/dbio.1996.0219. [DOI] [PubMed] [Google Scholar]
- Barnet ME, Peter IS, Davidson EH, Fraser SE. Dynamics of sea urchin gastrulation revealed by tracking cells of diverse lineage and regulatory state. Development. Submitted. [Google Scholar]
- Ben-Tabou de-Leon S, Davidson EH. Gene regulation: gene control network in development. Annu Rev Biophys Biomol Struct. 2007;36:191. doi: 10.1146/annurev.biophys.35.040405.102002. [DOI] [PubMed] [Google Scholar]
- Ben-Tabou de-Leon S, Davidson EH. Experimentally based sea urchin gene regulatory network and the causal explanation of developmental phenomenology. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2009;1:237–246. doi: 10.1002/wsbm.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Tabou de Leon S, Davidson EH. Information processing at the foxa node of the sea urchin endomesoderm specification network. Proc Natl Acad Sci U S A. 2010;107:10103–10108. doi: 10.1073/pnas.1004824107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossard P, Zaret KS. GATA transcription factors as potentiators of gut endoderm differentiation. Development. 1998;125:4909–4917. doi: 10.1242/dev.125.24.4909. [DOI] [PubMed] [Google Scholar]
- Boyle MJ, Seaver EC. Developmental expression of foxA and gata genes during gut formation in the polychaete annelid, Capitella sp. I. Evol Dev. 2008;10:89–105. doi: 10.1111/j.1525-142X.2007.00216.x. [DOI] [PubMed] [Google Scholar]
- Broitman-Maduro G, Owraghi M, Hung WW, Kuntz S, Sternberg PW, Maduro MF. The NK-2 class homeodomain factor CEH-51 and the T-box factor TBX-35 have overlapping function in C. elegans mesoderm development. Development. 2009;136:2735–2746. doi: 10.1242/dev.038307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtscher I, Lickert H. Foxa2 regulates polarity and epithelialization in the endoderm germ layer of the mouse embryo. Development. 2009;136:1029–1038. doi: 10.1242/dev.028415. [DOI] [PubMed] [Google Scholar]
- Capo-Chichi CD, Rula ME, Smedberg JL, Vanderveer L, Parmacek MS, Morrisey EE, Godwin AK, Xu XX. Perception of differentiation cues by GATA factors in primitive endoderm lineage determination of mouse embryonic stem cells. Dev Biol. 2005;286:574–586. doi: 10.1016/j.ydbio.2005.07.037. [DOI] [PubMed] [Google Scholar]
- Chen D, Riddle DL. Function of the PHA-4/FOXA transcription factor during C. elegans post-embryonic development. BMC Dev Biol. 2008;8:26. doi: 10.1186/1471-213X-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. The regulatory genome: gene regulatory networks in development and evolution. San-Diego: Academic press; 2006. [Google Scholar]
- Davidson EH, Erwin DH. An integrated view of precambrian eumetazoan evolution. Cold Spring Harb Symp Quant Biol. 2009;74:65–80. doi: 10.1101/sqb.2009.74.042. [DOI] [PubMed] [Google Scholar]
- Erwin DH, Davidson EH. The evolution of hierarchical gene regulatory networks. Nat Rev Genet. 2009;10:141–148. doi: 10.1038/nrg2499. [DOI] [PubMed] [Google Scholar]
- Fraser GJ, Hulsey CD, Bloomquist RF, Uyesugi K, Manley NR, Streelman JT. An ancient gene network is co-opted for teeth on old and new jaws. PLoS Biol. 2009;7:e31. doi: 10.1371/journal.pbio.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzenwanker JH, Saina M, Technau U. Analysis of forkhead and snail expression reveals epithelial-mesenchymal transitions during embryonic and larval development of Nematostella vectensis. Dev Biol. 2004;275:389–402. doi: 10.1016/j.ydbio.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Gao F, Davidson EH. Transfer of a large gene regulatory apparatus to a new developmental address in echinoid evolution. Proc Natl Acad Sci U S A. 2008;105:6091–6096. doi: 10.1073/pnas.0801201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, LeLay J, Vatamaniuk MZ, Rieck S, Friedman JR, Kaestner KH. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22:3435–3448. doi: 10.1101/gad.1752608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Hardin J. Local shifts in position and polarized motility drive cell rearrangement during sea urchin gastrulation. Dev Biol. 1989;136:430–445. doi: 10.1016/0012-1606(89)90268-6. [DOI] [PubMed] [Google Scholar]
