Abstract
Human serum albumin (HSA) and immunoglobulin G (IgG) represent over 75% of all proteins present in human plasma. These two proteins frequently interfere with detection, determination and purification of low abundance proteins that can be potential biomarkers and biomarker candidates for various diseases. Some low abundance plasma proteins such as clotting factors and inhibitors are also important therapeutic agents. In this paper, the characterization of ion-exchange monolithic supports under overloading conditions was performed by use of sample displacement chromatography (SDC). If these supports were used for separation of human plasma, the composition of bound and eluted proteins in both anion- and cation-exchange mode is dependent on column loading. Under overloading conditions, the weakly bound proteins such as HSA in anion-exchange and IgG in cation-exchange mode are displaced by stronger binding proteins, and this phenomenon was not dependent on column size. Consequently, small monolithic columns with a column volume of 100 and 200 μL are ideal supports for high-throughput screening in order to develop new methods for separation of complex mixtures, and for sample preparation in proteomic technology.
1 Introduction
Analysis of complex biological fluids such as serum, plasma, urine, and tissue homogenates is complicated by the large dynamic range of individual proteins that are present in these complex mixtures. This range is up to 108 to 1012 in serum and plasma, and up to 105 in cells [1–3]. In human plasma, 22 proteins account for 99% of the overall protein content [2]. Human serum albumin (HSA) and immunoglobulins are the most abundant ones and they represent over 75% of all proteins present in plasma, while the concentrations of low abundance proteins range from milli- to zeptomolar levels [1, 2, 4]. HSA and IgG hinder the detection, isolation and identification of other biopolymers present in trace amounts. The low abundance proteins are frequently potential biomarkers or biomarker candidates for various diseases [1]. After isolation and purification, some of biologically active proteins that are present in human plasma in very low concentrations such as clotting factors and inhibitors can be used for therapeutic purposes [5–7]. Both optimization industrial scale plasma fractionation and serum and plasma separation in order to isolate low abundance proteins in these complex biological fluids have alredy been topic of many studies [8, 9]. However, there is still need for further optimization, especially regarding the speed, high throughput and in case of plasma fractionation, optimization of the yield, purity and characterization of isolated therapeutic proteins [10. 11].
Already in very early stage of development monoliths made of polyglycidyl methacrylate polymers have been successfully used for separation of proteins from human plasma [12, 13]. Their good mechanical strength, high porosity and dynamic capacity for large molecules, high separation speed and high flow rates at a very low pressure drop enable rapid processing of large volumes of complex biological mixtures [14]. Additionally their pH resistance makes possible cleaning and sanitation under harsh conditions such as high and low pH, and repeating use of monolithic support also for isolation and high-throughput analysis of proteins for therapeutic use [13, 15].
Sample displacement chromatography (SDC) for preparative purification of peptides in reversed-phase mode was introduced by Hodges et al. [16, 17]. When this chromatographic separation mode is applied, during loading, there is competition among the sample components for the binding sites of the hydrophobic surface of the stationary phase. The more molecules compete for these sites, the more components with lower affinity to the surface will be displaced and eluted from the column. Veeraragavan et al. [18] applied the SDC method for purification of proteins in ion-exchange mode. The Hodges’ group further developed SDC for purification of synthetic peptides, and new system design for rapid, simple and cost-effective procedure for the purification of peptide mixtures was introduced [19]. The same group also modified the SDC procedure for preparative isolation of proteins from troponin, a rabbit skeletal multi protein complex [20]. Manseth et al. [21] applied SDC to purify active thrombin from plasma of Atlantic salmon on a Heparin Sepharose affinity matrix.
In this paper we demonstrate that if monolithic supports were used for separation of complex biological mixtures in SDC mode, the composition of bound and eluted proteins is dependent on column loading. Under overloading conditions, the weakly bound proteins are displaced by strongly binding ones, and this phenomenon was not dependent on column size. It could be demonstrated that small monolithic columns are ideal supports for development of new methods, especially for separation of complex biological fluids, and for sample preparation for further proteomic and glycomic analyses.
2. Materials and methods
2.1. Human plasma
The starting material was cryopoor, single donor human plasma (Rhode Island Blood Center, Providence, RI, USA). All plasma samples were screened to exclude the presence of blood-borne viruses (hepatitis A, B and C and HIV). Prior to use, the cryoglobulins were removed by precipitation at 4°C as described previously [22].
2.2. Ion-exchange chromatography
For anion-exchange chromatography monolithic, disk-shaped columns with a column volume of 100 and 340 μL respectively as well as 8 mL DEAE and QA CIM monoliths were used (BIA Separations, Ljubljana, Slovenia). For parallel experiments with columns packed with bulk supports, Toyopearl DEAE and Toyopearl Q gels pre-packed in 1 mL columns were used (Toyoscreen, Tosoh Bioscience GmbH, Stuttgart, Germany). After washing with HPLC water, the column was equilibrated with a low strength buffer (10 mM Tris-HCl, pH 7.4, Buffer A). The human plasma was four times diluted with Buffer A and applied to the column. Unbound proteins were collected and subsequently analyzed. After sample application, the column was washed with five column volumes of Buffer A, and the bound proteins were eluted with a step gradient of Buffer B (1 M NaCl in Buffer A).
For cation-exchange chromatography monolithic disk and cylinder-shaped columns (100 and 340 μL and 8 mL respectively, see above) with strong anion exchanger ligands (SO3, BIA Separations) were used. For parallel experiments with columns with bulk supports, Toyoscreen SO3 1 mL columns (Tosoh Bioscience) were used. The Buffer A was 10 mM Na citrate, pH 4.5, Buffer B was Buffer A containing 1 M NaCl. Further separation was performed as described above. After each chromatographic run, columns were sanitized with 0.5 M NaOH, and subsequently washed with HPLC water, 0.5 M Tris.HCl, pH 7.4 and re-equilibrated with the Buffer A. The flow rates for all chromatographic separation was between 0.5 and 10 mL/min, depending on column size. All runs were performed at 4°C, and a BioLogic Duo Flow chromatographic system (BioRad, Hercules, CA, USA) was used. Proteins were detected by UV absorption at 280, 260, and 210 nm. Each experiment was performed in triplicate.
For determination of column capacity and recovery, bovine serum albumin (BSA) for anion-exchange columns, and bovine IgG for cation-exchange columns were used (both from Sigma, St. Louis, MO, USA). The unbound material and eluted fractions were collected, and protein amounts were determined in all fractions and in the starting material.
2.3. Protein determination
Protein amounts in collected fractions and starting material were determined with the Bicinchoninic Acid Protein Assay kit (Pierce, Rockford, IL, USA) according to the manufacturer’s procedure.
2.4. Electrophoretic separations
About 15–20 μg protein of each sample were solubilized in NuPAGE sample buffer (Invitrogen, Carlsbad, CA, USA) and heated at 100 °C for 5 minutes. The electrophoretic separation was performed with precast 4–12% Bis-Tris gels in a Xcell Sure Lock Mini Cell (Invitrogen) according to the manufacturer’s procedure. The gels were stained with GelCodeBlue (Pierce) and visualized by a VersaDoc Imaging System (BioRad, Hercules, CA, USA). For further analysis, the band of interest were excised for “in-gel” digestion.
