Abstract
Biosensors based on luminescent bacteria may be valuable tools to monitor the chemical quality and safety of surface and drinking water. In this review, an overview is presented of the recombinant strains available that harbour the bacterial luciferase genes luxCDABE, and which may be used in an online biosensor for water quality monitoring. Many bacterial strains have been described for the detection of a broad range of toxicity parameters, including DNA damage, protein damage, membrane damage, oxidative stress, organic pollutants, and heavy metals. Most lux strains have sensitivities with detection limits ranging from milligrams per litre to micrograms per litre, usually with higher sensitivities in compound-specific strains. Although the sensitivity of lux strains can be enhanced by various molecular manipulations, most reported detection thresholds are still too high to detect levels of individual contaminants as they occur nowadays in European drinking waters. However, lux strains sensing specific toxic effects have the advantage of being able to respond to mixtures of contaminants inducing the same effect, and thus could be used as a sensor for the sum effect, including the effect of compounds that are as yet not identified by chemical analysis. An evaluation of the suitability of lux strains for monitoring surface and drinking water is therefore provided.
Keywords: Bioluminescence, Biosensors, Water monitoring, Toxicity, Reporter genes
Introduction
To ensure the chemical quality and safety of drinking water, it is essential to monitor the surface water sources as well as critical points in the distribution network. Currently, the presence of toxic chemicals in water is investigated by chemical analysis, by using aquatic organisms as biomonitors, and by in vitro toxicity assays [1]. Chemical analysis is quantitative, sensitive, and highly selective, but only target compounds are detected. The biomonitoring methods using mussels, Daphnia, algae, or natural bacteria are able to detect the total, mostly systemic, acute toxic effects of compounds such as herbicides and heavy metals. However, the toxic effects in these organisms have little predictive value for possible hazards for human individuals. In addition, these biomonitors do not react to non-systemic, specific toxic effects of compounds such as genotoxicants and endocrine disruptors. In vitro toxicity assays, using human or other mammalian cell lines, provide information on hazards relevant for human toxicity and can detect the sum effect of the whole mixture of toxicants present. For real-time monitoring of toxicants in water, there is currently no suitable system available that provides relevant information about human hazards. This gap may be filled by a type of biosensor that employs genetically modified luminescent bacteria which provide a rapid, easily measurable response in the presence of relevant toxic (mixtures of) compounds. A rapidly growing number of luminescent bacteria have already been constructed and described, and may be applicable for toxicity detection in water.
In this paper, an overview is provided of available bacterial luxCDABE strains and an evaluation and concurrent selection of strains which might be used in a biosensor for water quality monitoring. Lowe [2] defined a biosensor as “an analytical device, which converts the concentration of the target substance into an electrical signal through a combination of a biological recognition system associated with a physico-chemical transducer”. For a toxic compound to elicit a measurable response in bacterial cells in a biosensor, it first has to cross the cell wall and cell membrane. Then, it has to trigger a sensing element, in most cases a promoter linked to a reporter gene, leading to the production of easily measurable reporter proteins. Detailed reviews have been written by van der Meer et al. [3, 4] which explain the mechanisms involved in the cellular transport and activation mechanisms of analytes.
Currently, the most commonly used reporter proteins for optical detection in microbial systems are green fluorescent protein for fluorescence and bacterial luciferase for luminescence. Bioluminescence offers the advantages of faster response times and higher short-term sensitivity (seconds to minutes). Fluorescent proteins may keep accumulating for many hours and owing to their high stability, they allow detection even after cell death [5–7]. Green fluorescent protein also does not require a substrate or ATP, thereby lowering the burden on the cells [3]. For online monitoring of water, sensitivity and fast response times are more important factors than reporter stability. Therefore, luminescence is the detection method of choice for online monitoring, and this overview will thus focus on available luminescent bacterial reporter strains.
Bioluminescent bacteria express luminescence through the production of luciferase, either bacterial (lux) or firefly (luc). The latter has the advantage of a higher quantum yield, but requires the constant addition of luciferine. As a result, bacterial luciferase is favoured in most cases [8].
Bacterial luciferase catalyses the oxidation of a long-chain aliphatic aldehyde (RCHO) and a reduced flavin mononucleotide (FMNH2). In this reaction, free energy is emitted in the form of light with a wavelength of 490 nm:
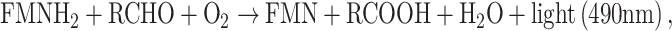 |
where FNM is flavin mononucleotide. As this reaction depends on a functional electron transport system, it only functions in viable cells [9].
Of the bacterial luciferase operon, only the luxAB genes are required for luminescence; however, in this case a substrate has to be administered externally. More practical for online monitoring is the use of the luxCDABE genes, in which luxCDE code for the (re)generation of the substrate [10]. Thus, no substrate addition is necessary and the luciferase reporter can operate independently. This overview will thus be limited to strains with the five bacterial luciferase genes, luxCDABE (lux strains).
Assays using bioluminescent bacteria can be divided into two groups, namely systems with constitutive expression and systems with inducible expression. Bacteria with constitutive expression normally have a high expression of luminescence, which decreases under toxic conditions (‘lights off’). They are usually natural bioluminescent bacteria, such as Aliivibrio fischeri (until recently known as Vibrio fischeri), and are often used to detect acute (cyto)toxicity, as the response is not compound-specific. The use of these bacteria is categorized under biomonitors, as it involves natural aquatic organisms displaying a cytotoxic response to the presence of toxic compounds. These organisms and assays are therefore not included in this overview.
Inducible systems, in contrast, have a low baseline luminescence, which increases after exposure to specific compounds (‘lights on’). In these systems, both promoter and reporter genes may be inserted from other bacteria to give an optimal response to the compounds of interest. It should be noted, however, that many inducible strains emit a low level of background luminescence when they are not induced. This enables the detection of high acute toxicity or other types of severe stress that may compromise the survival of the bacteria, as the background luminescence will then disappear [11]. However, when the background luminescence is on the high side, this tends to lower the sensitivity as it makes it harder to distinguish the signal [12].
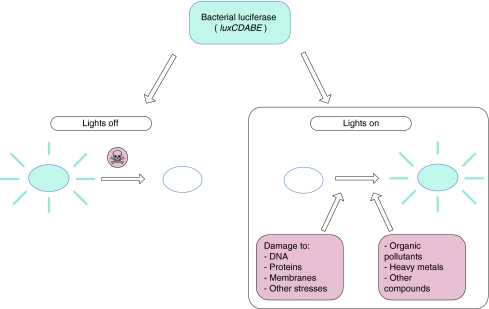
On the basis of the promoters used in the construction of such ‘lights-on’ bioreporters, the inducible systems can be divided into effect- and compound-specific strains. The former respond to a specific type of toxicity, for example DNA damage, oxidative stress, or protein damage (heat shock). The compound-specific strains detect a single compound or group of compounds with similar chemical characteristics or mode of action, such as specific metals, alkanes, or benzene, toluene, ethylbenzene, and xylene (BTEX) compounds (Fig. 1) [11, 13].
Fig. 1.
Overview of the luminescent bacterial reporter systems. The strains covered by this study are displayed in the lights on box
In summary, the aim of this review is to provide an overview of inducible, genetically engineered luminescent bioreporter bacteria harbouring the luxCDABE genes (lux strains), which can potentially be integrated into a biosensor device. Several strains of these luminescent bacteria will be categorized and compared on the basis of their inducers (and corresponding response). An evaluation is made of the characteristics that are important for water quality monitoring, for example sensitivity, response time, robustness, and pathogenicity.
Effect-specific lux strains
The effect-specific lux strains include strains that detect DNA, protein, and membrane damage, and oxidative stress. These strains are constructed by coupling the luxCDABE gene to a promoter that is involved in a specific stress response. As a result, the response of these strains is directly correlated with the total amount and potency of compounds that induce a specific type of stress.
DNA damage
Damage to DNA in bacteria can trigger at least two repair systems, the ada-controlled adaptive response specific to damage by alkylation, and the recA-dependent, lexA-controlled SOS response.
DNA damage by alkylation activates the ada gene, which leads to transcription of the ada, alkA, alkB, and aid genes [14]. The SOS response can be induced by many chemicals that damage DNA, arrest DNA synthesis, or arrest cell division, including mitomycin C (MMC), N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), and methyl methanesulfonate. More than 40 genes are activated in the SOS response, including recA, lexA, uvrA, and umuDC [14, 15]. When the promoters of these genes are coupled to the lux gene, the extent of DNA damage can be easily measured as an increase in luminescence. In most lux strains, recA is used as the promoter. An overview of the strains used in the studies described in the following can be found in Table 1.
Table 1.
