Abstract
Herein, we show that a naturally occurring RNA G-quadruplex element within the 5′ UTR of the human NRAS proto-oncogene is a target for a small molecule that inhibits translation in vitro. The present study provides a first demonstration that natural 5′ UTR mRNA G-quadruplexes have potential as molecular targets for small molecules that modulate translation.
Introduction
A major goal within biological science is to achieve manipulation of cellular events by using strategies that target nucleic acids and control gene expression. One such approach is to inhibit mRNA function(s). There has been considerable effort invested in the development of nucleic acid-based agents for sequence-directed silencing of mRNAs. Antisense oligonucleotides1 and small interfering RNAs2 have proved to be promising examples; however, they still face challenges that relate to poor pharmacological properties. A small molecule-based intervention strategy may provide some advantages, particularly with a view to ultimately translating chemical biology towards therapeutics.
The 5′ untranslated region (UTR) of mRNAs is generally important for the post-transcriptional control of gene expression,3 and thus of interest when considering strategies for interfering with mRNA functions. Small molecules that bind to structural elements within the 5′ UTR of mRNAs have been explored with a view to specifically interfering with translation initiation of the message. For prokaryotes, the discovery of riboswitch elements, which are RNA structures that interact specifically with natural metabolites and control the accessibility of the ribosome binding site, has driven recent investigations for finding synthetic metabolite analogs that could act as antibacterial agents.4 For eukaryotic translation, examples have been limited to unnatural RNA aptamers that have been in vitro selected to bind particular small molecules. Insertions of such aptamer sequences upstream of reporter genes have resulted in artificial systems that are responsive to the small molecules.5,6 These studies provided a proof of principle that small molecules that bind to structural elements within the 5′ UTR of eukaryotic mRNAs can form barriers to translation initiation, by blocking either ribosome formation on the mRNA template or the ribosome scanning process.6
An important consideration in targeting endogenous, cellular mRNAs with small molecules is to identify naturally occurring RNA structural elements that exhibit particular features amenable to selective recognition in the presence of numerous other nucleic acids and protein components. During the past decade there has been a growing interest in guanine (G)-rich nucleic acid sequences that can form non-canonical four-stranded structures, named G-quadruplexes.7 Intramolecular G-quadruplexes comprise planar G-tetrads connected through a Hoogsteen hydrogen bonded network, four grooves and three loops. Collectively these structural elements are very distinct from double helix-based nucleic acid secondary structures, and offer an attractive alternative for achieving selective molecular recognition by small molecules.8 There has been considerable focus on G-quadruplexes formed from genomic DNA sequences, their cellular functions, and their exploitation for biological intervention towards therapeutics.7,8 Nucleic acid chemists in particular have invested considerable effort in the design and synthesis of small molecules that interact with DNA G-quadruplexes.9 Bioinformatics studies have also indicated the presence of sequences with the potential to form RNA G-quadruplexes in the 5′ UTRs of many genes, including a larger number of proto-oncogene.10,11 We have recently reported on a naturally occurring RNA G-quadruplex (named NRQ; 5′ GGGAGGGGCGGGUCUGGG-3′)withinthe 5′ UTR of the NRAS proto-oncogene mRNA.10 Herein we show that the NRQ element is a target for small molecules that inhibit translation in vitro.The present study provides a first demonstration that 5′ UTR mRNA G-quadruplexes have potential as molecular targets for small-molecules that modulate translation.
Results
Design of the study
We devised a translational assay based on two 5′ capped reporter mRNA constructs: the first comprised the wild-type 254-nucleotide (nt) NRAS 5′ UTR, which includes the G-quadruplex element at +14 nt from the 5′ cap, placed upstream of the firefly luciferase coding sequence (NRAS UTR(+)Q, Fig. 1); and the second was a control, which comprised a 236-nt 5′ UTR derived from the wild-type NRAS 5′ UTR by removing the 18-nt NRQ element (NRAS UTR(−)Q, Fig. 1), to reveal any G-quadruplex-independent effects.
Fig. 1.