- Harvey RP. NK-2 homeobox genes and heart development. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- Hinman VF, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proc Natl Acad Sci U S A. 2007;104:19404–19409. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc Natl Acad Sci U S A. 2003;100:13356–13361. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland ND, Venkatesh TV, Holland LZ, Jacobs DK, Bodmer R. AmphiNk2-tin, an amphioxus homeobox gene expressed in myocardial progenitors: insights into evolution of the vertebrate heart. Dev Biol. 2003;255:128–137. doi: 10.1016/s0012-1606(02)00050-7. [DOI] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 1998;12:1947–1952. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Satoh N, Gojobori T. Brachyury-downstream gene sets in a chordate, Ciona intestinalis: integrating notochord specification, morphogenesis and chordate evolution. Evol Dev. 2008;10:37–51. doi: 10.1111/j.1525-142X.2007.00212.x. [DOI] [PubMed] [Google Scholar]
- Imai K, Takada N, Satoh N, Satou Y. (beta)-catenin mediates the specification of endoderm cells in ascidian embryos. Development. 2000;127:3009–3020. doi: 10.1242/dev.127.14.3009. [DOI] [PubMed] [Google Scholar]
- Istrail S, Ben-Tabou de-Leon S, Davidson EH. The regulatory genome and the computer. Dev Biol. 2007;310:187–195. doi: 10.1016/j.ydbio.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Istrail S, Davidson EH. Logic functions of the genomic cis-regulatory code. Proc Natl Acad Sci U S A. 2005;102:4954–4959. doi: 10.1073/pnas.0409624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, Epstein DJ. Distinct regulators of Shh transcription in the floor plate and notochord indicate separate origins for these tissues in the mouse node. Development. 2003;130:3891–3902. doi: 10.1242/dev.00590. [DOI] [PubMed] [Google Scholar]
- Kiefer JC, Smith PA, Mango SE. PHA-4/FoxA cooperates with TAM-1/TRIM to regulate cell fate restriction in the C. elegans foregut. Dev Biol. 2007;303:611–624. doi: 10.1016/j.ydbio.2006.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GJ, Kumano G, Nishida H. Cell fate polarization in ascidian mesenchyme/muscle precursors by directed FGF signaling and role for an additional ectodermal FGF antagonizing signal in notochord/nerve cord precursors. Development. 2007;134:1509–1518. doi: 10.1242/dev.02825. [DOI] [PubMed] [Google Scholar]
- Kimura-Yoshida C, Tian E, Nakano H, Amazaki S, Shimokawa K, Rossant J, Aizawa S, Matsuo I. Crucial roles of Foxa2 in mouse anterior-posterior axis polarization via regulation of anterior visceral endoderm-specific genes. Proc Natl Acad Sci U S A. 2007;104:5919–5924. doi: 10.1073/pnas.0607779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinuma S, Umesono Y, Watanabe K, Agata K. Planaria FoxA (HNF3) homologue is specifically expressed in the pharynx-forming cells. Gene. 2000;259:171–176. doi: 10.1016/s0378-1119(00)00426-1. [DOI] [PubMed] [Google Scholar]
- Kozmik Z. Pax genes in eye development and evolution. Curr Opin Genet Dev. 2005;15:430–438. doi: 10.1016/j.gde.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Kumano G, Yamaguchi S, Nishida H. Overlapping expression of FoxA and Zic confers responsiveness to FGF signaling to specify notochord in ascidian embryos. Dev Biol. 2006;300:770–784. doi: 10.1016/j.ydbio.2006.07.033. [DOI] [PubMed] [Google Scholar]
- Lemons D, Fritzenwanker JH, Gerhart J, Lowe CJ, McGinnis W. Co-option of an anteroposterior head axis patterning system for proximodistal patterning of appendages in early bilaterian evolution. Dev Biol. 2010;344:358–362. doi: 10.1016/j.ydbio.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Kutsch S, Kanzler B, Tamai Y, Taketo MM, Kemler R. Formation of multiple hearts in mice following deletion of beta-catenin in the embryonic endoderm. Dev Cell. 2002;3:171–181. doi: 10.1016/s1534-5807(02)00206-x. [DOI] [PubMed] [Google Scholar]
