Abstract
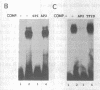
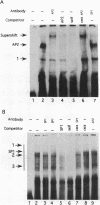
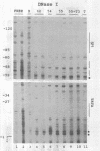
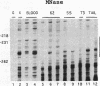
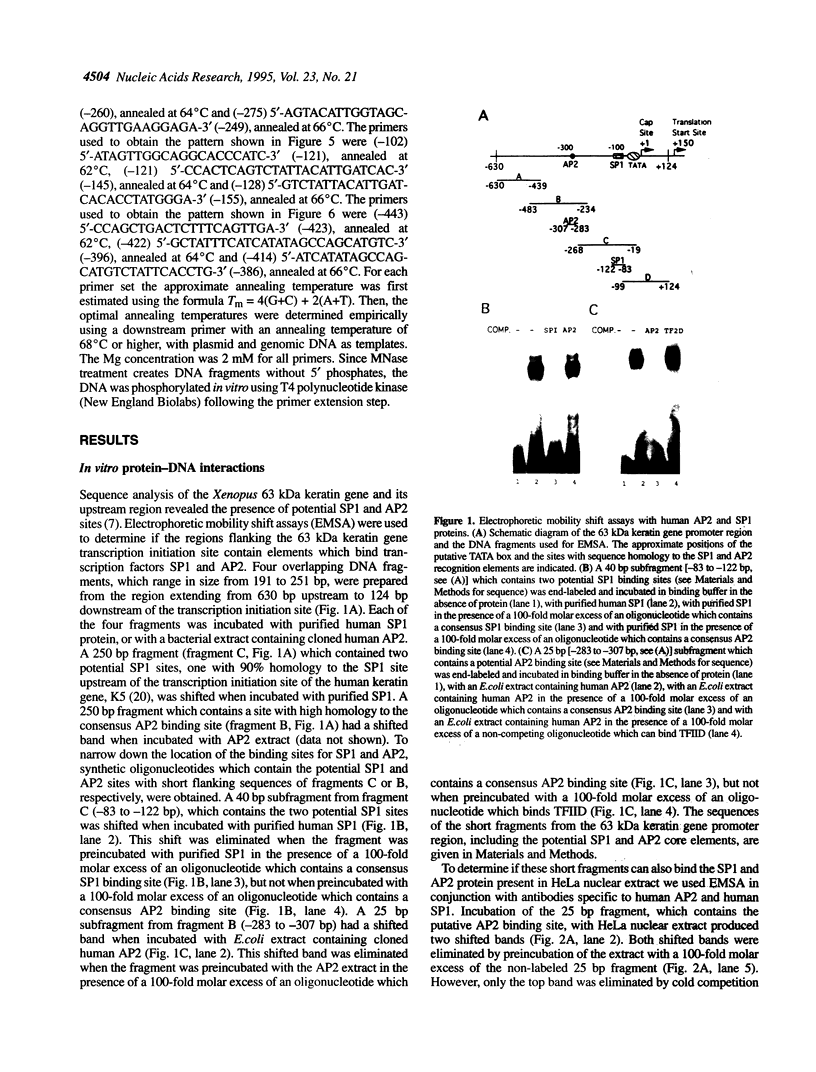
The Xenopus 63 kDa keratin gene is developmentally regulated and is expressed only in the epidermis. Full activation of the 63 kDa keratin gene requires two regulatory steps, the first independent and the second dependent on the thyroid hormone triiodothyronine (T3). Sequence analysis of a genomic clone of the 63 kDa keratin gene identified potential AP2 and SP1 binding sites upstream of the transcription initiation site. Electrophoretic mobility shift assays using purified or enriched proteins, as well as HeLa nuclear extract in conjunction with AP2- and SP1-specific antibodies, have been used to demonstrate that human AP2 and SP1 bind elements upstream of the transcription initiation site. In vivo footprinting with ligation mediated PCR revealed several footprints, within 350 bp upstream of the transcription initiation site, including those at the AP2 and SP1 sites, that are unique to epidermal cells which express the keratin gene. These footprints were absent in blood cells and XL177 cells which do not express the gene. Comparison of footprints between cells which express the 63 kDa keratin gene at low or high levels showed that the same binding sites are occupied, indicating that these sites are required for basal as well as T3-induced expression of the 63 kDa keratin gene.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brunvand M. W., Krumm A., Groudine M. In vivo footprinting of the human IL-2 gene reveals a nuclear factor bound to the transcription start site in T cells. Nucleic Acids Res. 1993 Oct 11;21(20):4824–4829. doi: 10.1093/nar/21.20.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne C., Tainsky M., Fuchs E. Programming gene expression in developing epidermis. Development. 1994 Sep;120(9):2369–2383. doi: 10.1242/dev.120.9.2369. [DOI] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Ellison T. R., Mathisen P. M., Miller L. Developmental changes in keratin patterns during epidermal maturation. Dev Biol. 1985 Dec;112(2):329–337. doi: 10.1016/0012-1606(85)90403-8. [DOI] [PubMed] [Google Scholar]
- French R. P., Warshawsky D., Tybor L., Mylniczenko N. D., Miller L. Upregulation of AP-2 in the skin of Xenopus laevis during thyroid hormone-induced metamorphosis. Dev Genet. 1994;15(4):356–365. doi: 10.1002/dvg.1020150407. [DOI] [PubMed] [Google Scholar]