Samples for 2-D electrophoresis containing 70 μg protein were prepared using Ready Prep 2-D Cleanup kit (Bio Rad) according to manufacturer’s instruction. After this preparation, samples were re-dissolved in IPG strip rehydratation buffer containing 8 M urea, 2% CHAPS, 0.5% carrier ampholytes, 0.002% bromophenol blue and 20 mM DDT. The dry IPG strips were rehydrated overnight with 160 μL of the protein sample. 2-D electrophoretic separation was performed as described previously [23]. Gel scanning was performed by use of a VersaDoc Imaging System (BioRad).
2. 5. Sample preparation for MS analysis
For “in-gel” digestion, the gel bands were excised by extracting six to ten gel particles with clean glass Pasteur pipettes and digested with trypsin as described preciously [23, 24].
For “in-solution” digestion, 50 μg of the acetone-precipitated and denatured protein pellet was resolubilized in 100 μl of NH4HCO3 (pH 8.0)/8 M urea. The resolubilized proteins were reduced with 20 mM dithiothreitol (37°C, 45 min) and then alkylated with 50 mM iodoacetamide at room temperature for 30 min in the dark. Before tryptic digestion, 100 mM ammonium bicarbonate buffer was added to reduce the concentration of urea. Trypsin was added to the protein mixture at an enzyme to substrate ratio of 1: 60 (w/w). After incubating at 37 °C overnight, the tryptic peptides were dried in a vacuum centrifuge (Vacufuge, Eppendorf, Hamburg, Germany). The material was then redissolved in a solution of 0.5% (v/v) formic acid and 20% (v/v) acetonitrile, with vacuum drying again. Subsequently, the peptides were isolated using a strong cation exchange TipTopTM (PolyLC, Inc., Columbia, MD, USA) according to the manufacturer’s instructions after resuspending in the same solvent and confirming the pH value. The resulting tryptic peptides were dried once more and were subject to the LC-MS/MS analysis after being redissolved in formic acid:water:acetonitrile:trifluoroacetic acid mixture (0.1:95:5:0.01).
2. 6. Identification of proteins with LC-MS/MS
Tryptic digests of whole fractions obtained by ion-exchange chromatography (“in-solution” digestion), or of proteins extracted from the gels after SDS-PAGE (“in-gel” digestion), were separated with a reversed-phase column (C-18 PepMap 100, LC Packings/Dionex, Synnyvale, CA, USA) as previously described [24]. Briefly: The column eluate was introduced directly onto a QSTAR XL mass spectrometer (Applied Biosystems and Sciex, Concord, Ontario, Canada) via electrospray ionization (ESI). Half-second MS scans (300–1500 Thompson, Thompson(Th) = Da/z) were used to identify candidates for fragmentation during MS/MS scans. Up to five 1.5 s MS/MS scans (65–1500 Th) were collected after each scan. An ion had to be assigned a charge in the range of +2 to +4. The dynamic exclusion was 40. Protein identifications were completed with ProteinPilot (Applied Biosystems and Sciex), setting with 1.5 Da mass tolerance for both MS and MS/MS and using the human and “RefSeq” databases from NCBI (http://www.ncbi.nlm.nih.gov/RefSeq/). ProteinPilot is the successor to ProID and ProGroup, and uses the same peptide and protein scoring method. Scores above 2.0 require that at least two sequence-independent peptides will be identified [24].
In parallel experiments, additional LC-MS/MS system was used (Agilent Technologies, Paolo Alto, CA, USA, and Thermo Electron Corporation, San Jose, CA, USA). When this system was used, tryptic peptides were separated on a 12 cm (75 μm I.D.) analytical column with 5 μm Monitor C18 resin (Column Engineering, Inc., Ontario, CA, USA) and containing an integrated ~4 μm ESI emitter tip. Solvent A was 0.1 M acetic acid in water, solvent B was 0.1 M acetic acid in acetonitrile. Peptides were eluted using a linear acetonitrile gradient (0–70% solvent B over 30 min). Peak parking during the time when peptides were expected to elute was accomplished by reducing the flow rate from 200 nL/min to ~20 nL/min.
Eluting peptides were introduced onto an LTQ linear ion trap mass spectrometer (Thermo Electron Corporation, San Jose, CA) with a 1.9 kV electrospray voltage. Full MS scans in the m/z range of 400–1800 were followed by data-dependent acquisition of MS/MS spectra for the five most abundant ions, using a 30-second dynamic exclusion time. Protein identification was performed in, at least, two independent experiments.
Peptide and protein identifications were performed with software contained BioWorks version 3.2 (Thermo Electron). Peak list files were created by the program extract_msn.exe, using the following settings: The mass had to fall in the range of 600 to 4500 Daltons. The minimum total ion current for the scan had to be over 1000. The precursor tolerance for grouping was 1.5 Daltons, with no differing intermediate scans allowed and only a single scan required to create a peak file. The minimum signal-to-noise for a peak to be written to the peak file was 3, and 25 such peaks had to be found for a peak file to be created. The program calculated charge states. However, in case of ambiguity, peak files for both the +2 and +3 charge states were created.
Database searching using the peak lists was performed by the program SEQUEST [25]. The precursor-ion tolerance was 2.0 Daltons and the fragment-ion tolerance was 0.8 Daltons. Enzymatic digestion was specified as trypsin, with up to 2 missed cleavages allowed.
3. Results
3.1. Anion-exchange chromatography
All chromatographic supports (DEAE and QA monoliths and Toyopearl DEAE bulk material) used for anion-exchange chromatography had a similar capacity of 23–30 mg/mL support for BSA. The capacity of both SO3 monoliths and bulk support (Toyopearl S) was about 20–25 mg IgG/ml.