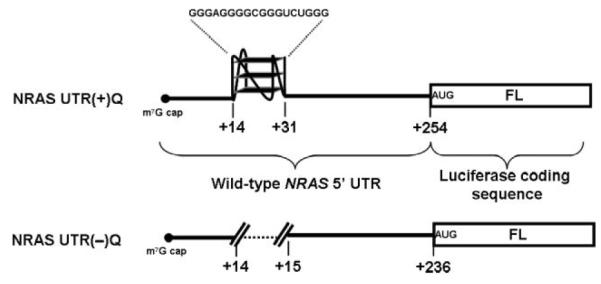
Schematic representation of NRAS UTR(−)Q and NRAS UTR(+)Q firefly luciferase reporter mRNAs.
The translation efficiency of these constructs in the presence of small molecule G-quadruplex ligands was evaluated in a eukaryotic cell-free system consisting of extracts from rabbit reticulocyte lysates (RRL), an established system to study translation independently of the other events of gene expression.
Differential effect of RR82 G-quadruplex ligand on the translation efficiency of NRAS UTR(−)Q and NRAS UTR(+)Q mRNAs
During the past decade, we have synthesised a number of distinct small molecule G-quadruplex ligands, each of which have demonstrated preferential binding to DNA G-quadruplexes rather than the DNA double-helix, and also show diversity in their selectivity for particular DNA G-quadruplex-forming-sequences.12,13 We started by screening some of these G-quadruplex ligands at 1.25 μM and 10 μM concentration in our translation assay. From these initial experiments, we identified a pyridine-2,6-bis-quinolino-dicarboxamide derivative (RR82, Fig. 2A) that demonstrated some selective inhibition of the translation of NRAS UTR(+)Q mRNA compared to the control. In the presence of 1.25 μM of RR82, the translation efficiency of NRAS UTR(+)Q was reduced to about 50%, as compared to a mocked-treated reaction in which water was added; whereas under the same conditions the control NRAS UTR(−)Q mRNA exhibited about 80% efficiency (Fig. 2B). Upon gradually increasing the concentration of RR82 from 1.25 μM to 10 μM, translation efficiencies of both the NRAS UTR(−)Q and the NRAS UTR(+)Q mRNAs were inhibited in a dose-dependent manner (Fig. 2B). However, at each of the ligand concentrations used, translation of the G-quadruplex-containing mRNA, i.e. NRAS UTR(+)Q, was inhibited to a higher level than that of the control NRAS UTR(−)Q mRNA, indicative of some degree of RNA G-quadruplex selectivity.
Fig. 2.
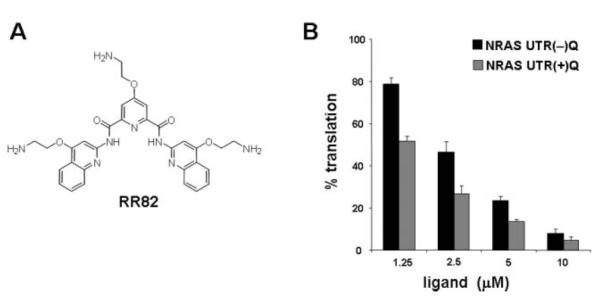
Effect of RR82 on the in vitro translation efficiency of NRAS UTR(−)Q and NRAS UTR(+)Q mRNAs. (A) Chemical structure of RR82. (B) Relative translation efficiencies of NRAS UTR(−)Q and NRAS UTR(+)Q mRNAs in the presence of the indicated amounts of RR82, as measured by luciferase activities. For each construct, the measured luciferase activity was normalized to the activity obtained in the absence of the ligand, which was set as 100%. Experiments were performed at least three times using at least two separate batches of RNA. The average values are presented along with the standard error on the mean.
Specific inhibition of NRAS UTR(+)Q mRNA translation by the G-quadruplex ligand RR110
Following these initial experiments, we next evaluated some structural variants of RR82 in our translation assay. A derivative that lacks the alkyl-amine appendage group at pyridine C4 exhibited similar translation repression trend than that of RR82 (data not shown). However, we then found a derivative with a para-fluorophenyl substituent at pyridine C4 (RR110, Fig. 3A) that showed considerably improved G-quadruplex selectivity. For concentrations up to 10 μM, RR110 had no effect on the translation efficiency of the control, while it inhibited translation of the G-quadruplex containing mRNA, i.e. NRAS UTR(+)Q, in a dose-dependent fashion (Fig. 3B). At 10 μM ligand concentration, RR110 inhibited NRAS UTR(+)Q translation by about 40%, but did not affect translation efficiency of the control.