- Hogan M. E., Roberson M. W., Austin R. H. DNA flexibility variation may dominate DNase I cleavage. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9273–9277. doi: 10.1073/pnas.86.23.9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. R., Benyajati C. In vivo stage- and tissue-specific DNA-protein interactions at the D. melanogaster alcohol dehydrogenase distal promoter and adult enhancer. Nucleic Acids Res. 1992 Oct 25;20(20):5413–5422. doi: 10.1093/nar/20.20.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. D., Idler W. W., Zhou X. M., Roop D. R., Steinert P. M. Structure of a gene for the human epidermal 67-kDa keratin. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1896–1900. doi: 10.1073/pnas.82.7.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leask A., Byrne C., Fuchs E. Transcription factor AP2 and its role in epidermal-specific gene expression. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):7948–7952. doi: 10.1073/pnas.88.18.7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leask A., Rosenberg M., Vassar R., Fuchs E. Regulation of a human epidermal keratin gene: sequences and nuclear factors involved in keratinocyte-specific transcription. Genes Dev. 1990 Nov;4(11):1985–1998. doi: 10.1101/gad.4.11.1985. [DOI] [PubMed] [Google Scholar]
- Mathisen P. M., Miller L. Thyroid hormone induces constitutive keratin gene expression during Xenopus laevis development. Mol Cell Biol. 1989 May;9(5):1823–1831. doi: 10.1128/mcb.9.5.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathisen P. M., Miller L. Thyroid hormone induction of keratin genes: a two-step activation of gene expression during development. Genes Dev. 1987 Dec;1(10):1107–1117. doi: 10.1101/gad.1.10.1107. [DOI] [PubMed] [Google Scholar]
- McPherson C. E., Shim E. Y., Friedman D. S., Zaret K. S. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell. 1993 Oct 22;75(2):387–398. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- Mueller P. R., Salser S. J., Wold B. Constitutive and metal-inducible protein:DNA interactions at the mouse metallothionein I promoter examined by in vivo and in vitro footprinting. Genes Dev. 1988 Apr;2(4):412–427. doi: 10.1101/gad.2.4.412. [DOI] [PubMed] [Google Scholar]
- Mueller P. R., Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989 Nov 10;246(4931):780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- Nishikawa A., Shimizu-Nishikawa K., Miller L. Isolation, characterization, and in vitro culture of larval and adult epidermal cells of the frog Xenopus laevis. In Vitro Cell Dev Biol. 1990 Dec;26(12):1128–1134. doi: 10.1007/BF02623689. [DOI] [PubMed] [Google Scholar]
- Nishikawa A., Shimizu-Nishikawa K., Miller L. Spatial, temporal, and hormonal regulation of epidermal keratin expression during development of the frog, Xenopus laevis. Dev Biol. 1992 May;151(1):145–153. doi: 10.1016/0012-1606(92)90222-3. [DOI] [PubMed] [Google Scholar]
- Ohtsuki M., Flanagan S., Freedberg I. M., Blumenberg M. A cluster of five nuclear proteins regulates keratin gene transcription. Gene Expr. 1993;3(2):201–213. [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Mueller P. R., Wold B., Riggs A. D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989 Nov 10;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Ranjan M., Wong J., Shi Y. B. Transcriptional repression of Xenopus TR beta gene is mediated by a thyroid hormone response element located near the start site. J Biol Chem. 1994 Oct 7;269(40):24699–24705. [PubMed] [Google Scholar]