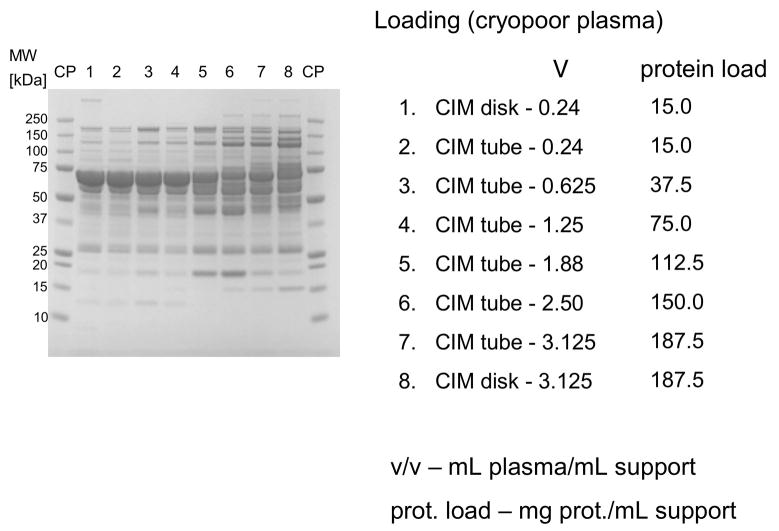
In further experiments, four times diluted human plasma was loaded to monolithic disk- (340 μL) and cylinder-shaped 8 mL columns in amounts of 0.2–5.0 mL plasma/mL support. The bound proteins were eluted in two steps with 0.155 and 1.0 M NaCl (see Figure 1). As shown in Figure 1, under overloading conditions, protein loading was 112.5 mg/mL support and higher (about 4–5 times higher than the column capacity for BSA, see above), HSA and other lower abundant weakly binding proteins were displaced by other, more tightly bound proteins (see lanes 5–8 in Figure 1). As mentioned above, 340 μL disk-shaped and 8 mL cylinder-shaped columns were used, and this phenomenon was independent on column shape and size respectively (see Figure 1).
Figure 1.
Separation of proteins from human plasma on DEAE CIM monolithic supports under overloading conditions. CIM DEAE monolithic disk (column volume 340 μL) and tube (column volume 8 mL) were used. Human plasma was four times diluted with Buffer A, and bound proteins were eluted with 0.155 (A) by use of a step gradient. The flow rate was 2 mL/min for the CIM disk, and 10 mL/min for CIM tube. Collected fractions were analyzed by SDS-PAGE. For other separation conditions see Material and methods.
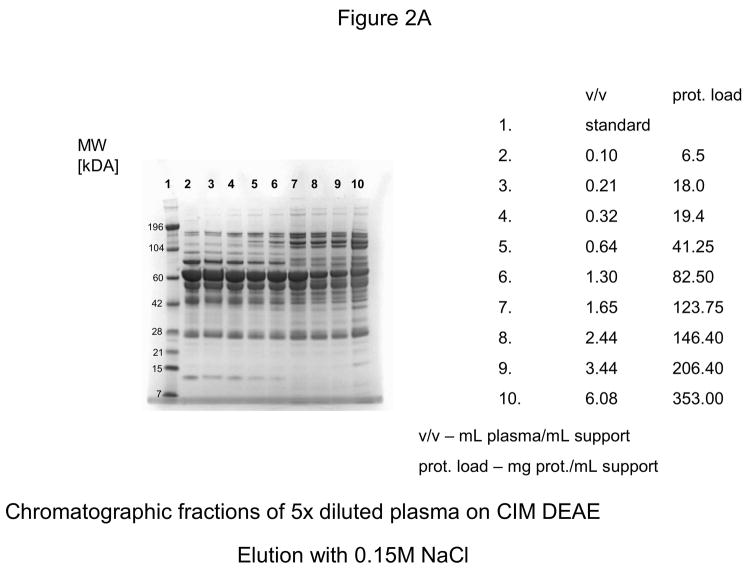
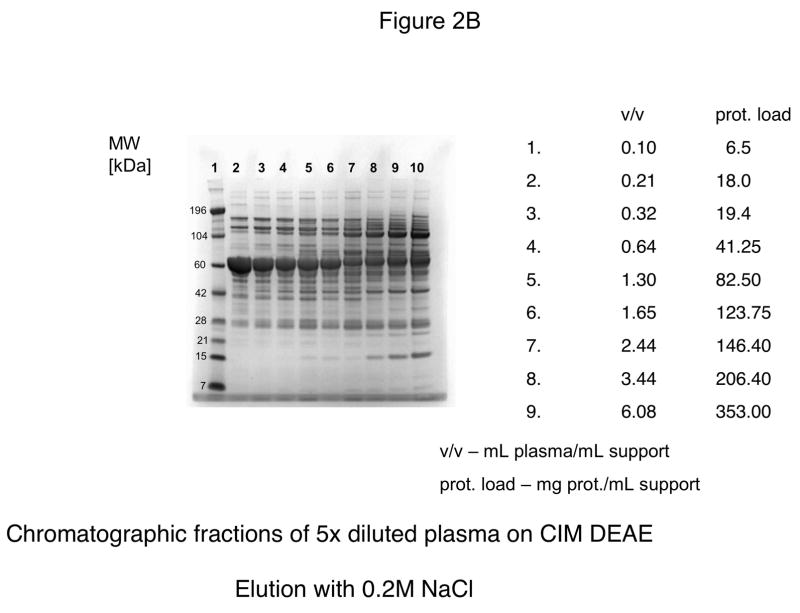
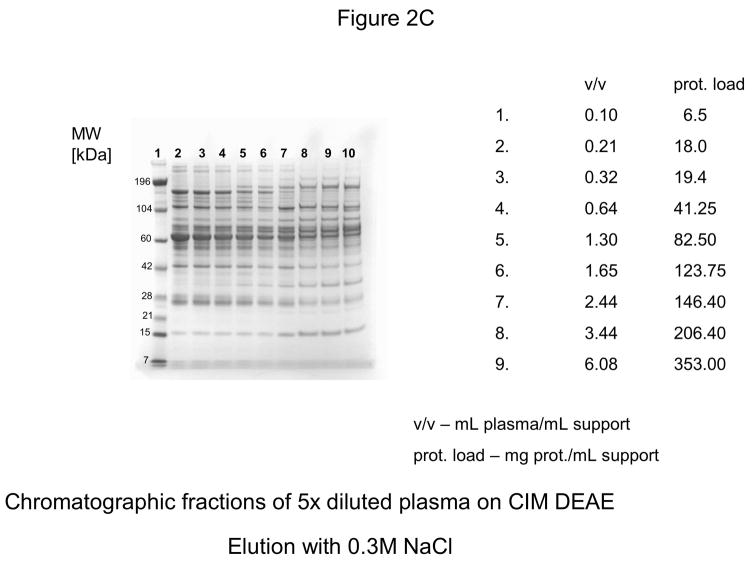
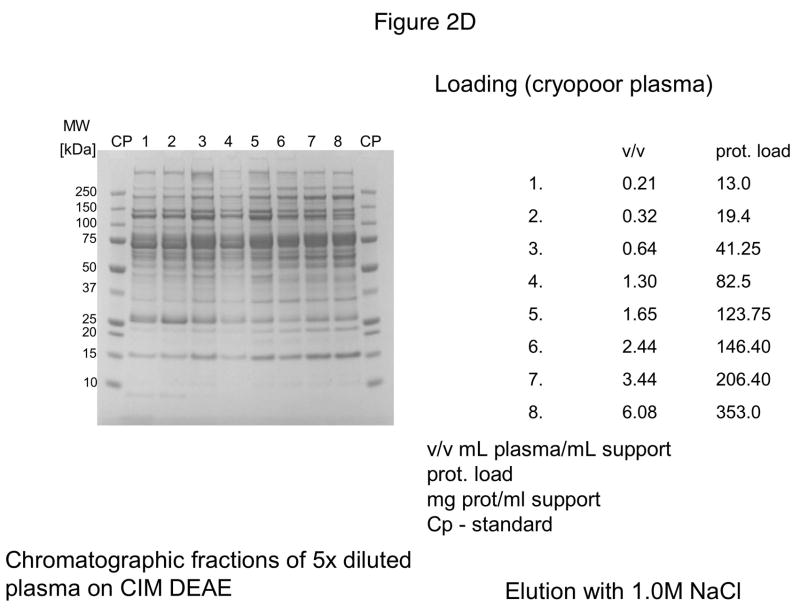
For further experiments, CIM DEAE disk with a bed volume of 340 μL was used, and four-steps-elution, a gradient with 0.155, 0.2, 0.3 and 1 M NaCl was applied. The results of these experiments are shown in Figures 2A–D. As shown in Figure 2D, the elution pattern with 1.0 M NaCl did not change under overloading conditions.