Fig. 3.
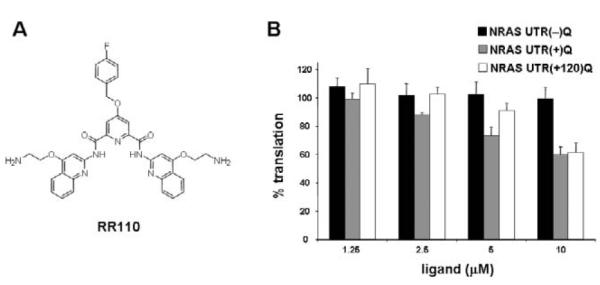
Effect of RR110 on the in vitro translation efficiency of mRNAs comprising a G-quadruplex motif in their 5′ UTR. (A) Chemical structure of RR110. (B) Relative in vitro translation efficiencies of NRAS UTR(−)Q, NRAS UTR(+)Q and NRAS UTR(+120)Q mRNAs in the presence of the indicated amounts of RR110, as measured by luciferase activities. For each construct, the measured luciferase activity was normalized to the activity obtained in the absence of the ligand, which was set as 100%. Experiments were performed at least three times using at least two separate batches of RNA. The average values are presented along with the standard error on the mean.
Above 10 μM, we started to observe G-quadruplex-independent inhibitory effects, which could be due to interactions between the small molecule and the others nucleic acids and/or protein components in the lysate (Fig. S1 in the ESI†). It is noteworthy that a slight variation of the molecular structure of the ligand, such as the replacement of a positively charged side chain by a more apolar fluorophenyl group, improved the specificity of the molecule for the G-quadruplex motif, providing an early insight into the scope for future optimization of such a ligand. To challenge the specificity of the ligand we also performed translation experiments with RR110 (10 μM) in the presence of a large (30- or 300-fold) excess of either a DNA duplex or an RNA hairpin competitor (i.e. 1 or 10 μM of competitor as compared to ca. 35 nM NRAS UTR(+)Q reporter mRNA). The results (Fig. 4) clearly demonstrate that neither the DNA duplex nor the RNA hairpin competitors are able to suppress the translation inhibitory effect of the RR110 molecule. However, when the NRQ RNA G-quadruplex was used as a competitor, RR110-mediated translation inhibition was considerably alleviated (Fig. 4); at 10 μM NRQ competitor, NRAS UTR(+) mRNA is translated as efficiently as in the absence of RR110. Taken together these results indicated selectivity of RR110 for RNA G-quadruplex.
Fig. 4.
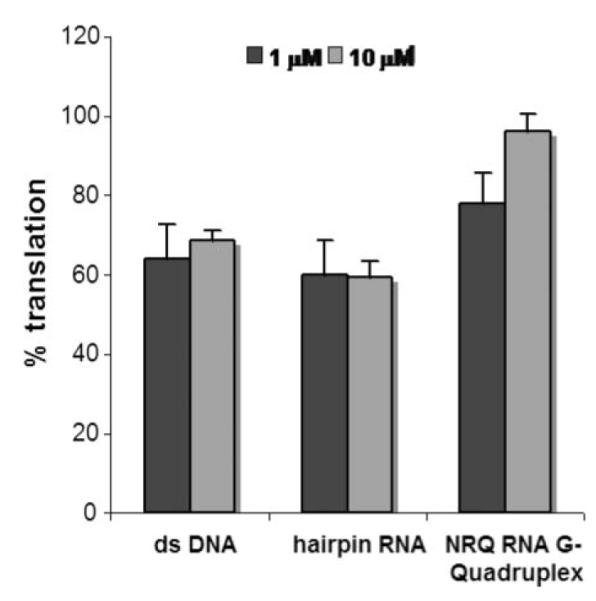
Effect of RR110 (10 μM) on the in vitro translation efficiency of NRAS UTR(+)Q mRNA in the presence (1 or 10 μM) of double-stranded DNA, hairpin RNA and NRQ RNA G-quadruplex nucleic acid competitors (see Experimental section for sequences). Measured luciferase activities were normalized to activities obtained in the absence of the ligand, which were set as 100%. Experiments were performed at least three times using at least two separate batches of RNA. The average values are presented along with the standard error on the mean.