Literature overview of lux strains for the detection of DNA damage and their detection limits
| Strain | References | Promoter | Receiving strain | LOD | Setup | Notes |
|---|---|---|---|---|---|---|
| DPD2794 | [21] | recA | RFM443 | 480 μg/l H2O2 | Culture plate | |
| [22] | Portable sensor | |||||
| [12] | 2 μg/l MMC | Culture plate | ||||
| [23] | 500 μg/l trimethroprin | Culture plate | ||||
| [24] | Optical fibre | |||||
| [25] | 10 μg/l MMC | Bioreactor | ||||
| [26] | Bioreactor | |||||
| [27] | 50 μg/l MMC | Bioreactor | ||||
| [28] | 5 μg/l MMC, 0.013 μg/l benzo[a]pyrene | Flasks | ||||
| [29] | Antibody plates | |||||
| [16] | 100 μg/l MMC | Culture plate | ||||
| DPD2797 | [12] | recA | DE112 | 0.1 μg/l MMC | Culture plate | tolC mutant |
| [30] | Optical fibre | |||||
| DPD1718 | [12] | recA | DPD1692 | Culture plate | Chr | |
| [31] | 100 μg/l MMC | Optical fibre | ||||
| [32] | 25 μg/l MMC | Optical fibre | ||||
| DPD1710 | [33] | recA | RFM443 | 1 μg/l MMC | Sol–gel wells | Chr |
| DPD1714 | [12] | recA | DM800 | Culture plate | Chr | |
| DPD1709 | [12] | recA | DM803 | Culture plate | Chr | |
| DPD3063 | [12] | recA | W3110 | 0.1 μg/l MMC | Culture plate | Chr |
| Sal94 | [12] | recA | WG49 | 250 μg/l 4-nitrophenol | Culture plate | Salmonella typhimurium |
| RM4440 | [17] | recA | FRD1 | Culture plate | Pseudomonas aeruginosa | |
| [18] | Alginate beads | |||||
| DPD2818 | [16] | uvrA | RFM443 | 10 μg/l MMC | Culture plate | |
| DPD2844 | [16] | alkA | RFM443 | < 10 μg/l MMC | Culture plate | |
| BBTNrdA | [20] | NrdA | 156.3 μg/l MNNG | Culture plate |
All strains are Escherichia coli based with luxCDABE as the reporter, unless stated otherwise under ‘Notes’.
LOD limit of detection, MMC mitomycin C, MNNG N-methyl-N′-nitro-N-nitrosoguanidine, Chr genes are inserted in the chromosome, instead of plasmids
Vollmer et al. [16] compared three lux strains, DPD2794, DPD2818, and DPD2844, containing the promoters of the recA, uvrA, or alkA genes, respectively, fused to the A. fischeri luxCDABE genes. The best results were gained with DPD2844 (alkA), which showed the highest sensitivity (detection limit of less than 0.01 mg/l MNNG) and shortest response time (40-50 min). An important cause for the very high sensitivity was the very low background luminescence in this strain. It should be mentioned, however, that not all strains were tested with the same compounds. For DPD2844, only results with MNNG were given, whereas for DPD2794 and DPD2818, detection limits for MMC and UV light were provided.
Davidov et al. [12] conducted a study to improve the sensitivity of recA-based DNA damage strains. The use of luxCDABE genes of A. fischeri resulted in a more sensitive, but slower response than with luxCDABE from Photorhabdus luminescens. The sensitivity was also improved by insertion of a single copy of the recA::lux fusion into the chromosome, instead of in multicopy plasmids, and by a mutation in the tolC gene. Insertion in the chromosome resulted in a lower background luminescence, thus enhancing the ability to distinguish the signal. A tolC mutation impairs the ability of the cell to excrete toxicants, which leads to higher intracellular concentrations. These two adaptations resulted in strains with detection limits of 0.1 μg/l for MMC. In addition to the positive controls MMC, MNNG, and H2O2, a group of other genotoxic and non-genotoxic compounds was also tested. No false negatives were found, and except for one, all positives had been tested as potential genotoxicants in other bioassays.
As an alternative for the Escherichia coli based strains, Elasri and Miller [17] developed strain RM4440, based on Pseudomonas aeruginosa. P. aeruginosa was chosen because it is a natural resident of water and soil, and as such, it was expected to be more robust than E. coli for online water monitoring. Strain RM4440 contained the recA and luxCDABE genes and was exposed to UV light. No limit of detection was mentioned, but lag times were about 30 min.
In a follow-up study by the same group, the same strain was exposed to 17 chemicals, of which eight are known SOS inducers. The cells were exposed in alginate beads to fixate them, as well as to simulate their own natural alginate biofilms. The P. aeruginosa reporter strain reacted to all compounds known to induce SOS in E. coli except H2O2 [18]. Suggested explanations were protection by the alginate or the ability of P. aeruginosa to actively degrade H2O2 [19]. A drawback of P. aeruginosa for use in environmental monitoring is that it is a known opportunistic human pathogen.
Hwang et al. [20] constructed strain BBTNrdA, which is an E. coli based lux strain that has the nrdA gene as the promoter. The nrdA gene is activated in DNA synthesis, but is not regulated by the SOS response. The strain reacted to all four DNA damaging agents tested (nalidixic acid, MMC, MNNG, and 4-nitroquinoline N-oxide). Also four phenolic compounds and four oxidative compounds were tested, but only one of these (H2O2) induced luminescence in this strain.
In conclusion, a large variety of DNA damage strains have been developed in the last few decades, based upon a number of different promoters involved in DNA damage repair. The most sensitive lux strains found in this study were two modified recA strains, namely DPD2797 and DPD3063 [12].
Protein damage
A mechanism found in cells of all organisms to counteract protein damage is the heat-shock response. This response can be triggered by high temperatures, viral infections, exposure to various chemicals that react with proteins, and abnormal proteins resulting from other processes.
Van Dyk et al. [34] used two promoters to develop lux strains for the detection of protein damage, namely the grpE gene and the dnaKp gene. The dnaK gene encodes for Hsp70, a heat-shock protein that has an important cellular function in protein folding and renaturation. The grpE gene encodes for Hsp60, which has a function similar to that of Hsp70.
Both promoters responded to generally the same compounds, namely ethanol, methanol, copper sulphate, phenol and derivates, and 2,4-dichlorophenoxyacetic acid, but grpE proved to be about fivefold to tenfold more sensitive. The grpE strain also had a higher basal luminescence level, and therefore it showed a lights-off effect at higher concentrations. Both lights-on and lights-off effects at high concentrations were enhanced in cells with a tolC mutation.
The grpE strain TV1061 was used in later studies, often incorporated in various setups (see Table 2). It has been shown to respond to a wider range of toxicants than any other lux strain, including phenols, halomethanes, oxidants, phosdrin, chlordimeform, sodium cyanide, and MNNG. However, it is less sensitive than, for example, DNA damage strains, with detection limits in the milligram per litre range. Human toxicants that did not induce TV1061 include colchincine, trimethylolpropane phosphate, nickel chloride, sodium selenite, and lindane [21, 30].
Table 2.
Summary of strains for the detection of protein damage and their corresponding detection limits
| Strain | References | Promoter | Receiving strain | LOD | Setup | Notes |
|---|---|---|---|---|---|---|
| TV1061 | [21] | grpE | RFM443 | 0.1 mg/l 4-bromophenol | Culture plate | |
| [22] | < 1% ethanol | Portable sensor | ||||
| [23] | 5 mg/l rifampicin | Culture plate | ||||
| [24] | Optical fibre | |||||
| [25] | 300 mg/l phenol | Bioreactor | ||||
| [26] | Bioreactor | |||||
| [35] | Bioreactor | |||||
| [33] | 0.3% ethanol | Sol–gel wells | ||||
| [30] | Optical fibre | |||||
| [29] | Antibody glass/gold plates | |||||
| [34] | ±12 mg/l pentachlorophenol, 1% ethanol | Culture plate | ||||
| WM1202 | [34] | dnaKp | RFM443 | 4% ethanol, 19 mg/l pentachlorophenol | Culture plate | |
| WM1302 | [34] | dnaKp | DE112 | 50 mg/l phenol | Culture plate | tolC mutant |
| TV1076 | [34] | grpE | DE112 | ±0.07 mg/l pentachlorophenol | Culture plate | tolC mutant |
All strains are E. coli with luxCDABE as the reporter
In conclusion, two promoter genes have been used to generate lux strains that are sensitive to protein damage. Of these, grpE gave the most sensitive strains (TV1061/TV1076), responded to a wide range of toxicants, and has been used more often.
Cell membrane damage
For compounds that cause cell membrane damage, bacteria can be used that contain the fabA gene, coupled to the full lux gene. The fabA gene codes for β-hydroxydecanoyl–acyl carrier protein dehydratase and is responsible for the formation of double bonds in fatty acids which are used in the cell membrane. The fabA gene is activated by binding of the FadR protein, a process that is inhibited by long-chain acyl-CoA thioesters. In the case of membrane damage, fatty acid starvation will occur and long-chain acyl-CoA thioester levels will be low, resulting in a high induction of fabA.
Strain DPD2540 containing fabA and luxCDABE was used in a study by Choi and Gu [36] and was tested with several compounds. As expected, fabA was induced by membrane-damaging agents, such as ethanol, phenol, and cerulenin. More surprisingly however, DPD2540 also responded to the DNA-damaging agent MMC and the oxidative agent H2O2. The reason for this is probably that these agents also cause damage to the membrane as a secondary effect. In these cases the response was delayed from around 60 min for ethanol to 150 min for MMC and H2O2. Unfortunately, no detection limits were given in this study. For more studies that used this strain and other fabA strains, see Table 3.