Figure 2.
Separation of proteins from human plasma on DEAE weak anion-exchanger CIM monolithic supports under overloading conditions. CIM DEAE monolithic disk (column volume 340 μL) was used. Human plasma was four times diluted with Buffer A, and bound proteins were eluted with 0.155 (A), 0.2 (B), 0.3 (C) and 1 M NaCl (D)* by use of a step gradient. The flow rate was 2 mL/min. Collected fractions were analyzed by SDS-PAGE. For other separation conditions see Material and methods.
*After injection of 37 μL plasma (6.5 mg protein/mL support), the amount of material eluted with 1M NaCl was not sufficient for subsequent SDS-PAGE
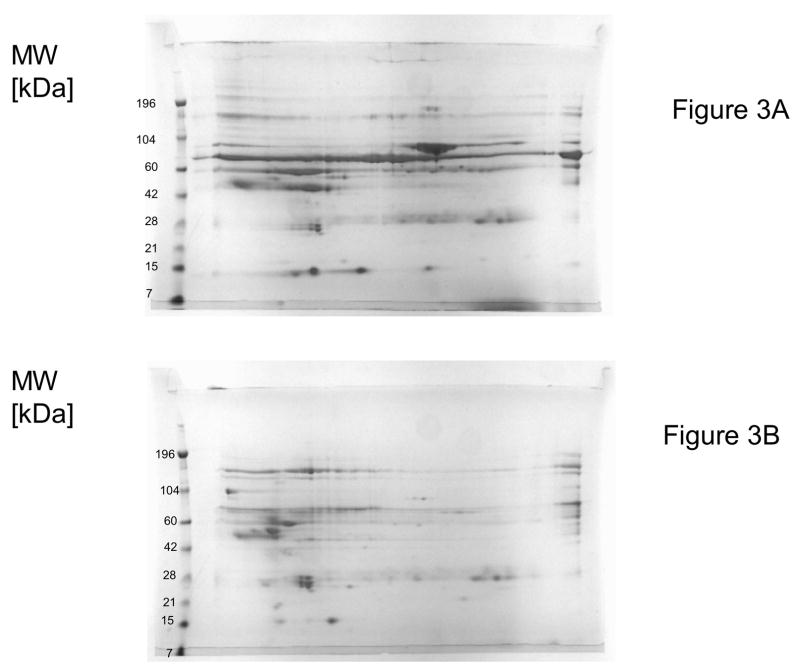
In Figure 3A and 3B, 2D electrophoretic separations of fractions eluted with 0.155 M NaCl after loading of 0.25 mL plasma/mL support (Figure 3A) and 12.5 mL plasma /mL (Figure 3B) support respectively were shown. Again, under overloading conditions (Figure 3B) HSA was almost completely displaced. Consequently, the absence of this highly abundant protein enabled a much better loading and 2D electrophoretic separation of less abundant components.
Figure 3.
Two dimensional electrophoretic separation of fractions eluted with 0.155 M NaCl from CIM DEAE disk-shaped monolithic column (column volume 340 μL) and displacement of HSA under overloading conditions.
A–0.25 mL plasma containing about 15 mg protein was loaded on the column (for comparison, see lane No. 1 in Figure 1A);
B–1.06 mL plasma containing 187.5 mg protein was loaded on the column (for comparison, see lane No. 8 in Figure 1A).
For separation conditions, see Figure 1 and Material and methods.
Parallel experiments with columns packed with Toyopearl DEAE, a bulk support that had similar capacity for BSA (about 25–30 mg protein/mL gel) did not show these phenomena, and no displacement of HSA and other weakly bound components was observed under identical loading conditions (0.2–5.0 mL plasma/mL support, not shown here).
When strong anion-exchange columns CIM QA disk-shaped columns were used, 0.2 M NaCl in Buffer A was applied for first elution step. Again, under overloading conditions, HSA was displaced by other, more tightly binding proteins (see Figure 4) and the pattern of proteins eluted with higher salt concentration (1.0 M NaCl) did not change significantly (not shown here).
Figure 4.
Separation of proteins from human plasma QA strong anion-exchanger CIM monolithic supports under overloading conditions. CIM QA monolithic disk (column volume 340 μL) was used. Human plasma was four times diluted with Buffer A, and bound proteins were eluted with 0.2, 0.3, 0.5 and 1 M NaCl by use of a step gradient. The flow rate was 2 mL/min. Collected fractions were analyzed by SDS-PAGE. Only the profiles of fractions eluted with 0.2 M NaCl are shown. For other separation conditions see Material and methods.
Identical results were obtained, when mini-disks with a column volume of 100 μL were used (not shown here). Again, in parallel experiments with columns packed with Toyopearl Q, these phenomena under identical loading conditions were not observed (not shown here).