- Lin W, Metzakopian E, Mavromatakis YE, Gao N, Balaskas N, Sasaki H, Briscoe J, Whitsett JA, Goulding M, Kaestner KH, Ang SL. Foxa1 and Foxa2 function both upstream of and cooperatively with Lmx1a and Lmx1b in a feedforward loop promoting mesodiencephalic dopaminergic neuron development. Dev Biol. 2009;333:386–396. doi: 10.1016/j.ydbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- Mango SE. The molecular basis of organ formation: insights from the C. elegans foregut. Annu Rev Cell Dev Biol. 2009;25:597–628. doi: 10.1146/annurev.cellbio.24.110707.175411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango SE, Lambie EJ, Kimble J. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development. 1994;120:3019–3031. doi: 10.1242/dev.120.10.3019. [DOI] [PubMed] [Google Scholar]
- Marcellini S, Technau U, Smith JC, Lemaire P. Evolution of Brachyury proteins: identification of a novel regulatory domain conserved within Bilateria. Dev Biol. 2003;260:352–361. doi: 10.1016/s0012-1606(03)00244-6. [DOI] [PubMed] [Google Scholar]
- McCauley BS, Weideman EP, Hinman VF. A conserved gene regulatory network subcircuit drives different developmental fates in the vegetal pole of highly divergent echinoderm embryos. Dev Biol. 2010;340:200–208. doi: 10.1016/j.ydbio.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Munro EM, Odell GM. Polarized basolateral cell motility underlies invagination and convergent extension of the ascidian notochord. Development. 2002;129:13–24. doi: 10.1242/dev.129.1.13. [DOI] [PubMed] [Google Scholar]
- Murakami R, Okumura T, Uchiyama H. GATA factors as key regulatory molecules in the development of Drosophila endoderm. Dev Growth Differ. 2005;47:581–589. doi: 10.1111/j.1440-169X.2005.00836.x. [DOI] [PubMed] [Google Scholar]
- Murray JI, Bao Z, Boyle TJ, Boeck ME, Mericle BL, Nicholas TJ, Zhao Z, Sandel MJ, Waterston RH. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat Methods. 2008;5:703–709. doi: 10.1038/nmeth.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveri P, Walton KD, Davidson EH, McClay DR. Repression of mesodermal fate by foxa, a key endoderm regulator of the sea urchin embryo. Development. 2006;133:4173–4181. doi: 10.1242/dev.02577. [DOI] [PubMed] [Google Scholar]
- Owraghi M, Broitman-Maduro G, Luu T, Roberson H, Maduro MF. Roles of the Wnt effector POP-1/TCF in the C. elegans endomesoderm specification gene network. Dev Biol. 2010;340:209–221. doi: 10.1016/j.ydbio.2009.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Passamaneck YJ, Katikala L, Perrone L, Dunn MP, Oda-Ishii I, Di Gregorio A. Direct activation of a notochord cis-regulatory module by Brachyury and FoxA in the ascidian Ciona intestinalis. Development. 2009;136:3679–3689. doi: 10.1242/dev.038141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. The endoderm gene regulatory network in sea urchin embryos up to mid-blastula stage. Dev Biol. 2010;340:188–199. doi: 10.1016/j.ydbio.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichaud F, Desplan C. Pax genes and eye organogenesis. Curr Opin Genet Dev. 2002;12:430–434. doi: 10.1016/s0959-437x(02)00321-0. [DOI] [PubMed] [Google Scholar]