Table 3.
Literature overview of lux strains for the detection of cell membrane damage and their detection limits
| Strain | References | Promoter | Receiving strain | LOD | Setup | Notes |
|---|---|---|---|---|---|---|
| DPD2544 | [37] | fabA | W3110 | 0.19 mg/l Triton X-100 | Culture plate | |
| [29] | Antibody glass/gold plates | |||||
| DPD2540 | [37] | fabA | RFM443 | Culture plate | ||
| [21] | RFM444 | 0.14 mg/l 4-bromophenol | Culture plate | |||
| [36] | Culture plate | |||||
| [22] | Portable sensor | |||||
| [25] | 100 mg/l phenol | Bioreactor | ||||
| [26] | Bioreactor | |||||
| [35] | Bioreactor | |||||
| DPD2543 | [37] | fabA | DE112 | 0.08 mg/l 4-nitrophenol | Culture plate | tolC mutant |
| [30] | Optical fibre | |||||
| DPD2546 | [37] | fabA | DC530 | Culture plate | ||
| DPD2549 | [37] | fabA | MH163 | Culture plate | ||
| DPD1674 | [37] | fabA | W3110 | Culture plate | Chr |
All strains are E. coli with luxCDABE as the reporter
A comparison of several fabA-based lux strains was made by Bechor et al. [37]. The strains used included a tolC mutant (DPD2543), a fadR mutant (DPD2549), and a strain with fabA::lux inserted in the chromosome (DPD1674). The fadR mutant gave almost no response, proving that the luminescence was indeed induced via fabA. The tolC mutant strain (DPD2543) was more sensitive for most of the chemicals tested than the non-mutant strain (DPD2544). However, this was not true for all membrane-damaging substances. For example, DPD2543 was more sensitive to many phenol derivatives, but not to phenol itself.
The chromosomal insertion resulted in a drop in background luminescence by about a factor of 100. DPD1674 had a higher response ratio for ethanol than DPD2544, but this was slightly lower for phenol. Whether the detection limit also differed was not clarified. Compounds that induced luminescence in fabA strains included alcohols, phenol and derivatives, halomethanes, aromatics, and detergents.
In comparisons between effect-specific lux strains, the fabA strains showed a close similarity to the protein damage strain (TV1061) in the compounds it responded to. Only in the cases of bromodichloromethane, paraquat, cumene hydroperoxide, and sodium cyanide was TV1061 induced, whereas the fabA strains failed to respond [21, 30].
In conclusion, several strains have been developed for the detection of membrane damage, all based on the fabA promoter. The amount of information on the sensitivity and specificity of these strains is limited, thus no preference for a strain could be entertained. Like the protein damage strains, they react to a broad range of compounds with detection limits in the milligrams per litre range.
Oxidative stress
Active oxygen species are a serious threat to cells as they are capable of damaging proteins, nucleic acids, lipids, and membranes. Oxidative stress occurs naturally in cells as a result of respiration, or may be caused by toxic compounds. These toxicants can generate peroxides (H2O2), superoxides (O•-2), or hydroxyl radicals (OH•). Peroxides lead to the activation of the E. coli OxyR regulon, whereas superoxides induce SoxRS. Despite the fact that O•-2 can also trigger the formation of OH•/H2O2, most promoters are only activated by one of the two groups [38]. A large number of promoters from these pathways have been used to form lux strains that detect oxidative stress. In Table 4 a literature overview of these strains is given.
Table 4.
Literature overview of lux strains for the detection of oxidative stress and their detection limits
| Strain | References | Reacts on | Promoter | Receiving strain | LOD | Setup |
|---|---|---|---|---|---|---|
| DPD2511 | [40] | Peroxides | katG | RFM443 | Culture plate | |
| [21] | 0.1 mg/l H2O2 | Culture plate | ||||
| [22] | 0.0006% H2O2 ∼ 6 mg/l | Portable sensor | ||||
| [41] | 0.1 mg/l bisphenol A | Culture plate | ||||
| [30] | Optical fibre | |||||
| [29] | Antibody glass/gold plates | |||||
| DPD2515 | [21] | Superoxides | micF | W3110 | 0.01 mg/l paraquat | Culture plate |
| [29] | Antibody glass/gold plates | |||||
| DP1 | [27] | Superoxides | pqi-5 | RFM443 | 0.1 mg/l paraquat | Bioreactor |
| [33] | 2.5% ethanol, 7.8 mg/l paraquat | Sol–gel wells | ||||
| [42] | Cell chip | |||||
| PGRFM | [38] | Superoxides, | ||||
| peroxides | pgi | RFM443 | 0.6 mg/l paraquat | Culture plate | ||
| DK1 | [27] | Peroxides | katG | RFM443 | 10 mg/l H2O2 | Bioreactor |
| [33] | 0.15% 2-propanol, 0.8 mg/l H2O2 | Sol–gel well plate | ||||
| [45] | 0.88 μM (0.03 mg/l) H2O2 | Microfluid chip | ||||
| EBSoxS | [44] | Superoxides | soxS | RFM443 | Cell chip | |
| DS1 | [44] | Superoxides | sodA | RFM443 | Cell chip | |
| ZWF | [42] | Cell chip | ||||
| [43] | Superoxides | zwf | RFM443 | Culture plate | ||
| FPR | [42] | Cell chip | ||||
| [43] | Superoxides | fpr | RFM443 | Culture plate | ||
| EBFumC | [42] | Superoxides | fumC | RFM443 | Cell chip | |
| EBHmp | [42] | Superoxides | hmp | RFM443 | Cell chip | |
| EBHJ | [41] | Superoxides | sodA | RFM443 | 0.015 mg/l paraquat | Culture plate |
All strains are E. coli with luxCDABE as the reporter
One of the genes under control of the OxyR regulon is katG (catalase hydroperoxidase I) [39]. Belkin et al. [40] introduced the katG promoter coupled to the full luxCDABE gene of A. fischeri in E. coli, resulting in strain DPD2511. Luminescence induction was found for several oxidative compounds, including H2O2, organic peroxides, paraquat, menadione, xanthine, xanthine oxidase, and cigarette smoke. It also responded to ethanol and showed a synergistic response to the combination of ethanol and H2O2. Additionally, the same strain has been reported to respond to cadmium chloride, ethidium bromide, and bisphenol A. No response was seen after exposure to potassium dichromate [41].
DK1, a similar strain with katG as the promoter, also responded on H2O2 and menadione, but not on paraquat or structural analogues of paraquat [42].
The defence mechanism against superoxides, the SoxRS regulon, acts in two steps. Superoxide-generating compounds are first detected by SoxR, which then induces transcription of SoxS, a transcriptional activator of 16 other genes. These include sodA (manganese superoxide dismutase), nfo (DNA repair endonuclease IV), zwf (glucose 6-phosphate dehydrogenase), acnA (aconitase), fumC (stable fumarase), fpr (ferredoxin reductase), acrAB (efflux pumps), micF (antisense RNA for the ompF porin messenger RNA), and fur (repressor of iron transport) [39].
Several superoxide-sensitive lux strains have been reported that are based on promoters from the SoxRS regulon, including DPD2515 (micF), EBSoxS (soxS), EBHJ (sodA), ZWF (zwf), DP1 (pqi-5), DS1 (sodA), EBFumC (fumC), and FPR (fpr) [21, 41–44]. Reported inducers of these strains include paraquat (methyl viologen) and its structural analogues ethyl viologen, benzyl viologen, and heptyl viologen. Exposure to hydroxyl radical or peroxides, such as H2O2, gave much lower or no responses [21, 42, 43]. Unfortunately, little is known about the sensitivity of these strains, although for DPD2515 and EBSoxS detection limits of about 0.01 mg/l paraquat have been reported [21, 41].
In addition to the aforementioned strains, EBHmp (hmp) and PGRFM (pgi) have been developed; they contain promoters that belong neither to the OxyR pathway nor to the SoxRS pathway. PGRFM has been reported to respond to both superoxides and peroxides, with a lowest detection limit of 0.6 mg/l for paraquat [38]. However, in another study both strains responded only to superoxides, namely paraquat, ethyl viologen, and heptyl viologen [42].
In conclusion, for the detection of all oxidative agents, both a superoxide-sensing strain and a peroxide-sensing strain is needed. For peroxides, only katG-based strains are available and relatively well documented. For superoxides, a large array of strains have been constructed, but there is very limited information on both their sensitivity and their selectivity.
Compound-specific strains
In this section an overview is given of lux strains that respond specifically to certain types of compounds. These strains have been constructed by combining the luxCDABE reporter with promoter genes from bacteria that have developed an enhanced resistance to specific toxic compounds. Included are strains that respond to several types of organic pollutants and heavy metals. An overview of the strains for the detection of organic compounds is given in Table 5, and for metal sensing strains in Table 6.
Table 5.