3.2. Cation-exchange chromatography
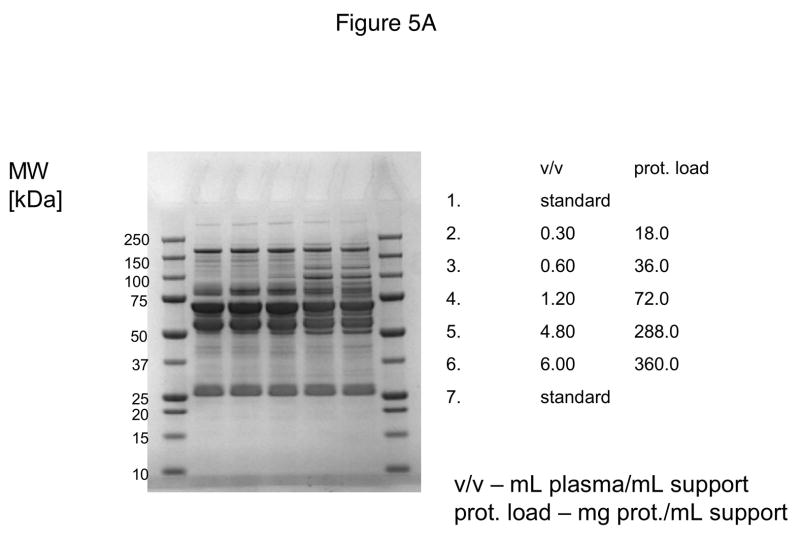
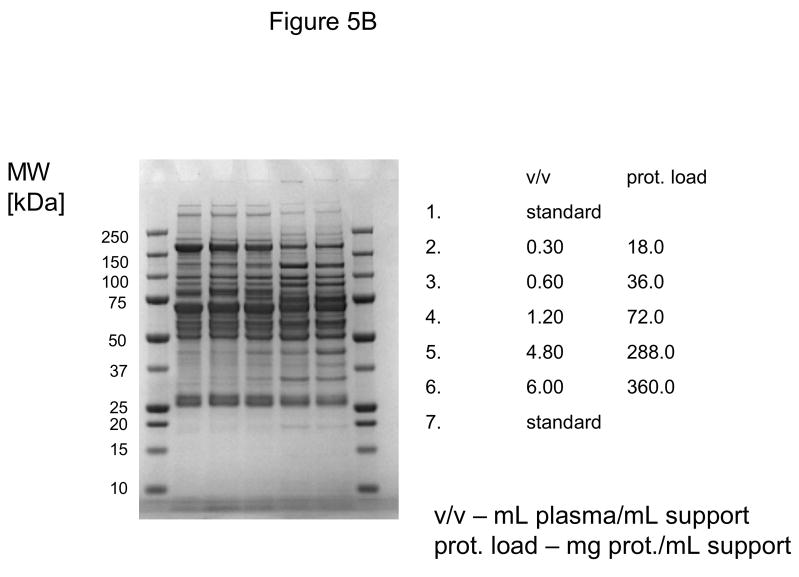
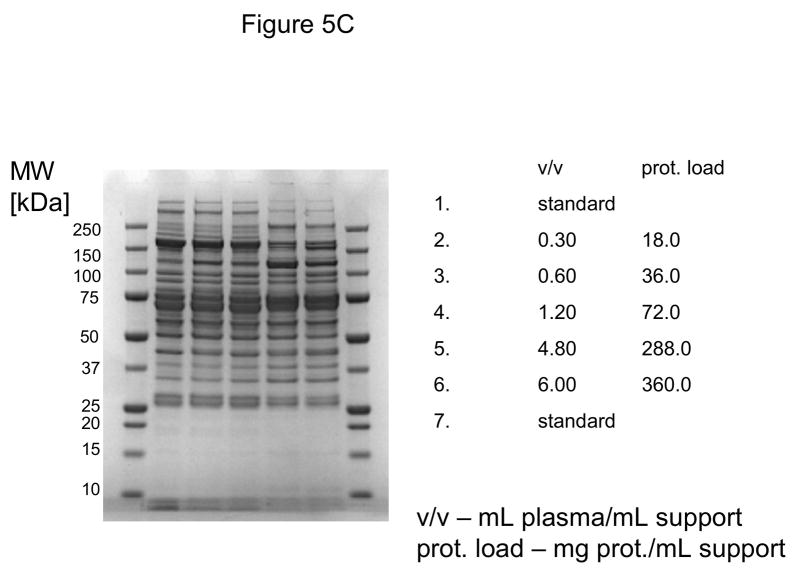
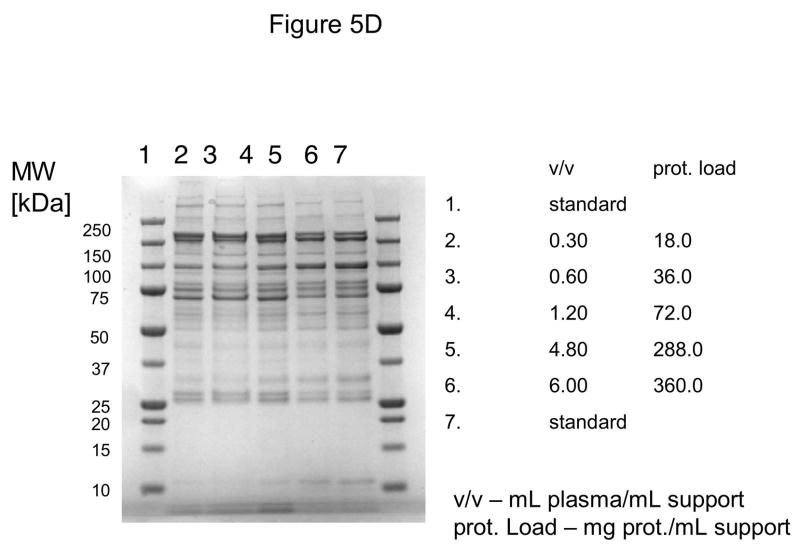
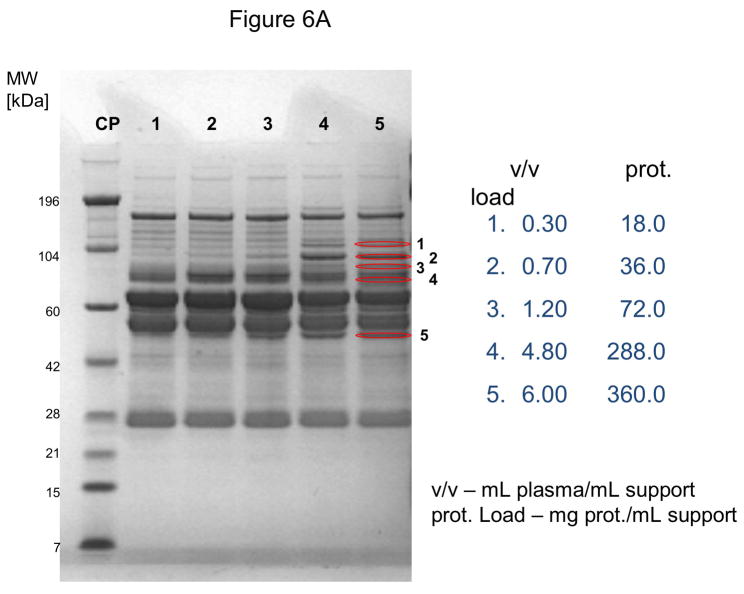
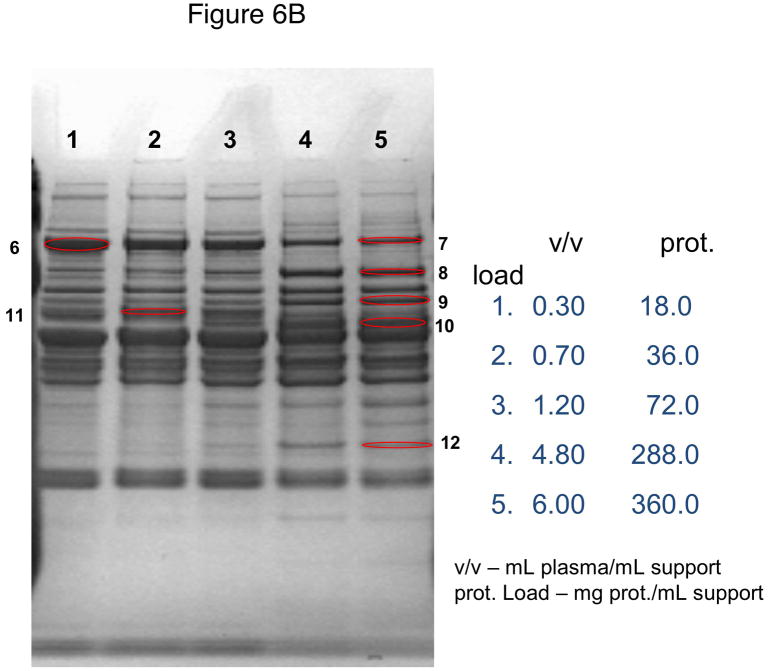
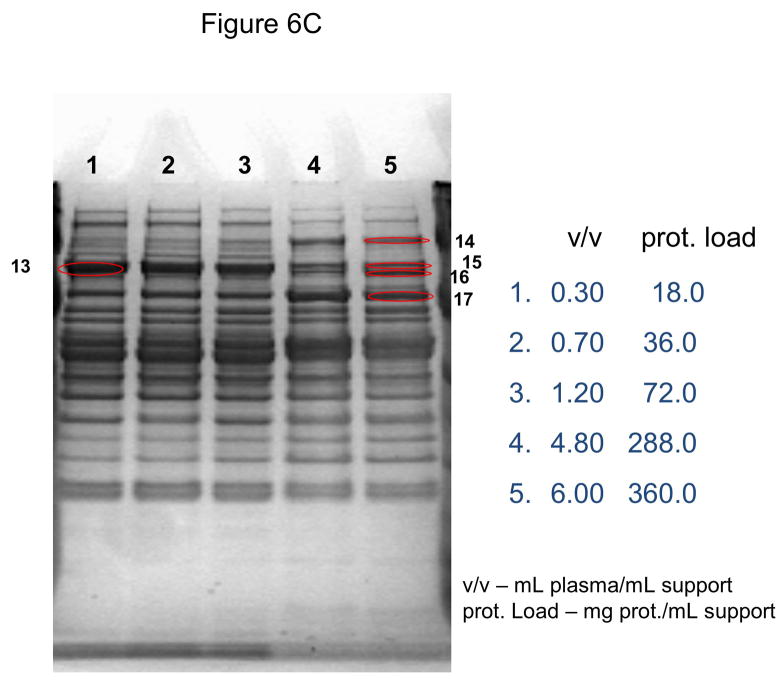
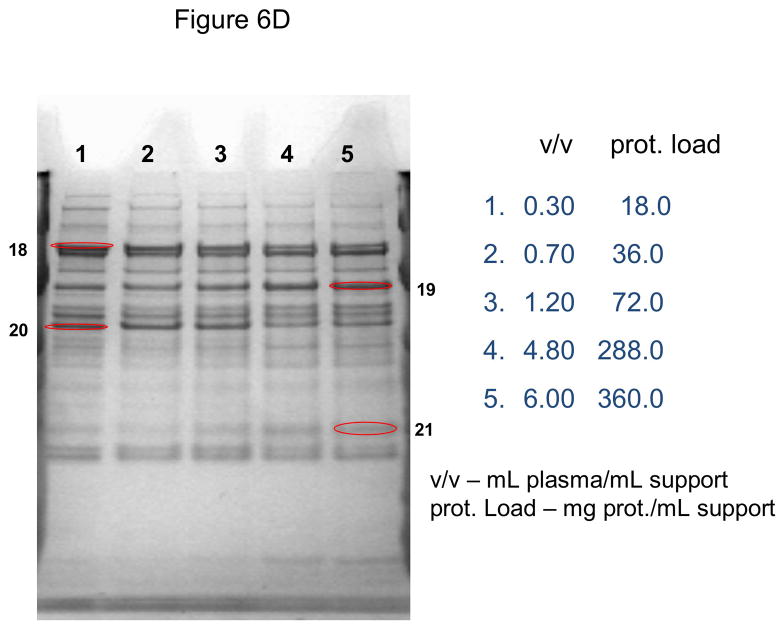
In these investigations, CIM SO3 monolithic, disk-shaped columns with a column volume of 100 and 340 μL respectively were used. In parallel experiments, 1 mL columns packed with bulk material (strong cation-exchanger with SO3 ligands) were applied. Because of lower concentration of IgG, the other high-abundant protein that binds to cation-exchangers, slightly different scheme for protein elution was applied (0.2, 0.3, 0.5 and 1.0 NaCl were used). As shown in Figures 5A–D, similar phenomenon as in anion-exchange chromatography can be observed. Again, weakly bound proteins such as IgG (two bands with the apparent molecular weight in SDS-PAGE of about 55 and 27 kDa respectively, see Figure 5A), apolipoprotein B (~300 kDa band, see Figure 4B) and alpha-2-macroglobulin (~175 kDa band, see Figure 5B and 5C) are displaced by other, tighter binding proteins. Again, the SDS-PAGE pattern of very tightly bound proteins that can be eluted only by high salt concentration (1.0 M NaCl) did not change with the amount of loaded sample (see Figure 5D). Identical results were also obtained, when mini SO3 disks with a column volume of 100 μL were used (see Figures 6A–D). Both above mentioned proteins apolipoprotein B (gi 105990532) and alpha-2-macroglobulin (gi 113215010) were identified by LC-MS/MS after excising the bands from SDS-PAGE gels and tryptic digestion of separated proteins.