G-quadruplex binding ligand RR110 stabilizes the NRQ element
To obtain further insights into the interaction between RR110 and NRQ element at a molecular level, we carried out ligand titration experiments followed by 1H NMR. The binding of RR110 to the NRQ was confirmed by a general line-broadening of the NRQ resonance signals upon addition of the ligand (Fig. 5A).14 Furthermore, there were marked upfield shifts in the imino proton region of the NRQ, indicative of π–π interactions between the aromatic surface of the ligand and the terminal G-tetrad(s) of the NRQ (Fig. 5A).14,15 For a number of proton resonances from the aromatic nucleobases, the line-broadening changes started to level after adding one equivalent of RR110 (Fig. 5A). The hydrogen-deuterium exchange (HDX) kinetics of the hydrogen-bonded imino protons of the G-tetrads provides a measure of the stability of the core of the folded RNA G-quadruplex structure. To obtain apparent HDX rates of the imino protons, the overall imino proton signals were integrated (over the range of δ1H 10.3–11.5 ppm) and normalized as a function of the exchange time following a non-linear regression procedure and assuming three single exponentially decaying components, corresponding to slow, medium and fast exchanging imino protons (see Experimental section). The three components were necessary to obtain reliable fitting results for all samples, judging from the fitting correlations and residuals (Fig. 5B). The presence of RR110 led to slower HDX kinetics, i.e. a higher degree of protection of the imino protons, suggesting that binding of RR110 strengthens the hydrogen bonding network and/or sequesters the imino protons of the G-tetrad from solvent exposure (Fig. 5B). We observed that RR110 binding induced a significant increase in the population of the slow phase (from 21% of the initial population to 43%; Table S1 in ESI) at the expense of the medium and fast phases. Collectively, these data support that RR110 binds the NRQ element to form a complex in which the G-quadruplex fold is stabilized.
Fig. 5.
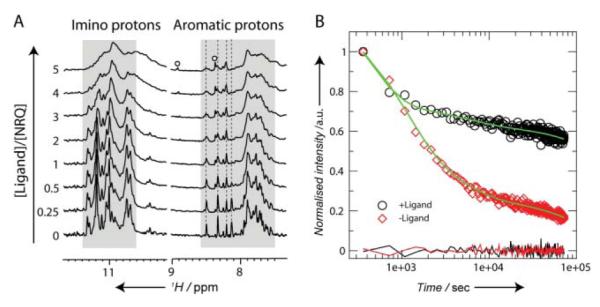
1H NMR spectroscopic analysis of the interaction between the RNA G-quadruplex (NRQ) and G-quadruplex ligand RR110. (A) 1H NMR titration of the NRQ element with RR110. The resonances arising from RR110 are indicated by open circles. While several aromatic protons from the RNA nucleobases exhibit negligible change in chemical shift (indicated by dashed lines), marked upfield shift changes are observed in the imino proton region at a ligand-to-NRQ ratio of three and higher. (B) HDX kinetics of the hydrogen-bonded imino proton resonances of the G-tetrads in the absence (open red diamond) and in the presence of 5 molar equivalent of RR110 (open black circle). The fitting results are shown in green lines with the residuals shown below.
Ligand-induced translation repression by interaction with a non-inhibitory cap distal RNA G-quadruplex target
The natural position of the NRQ element within the NRAS 5′ UTR is 14 nucleotides downstream from the 5′ cap. In this context, the NRQ element shows intrinsic translational inhibitory properties,10 whereas its inhibitory property are lost once it is moved sufficiently away from the 5′ end of the message.16 For example, the reporter construct NRAS UTR(+120)Q in which the NRQ element has been relocated to an unnatural position 120 nts downstream from the 5′ end of the NRAS 5′ UTR is translated as efficiently as the control NRAS UTR(−)Q.16 We performed a titration of RR110 on NRAS UTR(+120)Q in the translation assay. Upon addition of RR110, the translation of NRAS UTR(+120)Q showed a dose-dependent inhibition that is comparable to that observed for NRAS UTR(+)Q (Fig. 3B and Fig. S1 in ESI). At 10 μM ligand concentration, translation efficiencies of both NRAS UTR(+120)Q and the native construct are inhibited at the same level (ca. 40%) (Fig. 3B).