- Roop D. R., Huitfeldt H., Kilkenny A., Yuspa S. H. Regulated expression of differentiation-associated keratins in cultured epidermal cells detected by monospecific antibodies to unique peptides of mouse epidermal keratins. Differentiation. 1987;35(2):143–150. doi: 10.1111/j.1432-0436.1987.tb00162.x. [DOI] [PubMed] [Google Scholar]
- Shimizu-Nishikawa K., Miller L. Calcium regulation of epidermal cell differentiation in the frog Xenopus laevis. J Exp Zool. 1991 Nov;260(2):165–169. doi: 10.1002/jez.1402600205. [DOI] [PubMed] [Google Scholar]
- Shimizu-Nishikawa K., Miller L. Hormonal regulation of adult type keratin gene expression in larval epidermal cells of the frog Xenopus laevis. Differentiation. 1992 Mar;49(2):77–83. doi: 10.1111/j.1432-0436.1992.tb00771.x. [DOI] [PubMed] [Google Scholar]
- Snape A. M., Winning R. S., Sargent T. D. Transcription factor AP-2 is tissue-specific in Xenopus and is closely related or identical to keratin transcription factor 1 (KTF-1). Development. 1991 Sep;113(1):283–293. doi: 10.1242/dev.113.1.283. [DOI] [PubMed] [Google Scholar]
- Suck D., Oefner C. Structure of DNase I at 2.0 A resolution suggests a mechanism for binding to and cutting DNA. Nature. 1986 Jun 5;321(6070):620–625. doi: 10.1038/321620a0. [DOI] [PubMed] [Google Scholar]
- Törmänen V. T., Swiderski P. M., Kaplan B. E., Pfeifer G. P., Riggs A. D. Extension product capture improves genomic sequencing and DNase I footprinting by ligation-mediated PCR. Nucleic Acids Res. 1992 Oct 25;20(20):5487–5488. doi: 10.1093/nar/20.20.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshawsky D., Miller L. A rapid genomic walking technique based on ligation-mediated PCR and magnetic separation technology. Biotechniques. 1994 May;16(5):792-4, 796, 798. [PubMed] [Google Scholar]
- Williams T., Tjian R. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev. 1991 Apr;5(4):670–682. doi: 10.1101/gad.5.4.670. [DOI] [PubMed] [Google Scholar]
- Xu Q., Tata J. R. Characterization and developmental expression of Xenopus C/EBP gene. Mech Dev. 1992 Jul;38(1):69–81. doi: 10.1016/0925-4773(92)90039-m. [DOI] [PubMed] [Google Scholar]
- Yuspa S. H., Kilkenny A. E., Steinert P. M., Roop D. R. Expression of murine epidermal differentiation markers is tightly regulated by restricted extracellular calcium concentrations in vitro. J Cell Biol. 1989 Sep;109(3):1207–1217. doi: 10.1083/jcb.109.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Gralla J. D. In situ nucleoprotein structure involving origin-proximal SV40 DNA control elements. Nucleic Acids Res. 1990 Apr 11;18(7):1797–1803. doi: 10.1093/nar/18.7.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vliet P. C., Verrijzer C. P. Bending of DNA by transcription factors. Bioessays. 1993 Jan;15(1):25–32. doi: 10.1002/bies.950150105. [DOI] [PubMed] [Google Scholar]