- Range RC, Venuti JM, McClay DR. LvGroucho and nuclear beta-catenin functionally compete for Tcf binding to influence activation of the endomesoderm gene regulatory network in the sea urchin embryo. Dev Biol. 2005;279:252–267. doi: 10.1016/j.ydbio.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. cis-regulatory processing of Notch signaling input to the sea urchin glial cells missing gene during mesoderm specification. Dev Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Rojas A, Schachterle W, Xu SM, Martin F, Black BL. Direct transcriptional regulation of Gata4 during early endoderm specification is controlled by FoxA2 binding to an intronic enhancer. Dev Biol. 2010;346:346–355. doi: 10.1016/j.ydbio.2010.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Placzek M, Baldassare M, Dodd J, Jessell TM. Early stages of notochord and floor plate development in the chick embryo defined by normal and induced expression of HNF-3 beta. Dev Biol. 1995;170:299–313. doi: 10.1006/dbio.1995.1216. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hogan BL. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development. 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- Shimeld SM. Characterisation of amphioxus HNF-3 genes: conserved expression in the notochord and floor plate. Dev Biol. 1997;183:74–85. doi: 10.1006/dbio.1996.8481. [DOI] [PubMed] [Google Scholar]
- Smith PA, Mango SE. Role of T-box gene tbx-2 for anterior foregut muscle development in C. elegans. Dev Biol. 2007;302:25–39. doi: 10.1016/j.ydbio.2006.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Washington MK, Crawford HC. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 2010;70:2115–2125. doi: 10.1158/0008-5472.CAN-09-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudais C, Bielinska M, Heikinheimo M, MacArthur CA, Narita N, Saffitz JE, Simon MC, Leiden JM, Wilson DB. Targeted mutagenesis of the transcription factor GATA-4 gene in mouse embryonic stem cells disrupts visceral endoderm differentiation in vitro. Development. 1995;121:3877–3888. doi: 10.1242/dev.121.11.3877. [DOI] [PubMed] [Google Scholar]
- Stemple DL. The notochord. Curr Biol. 2004;14:R873–R874. doi: 10.1016/j.cub.2004.09.065. [DOI] [PubMed] [Google Scholar]
- Suri C, Haremaki T, Weinstein DC. Inhibition of mesodermal fate by Xenopus HNF3beta/FoxA2. Dev Biol. 2004;265:90–104. doi: 10.1016/j.ydbio.2003.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamplin OJ, Kinzel D, Cox BJ, Bell CE, Rossant J, Lickert H. Microarray analysis of Foxa2 mutant mouse embryos reveals novel gene expression and inductive roles for the gastrula organizer and its derivatives. BMC Genomics. 2008;9:511. doi: 10.1186/1471-2164-9-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Shu G, Yuan X, Jing N, Song J. FOXA2 functions as a suppressor of tumor metastasis by inhibition of epithelial-to-mesenchymal transition in human lung cancers. Cell Res. 2010 doi: 10.1038/cr.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technau U. Brachyury, the blastopore and the evolution of the mesoderm. Bioessays. 2001;23:788–794. doi: 10.1002/bies.1114. [DOI] [PubMed] [Google Scholar]
- Wan H, Dingle S, Xu Y, Besnard V, Kaestner KH, Ang SL, Wert S, Stahlman MT, Whitsett JA. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J Biol Chem. 2005;280:13809–13816. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- Weinstein DC, Ruiz i Altaba A, Chen WS, Hoodless P, Prezioso VR, Jessell TM, Darnell JE., Jr. The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. doi: 10.1016/0092-8674(94)90523-1. [DOI] [PubMed] [Google Scholar]
- Weitzel HE, Illies MR, Byrum CA, Xu R, Wikramanayake AH, Ettensohn CA. Differential stability of beta-catenin along the animal-vegetal axis of the sea urchin embryo mediated by dishevelled. Development. 2004;131:2947–2956. doi: 10.1242/dev.01152. [DOI] [PubMed] [Google Scholar]
- Wikramanayake AH, Peterson R, Chen J, Huang L, Bince JM, McClay DR, Klein WH. Nuclear beta-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis. 2004;39:194–205. doi: 10.1002/gene.20045. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Tamplin OJ, Beckers A, Gossler A, Rossant J. Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Dev Cell. 2007;13:884–896. doi: 10.1016/j.devcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Zorn AM, Wells JM. Molecular basis of vertebrate endoderm development. Int Rev Cytol. 2007;259:49–111. doi: 10.1016/S0074-7696(06)59002-3. [DOI] [PubMed] [Google Scholar]