Compound-specific lux strains for selective groups of organic chemicals and their detection limits
| Strain | Species | References | Reacts on | Promoter | LOD | Setup | Notes |
|---|---|---|---|---|---|---|---|
| B2 | Pseudomonas putida | [46] | Toluene and TCE | todC1C2BA | Vials and alginate beads | ||
| TVA8 | P. putida | [47] | All BTEX | tod | 30 μg/l toluene | Vials | Chr |
| P. putida | [56] | Vials | |||||
| HK44 | P. fluorescens | [49] | Naphthalene, salicylate | nahG | 45 μg/l naphthalene | Culture plate | |
| [51] | 550 μg/l naphthalene | Sensor probe | |||||
| [57] | Sol–gel beads | ||||||
| [50] | 1,200 μg/l naphthalene, 500 μg/l salicylic acid | Sol–gel plates | |||||
| NAGK-1768 | P. putida | [52] | Naphthalene, salicylate | nagR-nagAa | 4 μg/l salicylic acid | Culture plate | |
| DNT5 | E. coli | [53] | Salicylate | nagR-nagAa | 164 μg/l salicylic acid | Culture plate | |
| [52] | 331 μg/l salicylic acid | Culture plate | |||||
| ENV307 (pUTK60) | Ralstonia eutropha | [54] | PCBs | orf0-bphA1 | 150 μg/l 4-CB | Culture plate | |
| OS8 (pDNdmpRlux) | P. fluorescens | [55] | phenols | dmpR | 30 μg/l 2-methylphenol, 80 μg/l phenol | Culture plate |
All strains have luxCDABE as the reporter, unless stated otherwise under ‘Notes’
TCE trichloroethylene, BTEX benzene, toluene, ethylbenzene, and xylene, 4-CB 4-chlorobiphenyl
Table 6.
Overview of metal-specific strains and their detection limits
| Strain | Species | Reference | Reacts on | Promoter | LOD | Notes |
|---|---|---|---|---|---|---|
| DH5α (pJAMA-arsR) | E. coli | [59] | As, Sb | arsR | 4 μg/l As | luxAB |
| [60] | 7.5 μg/l As | +20 mg/l Fe | ||||
| MG1655 (pZNT-lux) | E. coli | [61] | Cd, Pb, Zn, Hg | zntA | 1.1 μg/l Cd | |
| MG1655 (pCOP-lux) | E. coli | [61] | Cu(II), Ag(I) | copA | 11 μg/l Ag | |
| HB101 (pRB28) | E. coli | [62] | Mercury | merR-merT' | 0.0136 μg/l HgCl2 | Latex immobilization |
|
MC1061 (pmerRluxCDABE) |
E. coli | [6] | Hg | merR | ||
| [63] | Optical fibre | |||||
| MC1061(pmerRBSBPmerlux) | E. coli | [64] | Hg, Cd | merRB | 0.002 μg/l CH3HgCl | |
| MC1061 (pSLzntR/pDNPzntAlux) | E. coli | [64] | Hg, Cd, Zn, Pb | zntRA | 3 μg/l CdCl2 | |
| MC1061 (pSLcueR/pDNPcopAlux) | E. coli | [64] | Cu, Ag | copA | 20 μg/l CuSO4 | |
| OS8 (pDNmerRBSBPmerlux) | P. fluorescens | [64] | Hg, Cd | merRB | 0.03 μg/l Ch3HgCl | |
| OS8 (pDNpbrRPpbrAlux) | P. fluorescens | [64] | Hg, Cd, Zn, Pb | pbrRA | 40 μg/l HgCl2 | |
| OS8 (pDNcadRPcadAlux) | P. fluorescens | [64] | Hg, Cd, Zn, Pb | cadRA | 10 μg/l CdCl2 | |
| OS8::KnmerRBSBPmerlux | P. fluorescens | [64] | Hg, Cd | merRB | 0.8 μg/l HgCl2 | Chr |
| OS8::KncueRPcopAlux | P. fluorescens | [64] | Cu | copA | 8,000 μg/l CuSO4 | Chr |
| OS8::KnzntRPzntAlux | P. fluorescens | [64] | Hg, Cd, Zn, Pb | zntRA | 20 μg/l CdCl2 | Chr |
| OS8::KnpbrRPpbrAlux | P. fluorescens | [64] | Hg, Cd, Zn, Pb | pbrRA | 8 μg/l HgCl2 | Chr |
| OS9::KncadRPcadAlux | P. fluorescens | [64] | Hg, Cd, Zn, Pb | cadRA | 5 μg/l HgCl2 | Chr |
| RN4220(pcadCPcadAlux) | Staphylococcus aureus | [64] | Hg, Cd, Zn, Pb | cadCA | 3 μg/l HgCl2 | |
| BR151(pcadCPcadAlux) | Bacillus subtilis | [64] | Hg, Cd, Zn, Pb | cadCA | 2 μg/l CdCl2 |
All strains have luxCDABE as the reporter, unless stated otherwise under ‘Notes’. With two exceptions, all experiments were performed on culture plates; for the exceptions, see ‘Notes”
BTEX compounds
For the monitoring of toluene and trichloroethylene (TCE) cometabolism, strain B2 based on Pseudomonas putida was constructed by Applegate et al. [46]. P. putida is a non-pathogenic soil bacterium that is capable of utilizing and degrading organic solvents. This strain harbours the tod-lux complex that enables the detection of toluene and cometabolized TCE. TCE is cometabolic in the sense that it cannot be used as a carbon source, but is degraded as a side effect. The tod operon encodes for a suite of enzymes that mediate the metabolism of toluene via the toluene dioxygenase complex.
The strain had a detection limit of 0.1 mg/l for toluene and a response time of 90 min. A strong response was also observed when the strain was exposed to jet fuel containing toluene [46].
In addition to the aforementioned strain, the same group also constructed a strain with the same tod-lux complex inserted in the chromosome (TVA8). This resulted in higher sensitivity with a detection limit of 30 μg/l, but a longer response time of 120 min. It was also responsive to benzene, ethylbenzene, m-xylene, and p-xylene, indicating that strains with the tod promoter can be used as a general BTEX monitor [47].
Naphthalene and salicylate
Naphthalene belongs to the polycyclic aromatic hydrocarbons and is a wide-spread environmental pollutant. For the detection as well as the catabolism of naphthalene and its degradation intermediate salicylate, Pseudomonas fluorescens strain HK44 was developed. P. fluorescens is a non-pathogenic bacterium that lives on plants, in water, and in soil. This strain harbours a fusion of the promoter of its own nahG gene for naphthalene degradation to luxCDABE. It responded in a dose-dependent manner to both naphthalene and salicylate, with a detection limit for naphthalene of 45 μg/l [48, 49].
In a comparative study by Trogl et al. [50], strain HK44 was exposed to 32 components other than naphthalene and salicylate. From the 32 compounds, it responded only to 2-aminobenzoic acid, salicylaldehyde, 4-methylsalicylic acid, and 4-chlorsalicylic acid.
The same strain, while immobilized in a sensor, also responded to jet fuel and contaminated soil extracts that contained naphthalene. Response times of 8–24 min were recorded in this setup. Toluene did not induce any measurable effects in these bacteria [51].
Two other strains based on a different naphthalene degradation pathway have been developed by Mitchell and Gu [52]. Both strains contain the nagR-nagAa gene promoters which are upregulated by salicylate, coupled to luxCDABE. The first strain, called DNT5, has E. coli as the host organism. The second strain, NAGK-1768, has P. putida as the host organism, which also possesses the ability to degrade naphthalene via the nah and sal operons. It was shown that of the two strains, NAGK-1768 had the more favourable response characteristics, with higher luminescence and a lower detection limit for salicylic acid (4 μg/l, versus 331 μg/l in DNT5). NAGK-1768 also responded on 13 of 25 tested salicylic acid derivatives (also called salicylates) and naphthalene. DNT5 reacted to five of 25 salicylic acid derivatives.
In an additional study on the performance of DNT5, a slightly lower detection limit was reached than before (164 μg/l). The strain was also responsive to benzoic acid and two of its derivatives. Benzene, naphthalene, and phenol failed to induce a response [53].
Polychlorinated biphenyls
Layton et al. [54] tested the polychlorinated biphenyl (PCB)-sensitive strain Ralstonia eutropha ENV307(pUTK60) containing the orf0-bphA1 genes coupled to luxCDABE.
R. eutropha is a non-pathogenic bacterium that lives in soil and water and can degrade chloroaromatic compounds. The bacteria were exposed to biphenyl, monochlorinated biphenyls, and Aroclor 1242 (PCB mixture) solutions. As the water solubility of PCBs is very low, non-ionic surfactants were used to achieve measurable concentrations in water. The highest sensitivity was reached with 4-chlorobiphenyl, which gave a detection limit of 0.15 mg/l. For Aroclor 1242, however, the detection limit was 1.5 mg/l, whereas the aqueous solubility without surfactants is less than 1 mg/l. As these results indicate that as the concentrations of PCBs in water will always be below the detection limit, the usefulness of this bacterial strain for in situ PCB monitoring in water is highly questionable.
Phenols
A strain for the detection of phenol and its methylated derivatives was developed by Leedjarv et al. [55]. The dmpR gene for catabolism of phenols was linked to luxCDABE and introduced in P. fluorescens. The lowest detection limits were found for 2-methylphenol (0.03 mg/l) and phenol (0.08 mg/l). Different phenols applied in a mixture caused an additive effect on the induction of the bacterial strain.