Discussion
The role of RNA G-quadruplexes in translation regulation has recently emerged as a new paradigm. We recently reported the example of a naturally occurring G-quadruplex in the 5′ UTR of a eukaryotic transcript that modulates translation.10 Specifically, we showed that a conserved intramolecular G-quadruplex structure in the 5′ UTR of the human NRAS transcript is involved in inhibiting protein translation in a eukaryotic cell-free translation system.10 Following this work, other RNA G-quadruplexes, identified within the 5′ UTRs of the human ZIC-117 and MP3-MMP18 mRNAs, have been demonstrated to inhibit translation in eukaryotic cells. Lately, an RNA G-quadruplex within the 5′ UTR of a ERS1 mRNA transcript transcribed from exon C has been shown to modulate the efficiency of translation in vitro.19 Futhermore, Halder et al. have establish general translational repression by synthetic G-quadruplex-forming sequences in 5′ UTRs in several mammalian cell lines.20 In addition, artificial G-rich elements that mask the ribosome binding site by folding into G-quadruplexes have been shown to modulate gene expression in bacteria.21 In a detailed computational analysis, we have identified about 2300 sequences with a potential to form G-quadruplexes in the 5′ UTRs of human genomic mRNAs.11 The associated genes includes several growth factors and oncogenes such as BCL2, JUN, MAF and FGR.
In this study, we have used the 5′ UTR of the human NRAS as an example to establish whether a naturally occurring 5′ UTR RNA G-quadruplex element can serve as a receptor for a small molecule that imparts translational control. We have employed luciferase reporter mRNAs that comprised either the native 254-nt NRAS 5′ UTR, including the G-quadruplex element located + 14 nts with respect to the 5′ cap, or a 236-nt control 5′ UTR derived from the NRAS 5′ UTR by deleting the quadruplex-forming sequence. The translation efficiency of the constructs in the presence of small molecule G-quadruplex ligands was evaluated in a translational system consisting of extracts of rabbit reticulocyte lysates. We first identified a pyridine-2,6-bis-quinolino-dicarboxamide derivative, RR82, that shows some selectivity for the G-quadruplex containing mRNA. For comparison, the commonly used G-quadruplex binding ligand TMPyP4, which we also included in our initial screening, showed no RNA G-quadruplex specificity and inhibited the translation of both the G-quadruplex-containing and the control mRNAs to the same extent (Figure S2 in the ESI†).
The challenge of specifically targeting a native RNA structure with a small molecule to inhibit translation of eukaryotic mRNA has been recognized as a major goal.22 Encouraged by our initial result, we next looked for molecules that show improved selectivity. We thus evaluated structural analogs of RR82 in the translation assay. We found a derivative, RR110, which exhibits markedly improved G-quadruplex selectivity. RR110 inhibits translation of the G-quadruplex-containing mRNA by approximately two-fold at 10 μM concentration, whereas it does not affect the translation efficiency of control mRNA. We also demonstrated that quadruplex-dependent translational inhibition by RR110 is maintained in the presence of a large excess of double-stranded DNA or hairpin RNA competitors. The outcomes of our study that used a native G-quadruplex structure within the wild-type NRAS 5′ UTR compare favorably with previous proof-of-concept experiments based on artificial in vitro selected RNA aptamers, such as an upstream biotin-binding aptamer that decreased the translational efficiency of a CAT reporter gene by about 50% in the presence of 1 mM biotin in RRL.6
Studies on hairpin RNA structures have demonstrated that the degree of translation repression by secondary structures within the 5′UTR of a mRNA is highly related to their thermodynamic stability, most stable structures being more inhibitory.23 Using 1H NMR and HDX experiments, we have demonstrated that RR110 bind to the NRQ element to form a complex in which the G-quadruplex fold is stabilized. In addition we have performed an mRNA stability analysis of NRAS UTR(+)Q translated in the absence or presence of RR110 (10 μM) (Fig. 6). The transcript showed the same stability, within experimental error, regardless of whether the small molecule was present or not. Thus the effect of RR110 on the translation efficiency of the G-quadruplex-containing mRNA reporter cannot be attributed to a difference in mRNA stability associated with its presence in the lysate. Taken together, these observations are consistent with RR110 inhibiting translation via interactions that stabilize the NRQ element in the NRAS 5′UTR. Studies on hairpin RNAs also showed that translation inhibition depends on the location of the structure within the 5′UTR.23 Even a relatively “weak” structure located proximal to the 5′ cap can be sufficient to interfere with the formation of the ribosomal pre-initiation complex (43S) at the 5′ end of the mRNA template, whereas a “strong” structure is required to block the migration of the 43S complex during the scanning process. When relocated at an unnatural position, 120 nts downstream from the 5′ cap, the NRQ element is not “strong” enough to interfere with the helicase activity of the scanning 43S complex and does not affect translation efficiency.16 Interestingly, upon addition of RR110 in the translation reaction, we did observe translation inhibition of the NRAS UTR(+120)Q construct. This observation is consistent with a mechanism whereby, upon binding of the ligand, the NRQ element at position +120 becomes sufficiently stable to perturb the migration of the small 43S ribosomal subunit during the scanning process of translation initiation.
