Abstract
Many types of cancer cells require a supply of fatty acids (FA) for growth and survival, and interrupting de novo FA synthesis in model systems causes potent anticancer effects. We hypothesized that, in addition to synthesis, cancer cells may obtain pre-formed, diet-derived fatty acids by uptake from the bloodstream. This would require hydrolytic release of FA from triglyceride in circulating lipoprotein particles by the secreted enzyme lipoprotein lipase (LPL), and the expression of CD36, the channel for cellular FA uptake. We find that selected breast cancer and sarcoma cells express and secrete active LPL, and all express CD36. We further demonstrate that LPL, in the presence of triglyceride-rich lipoproteins, accelerates the growth of these cells. Providing LPL to prostate cancer cells, which express low levels of the enzyme, did not augment growth, but did prevent the cytotoxic effect of FA synthesis inhibition. Moreover, LPL knockdown inhibited HeLa cell growth. In contrast to the cell lines, immunohistochemical analysis confirmed the presence of LPL and CD36 in the majority of breast, liposarcoma, and prostate tumor tissues examined (n = 181). These findings suggest that, in addition to de novo lipogenesis, cancer cells can use LPL and CD36 to acquire FA from the circulation by lipolysis, and this can fuel their growth. Interfering with dietary fat intake, lipolysis, and/or fatty acid uptake will be necessary to target the requirement of cancer cells for FA.
Keywords: fatty acids, breast cancer, liposarcoma, prostate cancer
Introduction
Many tumors, including those arising in breast, colon, ovary, and prostate, exhibit a “lipogenic” phenotype. This features brisk rates of saturated long-chain FA synthesis driven by enhanced expression of genes coding for the three enzymes required to produce palmitic acid from cytosolic citrate (ATP citrate-lyase, acetyl CoA-carboxylase, and fatty acid synthase (FASN)). Importantly, lipogenic tumor cell growth is slowed in vitro and survival is reduced by FA synthesis inhibitors, whereas nontransformed cells are unaffected (reviewed in (1, 2)). Moreover, blocking de novo lipogenesis with FASN inhibitors in vivo exerts potent antitumor effects in rodent models of breast (3) and prostate (4) cancer. These observations, coupled with the low rates of fatty acid synthesis in most normal human tissues (5), have spurred efforts to develop anticancer therapies based on inhibiting lipogenic enzyme activities or silencing the corresponding genes.
Attempts to exploit the metabolic requirements of lipogenic cancers have thus far focused solely on disrupting de novo fatty acid synthesis. Cytotoxicity following inhibition of lipid synthesis, however, may be obviated by the provision of exogenous fatty acids (6–8). This observation, and the improved outcome of breast cancer patients ingesting a low fat diet (9), led us to hypothesize that triglyceride in circulating lipoprotein particles could provide an additional, exogenous source of fatty acids for tumors. This would require triglyceride-rich chylomicrons or very low density lipoproteins (VLDL) as substrate, extracellular lipoprotein lipase (LPL) for hydrolysis, and fatty acid translocase (CD36) for cellular uptake of the free fatty acids (reviewed in (10)). As LPL is a secreted enzyme that is bound to the luminal surface of capillary endothelial cells, it could potentially be supplied by tumor cells or by nonmalignant cells in the tumor microenvironment.
Materials and Methods
cDNA microarray analysis
Production of the expression dataset has been previously described in detail, as have culture conditions for cell lines ((11), http://cancer.lbl.gov/breastcancer/data.php). RNA from 45 human breast cancer cell lines (ICBP45) grown at subconfluence was harvested, reverse transcribed, and hybridized to Affymetrix U133A gene chips. Resulting Affymetrix image files were normalized (RMA, (12)). Unsupervised average linkage cluster analysis of log2 signal intensities was performed using approximately 14,000 probeset IDs of highest variance, using the Cluster Software package, and the resulting dendrogram image produced with Treeview ((13), http://rana.lbl.gov/eisen/?page_id=7). Probeset IDs identifying CD36, LPL and FASN were identified, median centered, normalized and a heat map produced indicating the relative hybridization intensity for each sample.