Heavy metals
For the construction of metal-sensing strains, the operons for metal resistance that some naturally occurring bacteria possess are often used as promoters. For example, the mer operon enables cells to convert Hg(II) to the less toxic Hg(0). The ars operon, on the other hand, provides the ability to transport antimonite and arsenite out of the cell and to reduce arsenate to arsenite [58].
Unfortunately, no study was found in which an arsenic-sensing strain was used that contains the complete luciferase gene. Since it is a very relevant metal for water monitoring, it was decided to include some studies that used strains that contain luxAB instead.
Stocker et al. [59] used a regulatory gene from the ars operon, the arsR gene, combined with luxAB to construct a strain for the detection of arsenite. For this compound, it had a detection limit of 4 μg/l and a response time of 30 min. The strain also responded to arsenate and antimony. In a later study, the same strain was also used to determine arsenite in groundwater samples from Vietnam. The sensitivity in these samples was reduced somewhat by the presence of iron, which binds arsenite. It was determined that with 20 mg/l Fe the lowest detectable concentration was 7.5 μg/l, which is still below the World Health Organization (WHO) guideline value of 10 μg/l. In total, 194 samples were tested, of which 112 samples were deemed safe; 8.0% tested false-negative and 2.4% tested false-positive when compared with chemical analysis [60].
Two E. coli strains for the detection of various heavy metals were tested by Riether et al. [61]. The pZNT::lux strain proved to be sensitive to Cd(II), Pb(II), Zn(II), Hg(II), and to a lesser degree to Co(II), Ni(II), SbO-2, CrO2-4, and Cr2O2-7. The strongest inducer of this strain was cadmium, with a detection limit of 10 nM (1.1 μg/l). The other strain tested was pCOP::lux, which reacted only to copper and silver, with detection limits of 0.1 and 0.3-1 μM (11 and 19-64 μg/l), respectively. It was also verified that EDTA, and probably also other chelating components, lower the bioavailability of metals.
Three E. coli HB101 variants for the detection of Hg(II) were tested by Lyngberg et al. [62]. The largest dynamic range of detection for HgCl2 was found with strain HB101(pRB28), which contains merR and a truncated form of merT (merT’). The highest sensitivity was found in cells immobilized in latex, which gave a detection limit of 0.1 nM HgCl2 (27.2 ng/l) after 15 h of induction. The detection limit was a factor of 10 higher in suspended culture (1 nM or 0.27 μg/l), but the response time was reduced to 1 h for the lowest concentration. In all cases, the lag time was drastically shortened at higher concentrations, although the suspended cells remained the fastest responders. It was also possible to enhance the response and sensitivity by adding cysteine, which increases mercury uptake. This resulted in a detection limit of 0.05 nM (13.6 ng/l) in immobilized cells.
Another strain for the detection of mercury which contains the merR::luxCDABE genes was constructed by Hakkila et al. [6]. The response time of this strain was 30 min. A detection limit was not mentioned in this study. The same strain was also tested during the EILATox-Oregon Workshop while it was immobilized in an alginate gel on the tip of a fibre. It reacted to mercury chloride, a river sample, and slightly to sodium cyanide [63].
A large comparative study was performed by Ivask et al. [64] with 13 newly constructed metal-inducible strains with luxCDABE as the reporter. Also six unspecific constitutive strains were tested as controls, to be able to recognize general toxic effects of the compounds. Special attention was given to the effect of other types of bacteria as host organisms: three strains were E. coli, eight strains were P. fluorescens, one strain was Staphylococcus aureus, and one strain was Bacillus subtilis (Table 6). The detection limits were mostly in the microgram per litre range, with two mercury strains in the nanogram per litre range. Of the Gram-negative bacteria, E. coli was slightly more sensitive than P. fluorescens. Chromosomal insertion in the latter reduced background luminescence, but this did not lead to higher sensitivity. Gram-positive bacteria were more difficult to modify, requiring a helper plasmid to prevent loss of the plasmid. They yielded detection limits quite similar to those of the Gram-negative strains.
Discussion
Biosensors based on luminescent bacteria may prove to be a valuable additional tool for the monitoring of water quality and safety. By the combination of different stress-responsive promoters with the bioluminescence genes, strains have been constructed that react with differing degrees of specificity to toxic effects (e.g. DNA damage) or specific groups of chemicals (e.g. heavy metals). The main purpose of this review is to evaluate available lux strains and their perspective for use in online water quality and safety monitoring. A preliminary selection was made upfront for strains with the bacterial luxCDABE genes, as this enables a fast response without the need for substrate addition.
An important factor in the evaluation of the strains is whether the measured effect or compound(s) can be expected to cause toxic effects in humans and whether they have additional value above that of existing techniques. Since all strains mentioned in this review are genetically modified, strains should preferably be based on non-pathogenic bacteria to be allowed for use outside a laboratory. Another aspect that is taken into account is the sensitivity of the strains, which is compared with actual levels of contaminants that have been found in Dutch rivers, as well as drinking water standards. Unfortunately, in most cases little information is available on selectivity and specificity, which limits the possibility to compare strains for these parameters. The same lack of information exists for the robustness and stability of the bacteria when they are used over prolonged periods in natural water.
Effect-specific strains
Of all the effects that can be detected by bacterial strains, DNA damage is probably the most relevant for humans. Because it is an effect that causes little acute damage, existing biomonitors are not very sensitive to compounds causing this effect. However, it can have severe consequences in humans in the long term.
A number of strains have been developed for the detection of DNA damage by coupling DNA repair genes to luxCDABE. To improve performance, some strains also contain different adjustments, such as a tolC mutation or insertion of the genes in their chromosome. The most sensitive lux strains found in this study were two modified recA strains, namely DPD2797 and DPD3063 [12].
An important consideration when it comes to DNA damage that is rarely addressed is that many agents require metabolic activation before they become harmful to the DNA. This may be (partially) solved by adding rodent-derived cytochrome P450 (S9). However, in an online biosensor this is very impractical. Another option would be to incorporate some of these enzymes in the lux strains themselves as has been done in Salmonella typhimurium for the umu test (for an overview see [14]). These modifications are very appealing, but since a large variety of cytochrome P450 enzymes are involved and there is still very little experience with such strains, it is unlikely that they will be used in online biosensors soon.
Both protein and membrane damage strains are also referred to as strains for general damage or stress, as they are relatively unspecific and have overlapping target components. As a result, these strains are expected to have less additional value above that of the existing biomonitors than, for example, DNA damage strains. Additionally, compounds that also cause other types of damage (e.g. MNNG, paraquat) can usually be detected more sensitively with strains that react specifically to these effects. On the other hand, in a system that employs multiple strains, they may be very useful, as they detect toxicants that diminish the response in other, more specific strains [21, 30].
For the detection of protein damage, strain TV1061 (grpE::luxCDABE) has probably the most favourable combination of sensitivity, specificity, and user experience [65]. As all membrane damage strains have the fabA gene as the promoter, differences in sensitivity, selectivity, and response times between these strains are minor [37].
Oxidative stress differs somewhat from the aforementioned effects, as it causes damage in itself to DNA, proteins, and membranes. Thus, it can be expected that compounds that cause oxidative stress will be detected to some extent by the aforementioned strains. However, with the use of genes from specific defence regulons as promoters, strains have been generated that have enhanced sensitivity for oxidative compounds. These strains can be divided into peroxide- and superoxide-sensing strains. Both types showed detection limits of around 0.1 mg/l for H2O2 and for paraquat [44].
What might limit the usefulness of oxidative-stress-sensing strains in the field is the unstable nature of oxidative compounds. As no experimental field data are available on the occurrence of such compounds, it still has to be shown whether biosensors with these strains are of use.
Compound-specific strains
In addition to measuring a certain effect, one can also use a promoter that is activated in the presence of a specific compound or group of compounds. The advantage is that the detection limit for these compounds is usually lower in a compound-specific strain than in a strain that reports only the effect, often in the microgram per litre range. Because of their high specificity, they are generally less suitable for the monitoring of ‘normal’ surface or drinking water, as they are likely to miss many contaminations. However, in cases where only a specific type of contamination is of interest, they may be used as a fast and convenient detection method. Examples of such situations are after an incident or on (former) industrial sites [5]. The choice for a certain strain will in such a situation mainly depend on the toxicants involved.
General remarks: comparability of studies referenced
A comment that needs to be made when attempting to compare the performance of such a diverse selection of reporter strains is that it is practically impossible to directly compare levels of luminescence between studies. Depending on the instrumentation used, different units are used to express light intensity. These are often completely arbitrary, and presented as instrument-specific relative light units (RLUs), amperes, or specific bioluminescence (in nanoamperes divided by the optical density) [30, 36, 47, 66]. Another complicating factor is that the number of photons that corresponds to 1 RLU differs with the photomultiplier, even if the photomultipliers are of the same type and from the same manufacturer.