Fig. 6.
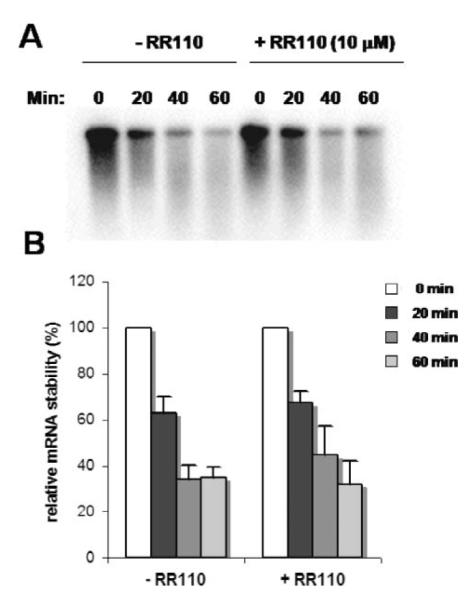
mRNA stability of the NRAS UTR(+)Q construct translated in the absence and in the presence of RR110 (10 μM). (A) A representative autoradiograph from one experiment. The time points and the absence or presence of RR110 is indicated above the panel. (B) Summary of the experiments. Values were normalized to the value at the beginning of the experiment (t = 0 min), which set at 100%. Experiments were performed at least three times using at least two separate batches of RNA. The average values are presented along with the standard error on the mean.
Conclusions
In conclusion, our study demonstrates proof-of-concept for small molecule-mediated regulation of translation by targeting a natural RNA G-quadruplex in a 5′ UTR. While there has been considerable attention on genomic DNA G-quadruplexes as potential drug targets, this current study suggests that RNA G-quadruplexes have appeal as targets for small molecule interference. Improvement in the selectivity and potency of the small molecule are now desirable goals that will be addressed in future studies.
Experimental section
Chemical synthesis
Synthesis of RR82 has been reported previously.13 A general scheme for the synthesis of RR110 is shown in Scheme S1 in the ESI.† A complete description of the synthesis and characterization of RR110 is given in the ESI.†
In vitro translation assays
Construction of the plasmids pSKC11, pSKC14 and pSKC17, which encode for reporter NRAS UTR(+)Q, NRAS UTR(−)Q and NRAS UTR(+120)Q transcripts, respectively, and conditions for in vitro transcription have been described previously.10,16 In vitro translation reactions of the mRNAs in the presence of ligands were carried out in a cell-free translation system consisting of extracts from nuclease-treated rabbit reticulocyte lysate (RRL) (Promega). Typically, 10 μL final volume translation mixtures were prepared that contain 70% (v/v) RRL, 10 μM amino acid mixtures minus methionine, 10 μM amino acid mixture minus leucine, 200 ng RNA and the indicated amount of small molecule ligand. The RNA was incubated with the small molecule [added as a 10× solution prepared by serial dilution from a 1 mM stock solution in water (RR82) or water/2.5% DMSO (RR110)] and the amino acids, in a total volume of 3 μL, for 30 min at room temperature, prior addition of 7 μLof RRL to start translation. Translation was carried out at 30 °C for 90 min. Translation efficiency was assessed by measuring firefly luciferase activity using Luciferase Assay Reagent (Promega) on an Orion II Microplate Luminometer (Berthold). Typically, 50 μL of luciferase assay reagent were added to 4.5 μLof in vitro translation mixture. The luciferase light intensity was measured for 10 s after a delay time of 2.05 s.