Figure 5.
Separation of proteins from human plasma SO3 strong cation-exchanger CIM monolithic supports under overloading conditions. CIM SO3 monolithic disk (column volume 340 μL) was used. Human plasma was four times diluted with Buffer A, and bound proteins were eluted with 0.2 (A), 0.3(B), 0.5(C) and 1 M (D) NaCl by use of a step gradient. The flow rate was 2 mL/min. Collected fractions were analyzed by SDS-PAGE. For other separation conditions see Material and methods.
Figure 6.
Use of 100 μL monolithic SO3 strong cation-exchanger CIM disk for separation of proteins from human plasma under overloading conditions. Human plasma was four times diluted with Buffer A, and bound proteins were eluted with 0.2 (A), 0.3(B), 0.5(C) and 1 M (D) NaCl by use of a step gradient. The flow rate was 1 mL/min. Collected fractions were analyzed by SDS-PAGE. For other separation conditions see Material and methods. Band labeled in the Figures were excised, digested by trypsin and used for identification by LC-MS/MS. Some identified proteins are listed in Table 1.
As is previous experiments a bulk support that had similar capacity for IgG (Toyopearl SO3) did not show these phenomena under similar loading conditions (up to 5 mL human plasma/mL support), and no displacement of IgG and other weakly bound components was observed (not shown here).