However, most performance criteria can be calculated regardless of the units of luminescence used. The most common way to do this is by using response ratios to quantify the signal; these are defined as the ratio of luminescence relative to that of the uninduced control [21]. The disadvantage of this method is that response ratios are strongly influenced by the background luminescence. As a result, the relationship between response ratios and actual luminescence differs with the strain, with, in general, lower maximal response ratios in strains with high background luminescence [64, 67]. Most studies with response ratios also give a graph with the luminescence levels in RLUs, to give some insight into the absolute difference between signal and background [12]. Another possibility is to use the absolute difference in luminescence between background and induced cells [67].
The detection limit has also been determined in several different manners. If response ratios are used, the detection limit is usually defined as the concentration that leads to a twofold induction of luminescence over the background, i.e. a response ratio of 2 [37]. In studies that express light intensity in RLUs, the detection limit is often defined as the concentration that leads to a significantly higher luminescence compared with the control [68]. It should be mentioned that the detection limit is not only dependent on the strain, but is also dependent on the inducer, cell concentration, bioavailability, measurement time, and operational protocol. These differences in determining induction and detection limits should be kept in mind when comparing studies, especially when the differences are relatively small. In this review, the detection limits given were determined by the original authors of the studies. However, for comparisons between studies, only differences in detection limits of a factor of 10 or more for the same component are considered relevant.
A similar observation may be made concerning the determination of response or lag times, which are highly dependent on the concentrations used. Nevertheless it can be said that, on average, most lag times are around 60 min, with a few fast responders of 30 min or less and slow responders of 90 min or more. In the case of an online biosensor that is used as an early warning system in surface water, a response time of around 1 h will usually be fast enough to detect passing contamination peaks. There will then be enough time to react to an alarm from the sensor before the contaminated water reaches the treatment plant inlet or ultimately the consumers as drinking water. For alarm monitoring of drinking water in distribution networks, naturally, response times will have to be much shorter.
Biosensing pollutants in surface water
The most important question that remains is whether these bacteria can actually detect the levels of toxicants that occur in reality. To truly answer this question, it would be necessary to perform field studies. However, an estimate can be made by comparing the detection limits with peak levels of compounds of concern that have been found recently in surface water, and with the target values for contaminants of drinking water.
A large number of studies have been performed to determine levels of pollutants in surface water, of which a few examples will be given. A survey was performed by Loos et al. [69] to determine the occurrence of polar organic pollutants in European rivers. The highest concentrations measured were 31 μg/l ibuprofen, 39.8 μg/l caffeine, 19.4 μg/l tolyltriazole, and 11.6 μg/l carbamazepine.
In the Netherlands, measurements have also been taken after large spills. There were 49 large spills reported in 2008 in the Rhine, of which most involved BTEX compounds, methyl tert-butyl ether, ethyl tert-butyl ether, or a combination of these substances. The highest concentration measured after an incident was 91 μg/l trichloromethane. The peak concentrations measured in routine measurements were generally lower, for example 6.0 μg/l methyl tert-butyl ether (Nieuwegein), 2.58 μg/l ethyl tert-butyl ether (Lobith), 1.3 μg/l toluene (Nieuwegein), and 7.8 μg/l lead (Nieuwegein) [70].
In two recent literature surveys, emerging contaminants and trace pollutants were evaluated for their occurrence and toxicity [71, 72]. The highest priority was given to compounds that are both a human health hazard and frequently detected in surface water. In Table 7 a summary of the highest-ranking compounds is given.
Table 7.
High-priority emerging contaminants and trace pollutants
| Compound | Common use | Maximum concentration (μg/l) | Location | Reference |
|---|---|---|---|---|
| Carbamazepine | Anticonvulsant | 12 | EU | [69] |
| 1,4-Dioxane | Solvent | 10 | EU | [72] |
| 17α-Ethinyl estradiol | Hormone | 0.83 | USA | [73] |
| 17β-Estradiol | Hormone | 0.2 | USA | [73] |
| Estrone | Hormone | 0.11 | USA | [73] |
| Perfluorooctanoic acid | Waterproofing | 19 | USA | [74] |
| Perfluorooctanesulfonic acid | Waterproofing | 1.4 | EU | [69] |
| Bis(2-ethylhexyl)phthalate | Plasticizer | 98 | EU | [75] |
| Diazinon | Insecticide | 1.1 | US | [76] |
| Methoxychlor | Herbicide | 1.7 | EU | [77] |
| Dieldrin | Insecticide | 0.21 | USA | [73] |
| N,N-Diethyl-m-toluamide | Insect repellent | 1.1 | USA | [73] |
| Triclosan | Antiseptic | 2.3 | US | [73] |
| Acetaminophen | Pain reliever | 10 | US | [73] |
The WHO has set guideline values for drinking water for some chemicals, including arsenic (10 μg/l), cadmium (3 μg/l), mercury (6 μg/l), and toluene (700 μg/l) [78]. These levels can be detected with strains specific for these compounds. However, for most of the detected compounds, no specific guideline values have been determined yet. With analytical methods becoming more and more sensitive, the number of contaminants detected in drinking water is increasing, but their levels are mostly not of concern for health. Recently, target values have been set by the Dutch drinking water industry to define what levels of contaminants are acceptable from both a human health perspective as well as an ethical or esthetical perspective. This latter perspective stems from the philosophy that contaminants do not belong in drinking water. On the basis of the thresholds of toxicological concern (TTCs) set for food additives and on the opinions of Dutch drinking water experts of what is ethically acceptable, target values of 0.01 μg/l for genotoxic contaminants and 0.1 μg/l for other contaminants have been derived [1, 72, 79].
When compared with the TTC-derived target values for drinking water, the detection thresholds of both effect- and compound-specific strains are generally too high to be used for monitoring. The only exceptions are the strains for the detection of mercury, with a lowest detection limit of 0.002 μg/l for methyl mercury [64].
However, the peak concentrations that have been measured in surface water are much higher, as is also expected to be the case after intentional water poisoning. In these cases, sensors based on lux strains may be very useful to detect spills at an early stage, provide an early warning of such events, and prevent the intake of contaminated water.
Additionally, various efforts are also being undertaken to improve the sensitivity of luminescent strains by using different or additional modifications. The insertion of the promoter-reporter genes in the chromosome and the addition of a tolC mutation have already been mentioned. In an article in this same special issue, Yagur-Kroll and Belkin [80] demonstrate that both the sensitivity and the response times can also be improved by splitting the luxCDABE genes into two separately controlled units. The best effect was gained when luxAB (the luciferase enzyme) was inducible by a promoter and luxCDE (the aldehyde substrate) was expressed constitutively.
What also should be noted is that contaminations often consist of mixtures of compounds. Compounds exerting a similar type of effect through a similar mode of action often behave additively in mixtures, but sometimes synergism and antagonism can also occur with compounds with a different mode of action, interfering with the effects observed. Depending on the strain and compounds involved, this can lead to either improvement or reduction of the ability of the bacteria to detect the collective activity of a complex mixture as a sum parameter. Currently, little is known about the significance of these mixture reactions for lux strains.
Another issue that is of great importance for the application of biosensors is the ability of the bacteria to remain alive and active in natural water. This is of course also dependent on the design of the sensor and the matrix in which the bacteria are contained. Nevertheless, differences in robustness between species of bacteria can be expected.
So far, most studies have been performed with E. coli K12, as these bacteria are non-pathogenic and relatively easy to modify. Since E. coli was originally an inhabitant of the intestine, it is not very robust when exposed to natural water. Several studies have shown that especially the presence of other microorganisms is detrimental for the survival of E. coli in unsterilized water [81–83].
However, very few other species have been used to construct lux strains. Only for the detection of organic pollutants such as toluene and naphthalene is it common to use different bacteria, such as the natural soil bacteria P. putida and P. fluorescens [46, 51, 84]. P. fluorescens has also been used in the development of metal-sensing strains, whereas cadA combined with luc or luxCDABE has been brought into B. subtilis and S. aureus; S. aureus is a common resident of human skin, and B. subtilis is a non-pathogenic soil bacterium [64, 85].
Although these bacteria are generally no more sensitive than E. coli, they might be more robust as water is often their natural habitat, which makes it likely they will remain active over a longer time. However, it still has to be confirmed whether they indeed yield better results in an online sensor.
Conclusion
A large number of luminescent bacteria have been developed for the detection of various toxic effects and compounds. Although most studies so far have been performed in the laboratory, implementation in a biosensor in the field holds the highest promise for these bacteria. The currently existing strains have detection thresholds that range from milligrams per litre to micrograms per litre. In general, more specific strains tend to be more sensitive. Since peak concentrations after incidents in Dutch rivers are generally in the low microgram per litre range, only a few strains will be capable of detecting individual compounds. Sensitivity improvement is therefore necessary. Nevertheless, these strains may have added value above that of existing techniques in the detection of mixtures of toxicants and in early warning systems. Other issues that need to be addressed in the case of field use include response times, robustness, signal quantification, and pathogenicity.
Acknowledgements
This work was performed in the TTIW-cooperation framework of Wetsus, Centre of Excellence for Sustainable Water Technology (http://www.wetsus.nl). Wetsus is funded by the Dutch Ministry of Economic Affairs. The authors would like to thank the participants of the Sensoring research theme for fruitful discussions and their financial support.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Published in the special issue Microorganisms for Analysis with Guest Editor Gérald Thouand.