Competition experiments were performed in the same conditions at 10 μM ligand concentration and in the presence of (i) a 26-mer self-complementary double-stranded DNA (5′-CAATCGGATCGAATTCGATCCGATTG-3′), (ii) an hairpin RNA (5′-CUACAGUACAGAUCUGUACUGUAG-3′), or (iii) the NRQ NRAS RNA G-quadruplex (5′- GGGAGGGGCGGGUCUGGG-3′).
1H NMR experiments
All NMR data were recorded at 298 K using a 700 MHz Bruker Advance NMR spectrometer, equipped with a TXI cryogenic probe. The water suppression was achieved by using the jump-and-return scheme. The NMR titration experiments were carried out by recording a series of 1D proton NMR spectra of the RNA G-quadruplex (NRQ, 5′-GGGAGGGGCGGGUCUGGG-3′) in the presence of aliquots of a stock solution of RR110. The RNA concentration of the sample was 200 μM in 7% D2O (v/v), buffered with 10 mM potassium phosphate at pH 7.0. The stock solution of molecule RR110 was prepared with d6-DMSO (99.9%) at a concentration of 42 mM. Aliquots of the stocks solution of RR110 were added to the NMR sample in steps to reach molar ratios of 0, 0.25, 0.5, 1, 2, 3, 4 and 5. The final NMR sample contained 2.5% of d6-DMSO. 1H NMR spectra of the RNA G-quadruplex in the presence of 2.5% DMSO confirmed that DMSO has little effect to the structure and dynamics of the folded G-quadruplex (Fig. S3 in the ESI†).
For the hydrogen-deuterium exchange (HDX) experiments, RNA NMR samples (200 μM, 200 μL) were flash frozen in liquid nitrogen and lyophilized. Equal volumes (200 μL) of D2O (99.9%) were then added to resuspend the NMR samples immediately before data acquisition. Three samples were prepared containing either no additional material, or 5 μL of the stock solution of RR110, or 5 μL of d6-DMSO (99.9%). A series of 1D proton spectra were recorded to follow the kinetics of HDX over 24 h with a dead-time of ca. 4 min. For each 1D spectrum, 256 transients were recorded and the experimental time, i.e., time resolution, is 6 min. The resulting spectra were processed and baseline corrected prior to analysis of the HDX rates. The HDX kinetics of observed resonances can be generally grouped into slow, medium and fast phases, as visualized by overlaying the 1D proton spectra at different time points (Fig. S3 in the ESI†). To obtain apparent HDX rates of the imino protons of the NRQ (Fig. S4 and Table S1 in the ESI†), the overall imino proton signals were integrated (over the range of δ1H 10.3–11.5 ppm) and normalized as a function of the exchange time and fitted to a sum of three exponentially decaying functions: I(t) = Asexp(−kst) + Amexp(−kmt) + Afexp(−kft), where As, Am and Af are the initial amplitudes of the slow, medium and fast phases, respectively, and ks, km and kf are the corresponding rate constants.
mRNA stability experiments
Translation experiments in the absence or presence (10 μM) of the small molecule were performed as described above in 80 μL final volume reaction mixtures with 32P-UTP in vitro transcribed mRNAs. Aliquots (20 μL) of the translation mixtures were taken out after 0, 20, 40 and 60 min, quickly frozen in dry-ice and stored at −20 °C, before being subjected to Trizol® extraction (500 μL) and isopropanol precipitation (90 min at −20 °C). Samples were run at 70 V for 40–45 min on 2% agarose gels. Gels were dried under vacuum at 60 °C and quantified on an Amersham Biosciences Typhoon Trio with a Amersham imaging screen.
Acknowledgements
We thank the BBSRC for project funding, Cancer Research UK for program funding, and the Cambridge Commonwealth Trust and Trinity College, Cambridge for studentship funding (S.K.). S-T.D.H. is a recipient of a Netherlands Ramsay Memorial Fellowship and a Human Frontier Long-term Fellowship (LT0798/2005). We thank the staff and the use of the Biomolecular NMR Facility, Department of Chemistry, University of Cambridge.