3.3. Protein identification
Additionally to the proteins that are displaced from the columns under overloading conditions (alpha-2-macroglobulin and apolipoprotein B), some proteins that are present in human plasma in trace amounts and that are enriched after separation under overloading conditions by use of amino- or cation-exchange monoliths, such as clotting factors and inhibitors and other physiologically active proteins were identified in eluted fractions after “in solution” digestion or after electrophoretic separation and “in gel” digestion (see Table 1, and Tables S1 and S2 in Supporting materials). Most of these proteins bound tightly on either the anion- or cation-exchange column and can be eluted only by use of high salt concentrations.
Table 1.
Some low abundance proteins identified by LC-MS/MS after separation on CIM ion-exchange monoliths under overloading conditions.
| Protein name | Concentration | Used monolith | Reference |
|---|---|---|---|
| Plasminogen | 10 ng/mL | CIM SO3 | [30] |
| Factor XII | 14 μg/mL | CIM SO3 | [31] |
| Gelsolin | 250 μg/mL | CIM SO3 | [10] |
| Factor II | 80 μg/mL | CIM DEAE | [22] |
| Factor IX | 4 μg/mL | CIM DEAE | [22] |
| Factor X | 6.4 μg/mL | CIM DEAE | [22] |
| Protein C | 4–6 μg/mL | CIM DEAE | [22] |
| Protein S | ~30 μg/mL | CIM DEAE | [22] |
| Protein z | 150 ng/mL | CIM DEAE | [22] |
4. Discussion
The use of small, short monolithic columns for removal of high abundant plasma proteins was already discussed. Cerk Petric et al. [26] use strong anion-exchanger QA monolithic QA disk, and after process optimization, HSA could be removed. Immunoglobulins were subsequently depleted by use of a monolithic CIM disk with immobilized protein G. Recently, Urbas et al. [27 ] used a CIM protein G disk in combination with a small Mimetic Blue pseudo-affinity column for effective and rapid depletion of HSA and IgG from human plasma. However, the topic of this work was not the use of ion-exchange columns under overloading conditions.
Anion-exchange resins, mostly DEAE and Q Sephadex are used for so-called solid-phase extraction of vitamin K dependent clotting factors and inhibitors from cryopoor human plasma [5, 6, 22]. In the industiral process, about 2–5 g resin/L cryopoor plasma was added. The resin is recovered by filtration, washed with a buffer containing 0.2–0.3 M NaCl to remove loosely bound proteins, and then eluted with up to 2 M NaCl in order to recover the fraction containing tightly bound fraction containing the vitamin K-dependent factors. HSA is barely present in the eluate, if solid-phase extraction was performed under such heavily overloading conditions (up to 500 mL cryopoor plasma/g support, [6, 22]). It was an early example for the use of SDC. Although relatively simple conditions were applied, the process is very reproducible, and for both final products prothrombin complex concentrate and clotting factor IX constant specifications are achieved [22 ]. In early applications of SDC in both reversed phase [16, 17] and ion-exchange mode [18], relatively simple mixtures of peptides and proteins were used for separations. Further development of SDC made possible separation of complex mixtures of synthetic peptides [19]. Preparative isolation of proteins from troponin complex by use of SDS in reversed-phase mode was a challenging task because of limited solubility of separated components and their tendency to aggregate. However, the number of proteins (altogether four different polypeptide chains) was still limited [20]. On the other hand, BaSO4 precipitate of plasma of Atlantic salmon was a relative complex protein mixture that was used as a starting material for isolation of highly pure and biologically active thrombin by use of SDS as a rapid, simple and cost-effective method [21]. Brown et al. [28] recently used ion-exchange membrane chromatography under overloading conditions for purification of monoclonal antibodies. The overloading of membranes results in binding of impurities, and breakthrough of purified antibody. In general, this approach was similar to the SDS on ion-exchangers, presented by Veerargavan et al. [18]. The significant advantage of membranes is that the binding of impurities and breakthrough of antibodies was practically flow rate independent [14, 28].
As presented here, if monolithic, polymethacrylate-based CIM supports were used in SDC mode, displacement of HSA, IgG and other weakly binding proteins can be achieved, 1.5 mL diluted human plasma/mL support, are used (see Figure 1A). These experiments presented her cannot be directly compared with the above discussed solid phase extraction of human plasma, because in industrial scale preparations, and in our earlier scale-down experiments undiluted plasma with a physiological ionic strength was used [29]. However, in our present experiments, up to 3.5 mL cryopoor plasma (about 180–200 mg protein)/mL bulk ion-exchange support was loaded on the column. This amount is up to 2–7 times higher than the column capacity (between 22–30 mg protein/mL), but under these overloading conditions no displacement of weaker binding proteins such as HSA was observed. Veeraragavan et al. [18] have clearly shown sample displacement in ion-exchange mode. The reason for the absence of sample displacement under above listed conditions, when human plasma was used, is very high concentration of both proteins HAS and IgG, and significantly lower concentration (more that 1 order of magnitude) of strongly binding proteins. Our further preliminary experiments show that this displacement also on “classical”, porous supports can be demonstrated, however, under extreme sample overloading (not shown here).
As shown in Figs. 6A–D, miniaturized, 100 μL monolithic columns could be used for these experiments. In order to identify plasminogen or clotting factor XII, low abundance proteins with a plasma concentration of about 10 ng/mL [30] and 15 μg/mL [31] respectively, a load of about 188 mg plasma protein/mL CIM SO3 monolith was used (see Table 1). Consequently, an amount of about 300 μL human plasma for separation and identification of these proteins is necessary, if a 100 μL monolithic column is used for sample preparation and proteomic analysis of this complex body fluid.
The high-throughput screening with miniaturized columns packed different types of chromatographic supports is an already routinely used method for optimization of industrial separations [32]. It is well known [33], and it could also be documented here, that the separation efficiency of monolithic supports is practically independent of column volume (see Figure 1). Pucic et al. [34] recently demonstrated, that small, 200 μL monolithic columns mounted in 96 well ELISA can be used for high-throughput analysis of plasma proteins, and further proteomic and glycomic investigations. These investigations also show that small monolithic columns with ion-exchange ligands can be optimally used for development of new methods for separation of complex mixtures and sample preparation for further high-throughput analyses and sample preparation in proteomic technology.
4. Conclusions
If glycidyl methacrylate monolithic supports with ion-exchange ligands were used for separation of human plasma, the composition of bound and eluted proteins is dependent on column load.
Under overloading conditions, in sample displacement chromatography mode, the weakly bound proteins seem to be displaced by stronger binding proteins.
This phenomenon was not dependent on column size.
Consequently, small monolithic columns are ideal supports for enrichment of low abundance proteins from complex biological mixtures and sample preparation for proteomic analyses.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH), Centers for Biochemical Research Excellence (COBRE), grant No. P20RR017695 and NIH grant No. 1S10RR025623-01 (Dj. Josic).