References
- 1.van Wezel A, Mons M, van Delft W. J Environ Monit. 2010;12:80–89. doi: 10.1039/b912979k. [DOI] [PubMed] [Google Scholar]
- 2.Lowe CR. In: Handbook of biosensors and biochips. Marks RS, Cullen DC, Karube I, Lowe CR, Weetall HH, editors. Chichester: Wiley; 2007. pp. 7–22. [Google Scholar]
- 3.van der Meer JR, Tropel D, Jaspers M. Environ Microbiol. 2004;6(10):1005–1020. doi: 10.1111/j.1462-2920.2004.00655.x. [DOI] [PubMed] [Google Scholar]
- 4.van der Meer JR, Belkin S. Nat Rev Microbiol. 2010;8(7):511–522. doi: 10.1038/nrmicro2392. [DOI] [PubMed] [Google Scholar]
- 5.Sagi E, Hever N, Rosen R, Bartolome AJ, Premkumar JR, Ulber R, Lev O, Scheper T, Belkin S. Sens Actuators B Chem. 2003;90(1–3):2–8. doi: 10.1016/S0925-4005(03)00014-5. [DOI] [Google Scholar]
- 6.Hakkila K, Maksimow M, Karp M, Virta M. Anal Biochem. 2002;301(2):235–242. doi: 10.1006/abio.2001.5517. [DOI] [PubMed] [Google Scholar]
- 7.Li YF, Li FY, Ho CL, Liao VHC. Environ Pollut. 2008;152(1):123–129. doi: 10.1016/j.envpol.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Keane A, Phoenix P, Ghoshal S, Lau PCK. J Microbiol Methods. 2002;49(2):103–119. doi: 10.1016/S0167-7012(01)00382-7. [DOI] [PubMed] [Google Scholar]
- 9.Daniel R, Almog R, Ron A, Belkin S, Diamand YS. Biosens Bioelectron. 2008;24(4):882–887. doi: 10.1016/j.bios.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 10.Billard P, DuBow MS. Clin Biochem. 1998;31(1):1–14. doi: 10.1016/S0009-9120(97)00136-7. [DOI] [PubMed] [Google Scholar]
- 11.Gu MB, Mitchell RJ, Kim BC. Adv Biochem Eng Biotechnol. 2004;87:269–305. doi: 10.1007/b13533. [DOI] [PubMed] [Google Scholar]
- 12.Davidov Y, Rozen R, Smulski DR, Van Dyk TK, Vollmer AC, Elsemore DA, LaRossa RA, Belkin S. Mutat Res Genet Toxicol Environ Mutagen. 2000;466(1):97–107. doi: 10.1016/S1383-5718(99)00233-8. [DOI] [PubMed] [Google Scholar]
- 13.Belkin S. Curr Opin Microbiol. 2003;6(3):206–212. doi: 10.1016/S1369-5274(03)00059-6. [DOI] [PubMed] [Google Scholar]
- 14.Biran A, Yagur-Kroll S, Pedahzur R, Buchinger S, Reifferscheid G, Ben-Yoav H, Shacham-Diamand Y, Belkin S. Microb Biotechnol. 2009 doi: 10.1111/j.1751-7915.2009.00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janion C. Int J Biol Sci. 2008;4(6):338–344. doi: 10.7150/ijbs.4.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vollmer AC, Belkin S, Smulski DR, VanDyk TK, LaRossa RA. Appl Environ Microbiol. 1997;63(7):2566–2571. doi: 10.1128/aem.63.7.2566-2571.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elasri MO, Miller RV. Appl Microbiol Biotechnol. 1998;50(4):455–458. doi: 10.1007/s002530051320. [DOI] [PubMed] [Google Scholar]
- 18.Elasri MO, Reid T, Hutchens S, Miller RV. FEMS Microbiol Ecol. 2000;33(1):21–25. doi: 10.1111/j.1574-6941.2000.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 19.Elkins JG, Hassett DJ, Stewart PS, Schweizer HP, McDermott TR. Appl Environ Microbiol. 1999;65(10):4594–4600. doi: 10.1128/aem.65.10.4594-4600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang ET, Ahn JM, Kim BC, Gu MB. Sensors. 2008;8(2):1297–1307. doi: 10.3390/s8021297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belkin S, Smulski DR, Dadon S, Vollmer AC, Van Dyk TK, Larossa RA. Water Res. 1997;31(12):3009–3016. doi: 10.1016/S0043-1354(97)00169-3. [DOI] [Google Scholar]
- 22.Choi SH, Gu MB. Biosens Bioelectron. 2002;17(5):433–440. doi: 10.1016/S0956-5663(01)00303-7. [DOI] [PubMed] [Google Scholar]
- 23.Eltzov E, Ben-Yosef DZ, Kushmaro A, Marks R. Sens Actuators B Chem. 2008;129(2):685–692. doi: 10.1016/j.snb.2007.09.054. [DOI] [Google Scholar]
- 24.Eltzov E, Marks RS, Voost S, Wullings BA, Heringa MB. Sens Actuators B Chem. 2009;142(1):11–18. doi: 10.1016/j.snb.2009.08.024. [DOI] [Google Scholar]
- 25.Kim BC, Gu MB. Environ Monit Assess. 2005;109(1–3):123–133. doi: 10.1007/s10661-005-5843-7. [DOI] [PubMed] [Google Scholar]
- 26.Kim BC, Gu MB, Hansen PD. In: Environmental chemistry: green chemistry and pollutants in ecosystems. Lichtfouse E, Schwarzbauer J, Didier R, editors. Springer: Berlin; 2005. [Google Scholar]
- 27.Lee JH, Gu MB. Biosens Bioelectron. 2005;20(9):1744–1749. doi: 10.1016/j.bios.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 28.Min J, Kim EJ, LaRossa RA, Gu MB. Mutat Res Genet Toxicol Environ Mutagen. 1999;442(2):61–68. doi: 10.1016/S1383-5718(99)00059-5. [DOI] [PubMed] [Google Scholar]
- 29.Premkumar JR, Lev O, Marks RS, Polyak B, Rosen R, Belkin S. Talanta. 2001;55(5):1029–1038. doi: 10.1016/S0039-9140(01)00533-1. [DOI] [PubMed] [Google Scholar]
- 30.Pedahzur R, Polyak B, Marks RS, Belkin S. J Appl Toxicol. 2004;24(5):343–348. doi: 10.1002/jat.1023. [DOI] [PubMed] [Google Scholar]
- 31.Polyak B, Bassis E, Novodvorets A, Belkin S, Marks RS. Water Sci Technol. 2000;42(1–2):305–311. [Google Scholar]
- 32.Polyak B, Bassis E, Novodvorets A, Belkin S, Marks RS. Sens Actuators B Chem. 2001;74(1–3):18–26. doi: 10.1016/S0925-4005(00)00707-3. [DOI] [Google Scholar]
- 33.Mitchell RJ, Gu MB. Biosens Bioelectron. 2006;22(2):192–199. doi: 10.1016/j.bios.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 34.Van Dyk TK, Majarian WR, Konstantinov KB, Young RM, Dhurjati PS, Larossa RA. Appl Environ Microbiol. 1994;60(5):1414–1420. doi: 10.1128/aem.60.5.1414-1420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JH, Song CH, Kim BC, Gu MB. Water Sci Technol. 2006;53(4–5):341–346. doi: 10.2166/wst.2006.139. [DOI] [PubMed] [Google Scholar]
- 36.Choi S, Gu M. Biotechnol Bioprocess Eng. 1999;4(1):59–62. doi: 10.1007/BF02931916. [DOI] [Google Scholar]
- 37.Bechor O, Smulski DR, Van Dyk TK, LaRossa RA, Belkin S. J Biotechnol. 2002;94(1):125–132. doi: 10.1016/S0168-1656(01)00423-0. [DOI] [PubMed] [Google Scholar]
- 38.Niazi JH, Kim BC, Ahn JM, Gu MB. Biosens Bioelectron. 2008;24(4):670–675. doi: 10.1016/j.bios.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 39.Asad NR, Asad L, de Almeida CEB, Felzenszwalb I, Cabral-Neto JB, Leitao AC. Genet Mol Biol. 2004;27(2):291–303. doi: 10.1590/S1415-47572004000200026. [DOI] [Google Scholar]
- 40.Belkin S, Smulski DR, Vollmer AC, VanDyk TK, LaRossa RA. Appl Environ Microbiol. 1996;62(7):2252–2256. doi: 10.1128/aem.62.7.2252-2256.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee HJ, Gu MB. Appl Microbiol Biotechnol. 2003;60(5):577–580. doi: 10.1007/s00253-002-1168-4. [DOI] [PubMed] [Google Scholar]
- 42.Lee JH, Youn CH, Kim BC, Gu MB. Biosens Bioelectron. 2007;22(9–10):2223–2229. doi: 10.1016/j.bios.2006.10.038. [DOI] [PubMed] [Google Scholar]
- 43.Niazi JH, Kim BC, Gu MB. Appl Microbiol Biotechnol. 2007;74(6):1276–1283. doi: 10.1007/s00253-006-0758-y. [DOI] [PubMed] [Google Scholar]