Footnotes
Electronic supplementary information (ESI) available: A general scheme for the synthesis of RR110 and a complete description of its synthesis and characterization. Additional translation and NMR data. See DOI: 10.1039/c002418j
Notes and references
- 1.Crooke ST. Annu. Rev. Med. 2004;55:61–95. doi: 10.1146/annurev.med.55.091902.104408. [DOI] [PubMed] [Google Scholar]
- 2.Zamore PD. Nat. Struct. Biol. 2001;8:746–750. doi: 10.1038/nsb0901-746. [DOI] [PubMed] [Google Scholar]
- 3.van der Velden AW, Thomas AA. Int. J. Biochem. Cell Biol. 1999;31:87–106. doi: 10.1016/s1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]; Pickering BM, Willis AE. Semin. Cell Dev. Biol. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]; Chatterjeeand S, Pal JK. Biol. Cell. 2009;101:251–262. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- 4.Blount KF, Breaker RR. Nat. Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]; Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Nat. Chem. Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 5.Werstuckand G, Green MR. Science. 1998;282:296–298. [Google Scholar]
- 6.Harvey I, Garneauand P, Pelletier J. RNA. 2002;8:452–463. doi: 10.1017/s135583820202633x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neidle S, Balasubramanian S. Quadruplex Nucleic Acids. RSC Biomolecular Sciences; Cambridge, UK: [Google Scholar]
- 8.Patel DJ, Phan AT, Kuryavyi V. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monchaud D, Teulade-Fichou M-P. Org. Biomol. Chem. 2008;6:627–636. doi: 10.1039/b714772b. M.-P. [DOI] [PubMed] [Google Scholar]
- 10.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schouten JA, Ladame S, Mason SJ, Cooper MA, Balasubramanian S. J. Am. Chem. Soc. 2003;125:5594–5595. doi: 10.1021/ja029356w. [DOI] [PubMed] [Google Scholar]; Jantos K, Rodriguez R, Ladame S, Shirude PS, Balasubramanian S. J. Am. Chem. Soc. 2006;128:13662–13663. doi: 10.1021/ja064713e. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bejugam M, Sewitz S, Shirude PS, Rodriguez R, Shahid R, Balasubramanian S. J. Am. Chem. Soc. 2007;129:12926–12927. doi: 10.1021/ja075881p. [DOI] [PMC free article] [PubMed] [Google Scholar]; Waller ZA, Shirude PS, Rodriguez R, Balasubramanian S. Chem. Commun. 2008:1467–1469. doi: 10.1039/b718854d. [DOI] [PMC free article] [PubMed] [Google Scholar]; Dash J, Shirude PS, Balasubramanian S. Chem. Commun. 2008:3055–3057. doi: 10.1039/b806042h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez R, Muller S, Yeoman JA, Trentesaux C, Riou J-F, Balasubramanian S. J. Am. Chem. Soc. 2008;130:15758–15759. doi: 10.1021/ja805615w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster MP, McElroy CA, Amero CD. Biochemistry. 2007;46:331–340. doi: 10.1021/bi0621314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumari S, Bugaut A, Balasubramanian S. Biochemistry. 2008;47:12664–12669. doi: 10.1021/bi8010797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris MJ, Basu S. Biochemistry. 2009;48:5313–5319. doi: 10.1021/bi900498z. S. [DOI] [PubMed] [Google Scholar]
- 19.Balkwill GD, Derecka K, Garner TP, Hodgman C, Flint APF, Searle MA. Biochemistry. 2009;48:11487–11495. doi: 10.1021/bi901420k. [DOI] [PubMed] [Google Scholar]
- 20.Halder K, Wieland M, Hartig JS. Nucleic Acids Res. 2009;37:6811–6817. doi: 10.1093/nar/gkp696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wieland M, Hartig JS. Chem. Biol. 2007;14:757–763. doi: 10.1016/j.chembiol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Thomas JR, Hergenrother PJ. Chem. Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. P. J. [DOI] [PubMed] [Google Scholar]
- 23.Kozak M. Proc. Natl. Acad. Sci. U. S. A. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kozak M. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]; Babendure JR, Babendure JL, Ding JH, Tsien RY. RNA. 2006;12:851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]