We thank BIA Separations for providing the CIM monoliths.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nice EC, Rothacker J, Weinstock J, Lim L, Catimel B. J Chromatogr A. 2007;1168:190. doi: 10.1016/j.chroma.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 2.Anderson NL, Anderson NG. Mol Cell Proteomics. 2002;1:845. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Issaq HJ. Electrophoresis. 2001;22:3629. doi: 10.1002/1522-2683(200109)22:17<3629::AID-ELPS3629>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 4.Ivanov YD, Govorun VM, Bykov VA, Archakov AI. Proteomics. 2006;6:1399. doi: 10.1002/pmic.200402087. [DOI] [PubMed] [Google Scholar]
- 5.Burnouf T. J Chromatogr B. 1995;664:3. doi: 10.1016/0378-4347(94)00532-a. [DOI] [PubMed] [Google Scholar]
- 6.Hoffer L, Schwinn H, Josic Dj. J Chromatogr A. 1999;844:119. doi: 10.1016/s0021-9673(99)00348-9. [DOI] [PubMed] [Google Scholar]
- 7.Basilico F, Nardini I, Mori F, Brambilla E, Benazzi L, De Palma A, Rosti E, Farina C, Mauri PL. J Pharm Biomed Anal. 2010;53:50. doi: 10.1016/j.jpba.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 8.Faca V, Pitteri SJ, Newcomb L, Glukhova V, Phanstiel D, Krasnoselsky A, Zhang Q, Struthers J, Wang H, Eng J, Fitzgibbon M, McIntosh M, Hanash S. J Proteome Res. 2007;6:3558–3565. doi: 10.1021/pr070233q. [DOI] [PubMed] [Google Scholar]
- 9.de Roos B, Duthie SJ, Polley ACJ, Mulholland F, Bouwman FG, Heim C, Rucklidge GJ, Johnson IT, Mariman EC, Daniel H, Elliott RM. J Proteome Res. 2008;7:2280. doi: 10.1021/pr700714x. [DOI] [PubMed] [Google Scholar]
- 10.Kovac S, Yang X, Huang F, Hixson D, Josic Dj. J Chromatogr A. 2008;1194:38. doi: 10.1016/j.chroma.2008.04.026. [DOI] [PubMed] [Google Scholar]
- 11.Yang X, Clifton J, Huang F, Kovac S, Hixson DC, Josic Dj. Electrophoresis. 2009;30:1185. doi: 10.1002/elps.200800501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Josic Dj, Reusch J, Löster K, Baum O, Reutter W. J Chromatogr A. 1992;590:59. doi: 10.1016/0021-9673(92)87006-t. [DOI] [PubMed] [Google Scholar]
- 13.Strancar A, Barut M, Podgornik A, Koselj P, Schwinn H, Raspor P, Josic Dj. J Chromatogr A. 1997;760:475. doi: 10.1016/s0021-9673(96)00675-9. [DOI] [PubMed] [Google Scholar]
- 14.Tennikova TB, Reusch J. J Chromarogr A. 2005;1065:13. doi: 10.1016/j.chroma.2004.10.074. [DOI] [PubMed] [Google Scholar]
- 15.Tetala KK, van Beek TA. J Sep Sci. 2010;33:422. doi: 10.1002/jssc.200900635. [DOI] [PubMed] [Google Scholar]
- 16.Hodges RS, Burke TWL, Mant CT. J Chromatogr. 1988;444:349. doi: 10.1016/s0021-9673(01)94036-1. [DOI] [PubMed] [Google Scholar]
- 17.Hodges RS, Burke TWL, Mant CT. J Chromatogr. 1991;548:267. doi: 10.1016/s0021-9673(01)88608-8. [DOI] [PubMed] [Google Scholar]
- 18.Veeraragavan K, Bernier A, Braendli E. J Chromatogr. 1991;541:207. [Google Scholar]
- 19.Husband DL, Mant CT, Hodges RS. J Chromatogr A. 2000;893:81. doi: 10.1016/s0021-9673(00)00751-2. [DOI] [PubMed] [Google Scholar]
- 20.Mant CT, Hodges RS. J Chromatogr A. 2002;972:101. doi: 10.1016/s0021-9673(02)01079-8. [DOI] [PubMed] [Google Scholar]
- 21.Manseth E, Skjervold PO, Flengsrud R. J Biochem Biophys Methods. 2004;60:39. doi: 10.1016/j.jbbm.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 22.Josic Dj, Hoffer L, Buchacher A. J Chromatogr B. 2003;790:183. doi: 10.1016/s1570-0232(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 23.Josic Dj, Brown MK, Huang F, Lim Y-P, Rucevic M, Clifton JG, Hixson D. Proteomics. 2006;6:2874. doi: 10.1002/pmic.200500563. [DOI] [PubMed] [Google Scholar]
- 24.Lawson E, Clifton JG, Huang F, Li X, Hixson D, Josic Dj. Electrophoresis. 2006;27:2747. doi: 10.1002/elps.200600059. [DOI] [PubMed] [Google Scholar]
- 25.Eng J, McCormack AL, Yates AL., III J Am Soc Mass Spectrom. 1994;5:976. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 26.Cerk Petric T, Brne P, Gabor B, Govednik L, Barut M, Strancar A, Zupancic Kralj L. J Pharm Biomed Anal. 2007;43:243. doi: 10.1016/j.jpba.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 27.Urbas L, Brne P, Gabor B, Barut M, Strlic M, Cerk Petric T, Strancar A. J Chromatogr A. 2009;1216:2689. doi: 10.1016/j.chroma.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 28.Brown A, Bill J, Tully T, Radhamohan A, Dowd C. Biotechnol Appl Biochem. 2010;56:59. doi: 10.1042/BA20090369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josic Dj, Hoffer L, Buchacher A, Schwinn H, Frenzel W, Biesert L, Klöcking H-P, Hellstern P, Rokicke-Milewska R, Klukowska A. Thromb Res. 2000;100:433. doi: 10.1016/s0049-3848(00)00339-x. [DOI] [PubMed] [Google Scholar]
- 30.Riker PM, Vaughan DE, Stampfer MJ, Glynn RJ, Hannekens CH. JAMA. 1994;272:929. doi: 10.1001/jama.1994.03520120039028. [DOI] [PubMed] [Google Scholar]
- 31.Ratnoff OD, Everson B, Donaldson VH, Mitchell BH. Blood. 1986;67:1550. [PubMed] [Google Scholar]
- 32.Wiendahl M, Schulze Wierling P, Nielsen J, Fomsgaard Christensen D, Krarup J, Staby A, Hubbuch J. Chem Eng Technol. 2008;31:893. [Google Scholar]
- 33.Strancar A, Koselj P, Schhwinn H, Josic Dj. Anal Chem. 1996;68:3483. doi: 10.1021/ac960292f. [DOI] [PubMed] [Google Scholar]
- 34.Pucic M, Vidic J, Rucevic M, Josic Dj, Lauc G. MSS. Portoroz, Slovenia: 2010. High throughput isolation of IgG from human plasma using 96-well protein G monolithic plate. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.