- 44.Lee JH, Mitchell RJ, Kim BC, Cullen DC, Gu MB. Biosens Bioelectron. 2005;21(3):500–507. doi: 10.1016/j.bios.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 45.Yoo SK, Lee JH, Yun SS, Gu MB. Biosens Bioelectron. 2007;22(8):1586–1592. doi: 10.1016/j.bios.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 46.Applegate B, Kelly C, Lackey L, McPherson J, Kehrmeyer S, Menn FM, Bienkowski P, Sayler G. J Ind Microbiol Biotechnol. 1997;18(1):4–9. doi: 10.1038/sj.jim.2900334. [DOI] [PubMed] [Google Scholar]
- 47.Applegate BM, Kehrmeyer SR, Sayler GS. Appl Environ Microbiol. 1998;64(7):2730–2735. doi: 10.1128/aem.64.7.2730-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb OF, Bienkowski PR, Matrubutham U, Evans FA, Heitzer A, Sayler GS. Biotechnol Bioeng. 1997;54(5):491–502. doi: 10.1002/(SICI)1097-0290(19970605)54:5<491::AID-BIT8>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 49.Heitzer A, Webb OF, Thonnard JE, Sayler GS. Appl Environ Microbiol. 1992;58(6):1839–1846. doi: 10.1128/aem.58.6.1839-1846.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trogl J, Ripp S, Kuncova G, Sayler GS, Churava A, Parik P, Demnerova K, Halova J, Kubicova L. Sens Actuators B Chem. 2005;107(1):98–103. doi: 10.1016/j.snb.2004.07.039. [DOI] [Google Scholar]
- 51.Heitzer A, Malachowsky K, Thonnard JE, Bienkowski PR, White DC, Sayler GS. Appl Environ Microbiol. 1994;60(5):1487–1494. doi: 10.1128/aem.60.5.1487-1494.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitchell RJ, Gu MB. Appl Biochem Biotechnol. 2005;120(3):183–197. doi: 10.1385/ABAB:120:3:183. [DOI] [PubMed] [Google Scholar]
- 53.Lee JH, Mitchell RJ, Gu MB. J Biotechnol. 2007;131(3):330–334. doi: 10.1016/j.jbiotec.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 54.Layton AC, Muccini M, Ghosh MM, Sayler GS. Appl Environ Microbiol. 1998;64(12):5023–5026. doi: 10.1128/aem.64.12.5023-5026.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leedjarv A, Ivask A, Virta M, Kahru A. Chemosphere. 2006;64(11):1910–1919. doi: 10.1016/j.chemosphere.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 56.Bhattacharyya J, Read D, Amos S, Dooley S, Killham K, Paton GI. Environ Pollut. 2005;134(3):485–492. doi: 10.1016/j.envpol.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Matrubutham U, Thonnard JE, Sayler GS. Appl Microbiol Biotechnol. 1997;47(5):604–609. doi: 10.1007/s002530050981. [DOI] [Google Scholar]
- 58.Ramanathan S, Ensor M, Daunert S. Trends Biotechnol. 1997;15(12):500–506. doi: 10.1016/S0167-7799(97)01120-7. [DOI] [PubMed] [Google Scholar]
- 59.Stocker J, Balluch D, Gsell M, Harms H, Feliciano J, Daunert S, Malik KA, Van der Meer JR. Environ Sci Technol. 2003;37(20):4743–4750. doi: 10.1021/es034258b. [DOI] [PubMed] [Google Scholar]
- 60.Trang PTK, Berg M, Viet PH, Van Mui N, Van Der Meer JR. Environ Sci Technol. 2005;39(19):7625–7630. doi: 10.1021/es050992e. [DOI] [PubMed] [Google Scholar]
- 61.Riether KB, Dollard MA, Billard P. Appl Microbiol Biotechnol. 2001;57(5–6):712–716. doi: 10.1007/s00253-001-0852-0. [DOI] [PubMed] [Google Scholar]
- 62.Lyngberg OK, Stemke DJ, Schottel JL, Flickinger MC. J Ind Microbiol Biotechnol. 1999;23(1):668–676. doi: 10.1038/sj.jim.2900679. [DOI] [PubMed] [Google Scholar]
- 63.Hakkila K, Green T, Leskinen P, Ivask A, Marks R, Virta M. J Appl Toxicol. 2004;24:333–342. doi: 10.1002/jat.1020. [DOI] [PubMed] [Google Scholar]
- 64.Ivask A, Rolova T, Kahru A. BMC Biotechnol. 2009;9:41. doi: 10.1186/1472-6750-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vandyk TK, Majarian WR, Konstantinov KB, Young RM, Dhurjati PS, Larossa RA. Appl Environ Microbiol. 1994;60(5):1414–1420. doi: 10.1128/aem.60.5.1414-1420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vijayaraghavan R, Islam SK, Zhang M, Ripp S, Caylor S, Bull ND, Moser S, Terry SC, Blalock BJ, Sayler GS. Sens Actuators B Chem. 2007;123(2):922–928. doi: 10.1016/j.snb.2006.10.064. [DOI] [Google Scholar]
- 67.Yagur-Kroll S, Bilic B, Belkin S. Microb Biotechnol. 2010;3(3):300–310. doi: 10.1111/j.1751-7915.2009.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sticher P, Jaspers MCM, Stemmler K, Harms H, Zehnder AJB, vanderMeer JR. Appl Environ Microbiol. 1997;63(10):4053–4060. doi: 10.1128/aem.63.10.4053-4060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Loos R, Gawlik BM, Locoro G, Rimaviciute E, Contini S, Bidoglio G. Environ Pollut. 2009;157(2):561–568. doi: 10.1016/j.envpol.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 70.Rijnwaterbedrijven R. Jaarrapport 2008 de Rijn. Vereniging van Rivierwaterbedrijven, Nieuwegein: RIWA-Rijn; 2008. [Google Scholar]
- 71.Murray KE, Thomas SM, Bodour AA. Environ Pollut. 2010;158(12):3462–3471. doi: 10.1016/j.envpol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 72.Schriks M, Heringa MB, van der Kooi MME, de Voogt P, van Wezel AP. Water Res. 2010;44(2):461–476. doi: 10.1016/j.watres.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 73.Kolpin DW, Furlong ET, Meyer MT, Thurman EM, Zaugg SD, Barber LB, Buxton HT. Environ Sci Technol. 2002;36(6):1202–1211. doi: 10.1021/es011055j. [DOI] [PubMed] [Google Scholar]
- 74.Fromme H, Tittlemier SA, Völkel W, Wilhelm M, Twardella D. Int J Hyg Environ Health. 2009;212(3):239–270. doi: 10.1016/j.ijheh.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 75.Fromme H, Kuchler T, Otto T, Pilz K, Müller J, Wenzel A. Water Res. 2002;36(6):1429–1438. doi: 10.1016/S0043-1354(01)00367-0. [DOI] [PubMed] [Google Scholar]
- 76.Kolpin DW, Skopec M, Meyer MT, Furlong ET, Zaugg SD. Sci Total Environ. 2004;328(1–3):119–130. doi: 10.1016/j.scitotenv.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 77.Badach H, Nazimek T, Kaminski R, Turski WA. Ann Agric Environ Med. 2000;7(1):25–28. [PubMed] [Google Scholar]
- 78.WHO . Guidelines for drinking-water quality incorporating first and second addenda to third edition, vol. 1. Recommendations. Geneva: World Health Organization; 2008. [PubMed] [Google Scholar]
- 79.Kroes R, Renwick AG, Cheeseman M, Kleiner J, Mangelsdorf I, Piersma A, Schilter B, Schlatter J, van Schothorst F, Vos JG, Wurtzen G. Food Chem Toxicol. 2004;42(1):65–83. doi: 10.1016/j.fct.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 80.Yagur-Kroll S, Belkin S (2010) Anal Bioanal Chem (in press). doi:10.1007/s00216-010-4266-7 [DOI] [PubMed]
- 81.Gordon C, Toze S. J Appl Microbiol. 2003;95(3):536–544. doi: 10.1046/j.1365-2672.2003.02010.x. [DOI] [PubMed] [Google Scholar]
- 82.Bogosian G, Sammons LE, Morris PJL, Oneil JP, Heitkamp MA, Weber DB. Appl Environ Microbiol. 1996;62(11):4114–4120. doi: 10.1128/aem.62.11.4114-4120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brettar I, Hofle MG. Appl Environ Microbiol. 1992;58(7):2201–2210. doi: 10.1128/aem.58.7.2201-2210.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Werlen C, Jaspers MCM, van der Meer JR. Appl Environ Microbiol. 2004;70(1):43–51. doi: 10.1128/AEM.70.1.43-51.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tauriainen S, Karp M, Chang W, Virta M. Biosens Bioelectron. 1998;13(9):931–938. doi: 10.1016/S0956-5663(98)00027-X. [DOI] [PubMed] [Google Scholar]