RT-PCR
RNA was isolated using the RNeasy Mini Kit (Quiagen, Valencia, CA). One μg RNA was reverse transcribed using random hexamer primers with M-MULV reverse transcription (New England Biolabs, Ipswich, MA). PCR was as described (14). Primers employed are described in Supplemental Table SI. Different primers were used for real time RT-PCR.
Quantitative real time RT-PCR
RNA was prepared using the PureLinkTM Total RNA purification system (Invitrogen, Paisley, UK). The purity and concentration of RNA were assessed using a NanoDrop DM-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA was converted to cDNA using Superscript II RT and random hexamer primers, according to the manufacturer’s protocol (Invitrogen). Primer sequences for LPL were 5'-TATCCGCGTGATTGCAGAGA-3' (forward) and 5'-GCCTTACTTGGATTTTCTTCATTCA-3' (reverse). Sybr green was used for detection and 18S rRNA was used as an internal control. Primer sequences for 18S were 5'-CGCCGCTAGAGGTGAAATTC-3' (forward) and 5'-TTGGCAAATGCTTTCGCTC-3' (reverse). PCR was in the 7500 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA). The program used included 2 min at 50°C, 1 min at 95°C and 40 cycles of 3 sec at 95°C and 30 sec at 60°C. The average of the Ct-values for each triplicate reaction was expressed relative to the amount of 18S rRNA in the sample.
Tissue culture
LiSa-2 liposarcoma cells were from Martin Wabitsch, University of Ulm, Germany, and we confirmed their identity by the ability to produce lipid droplets that stained with oil red-O upon confluence. All other lines were from ATCC except VCaP, which was from ECACC, and these lines were acquired recently and were of low passage number (< 10). Cells were grown in DMEM:F12 supplemented with 10% fetal calf serum (Atlanta Biologicals, Lawrenceville, GA), 1% penicillin/streptomycin, and 2 mM L-glutamine, in 5% CO2 at 370 C. Cell growth was monitored in an MTT assay (15). Lipoprotein deficient fetal calf serum was prepared by the method of Goldstein and coworkers (16).
Knockdown of LPL
HeLa cells were transfected with 10 nmol/L siRNA targeting LPL ( siRNA A : 5’-GGUAGAUAUUGGAGAACUA ; siRNA B : 5 ’-GGAUGGAGGAGGAGUUUAA; Dharmacon) with the use of Lipofectamine RNAiMAX (Invitrogen) . A siRNA targeting luciferase (5’-CGUACGCGGAAUACUUCGA) was used as control. RNA was harvested 48 h later, and cell viability was assessed 96 h after transfection.
Fluorescent labeling of VLDL particles
Dialyzed VLDL (0.5 mg; Kalen Biomedical) were incubated at 37 C for 15 h with 25μl diI D282 (Invitrogen; 3mg/ml in DMSO). Free dye was removed by dialysis against PBS. Prior to observation by confocal microscopy, 25 μl labeled VLDLs were added to slide chambers (Tissue Tek #177402) containing 106 cells grown for 3 days in delipidated DMEM. Excitation was at 549 nm, and fluorescence was detected at 565 nm.
Production of anti-human LPL antibodies
Mice were immunized with a peptide (Sigma) representing human LPL residues 21–36 (CASRGGVAAAQRRDFID) coupled to keyhole limpet hemocyanin. After fusion of splenocytes to mouse multiple myeloma cells, media from candidate clones were screened for reactivity to bacterially expressed LPL in an enzyme linked immunosorbant assay. Positive clones were further screened by western blot of skeletal muscle from transgenic mice expressing a human muscle-specific LPL transgene (MCK-LPL, kindly supplied by Ira Goldberg, Columbia School of Medicine, New York, NY) and against human breast milk. Mouse tissues were homogenized in RIPA buffer containing 10 μg/ml PMSF. Samples were centrifuged at 10,000 x g for 10 min x 2. Protein content was determined using the BCA assay (Pierce). Samples were boiled in 2X sample buffer and fractionated through 15% acrylamide. Following transfer to PVDF membranes (Immobilon–FL, Millipore), blocking was with SuperBlock (Pierce). Incubation with 1:200 dilution of the primary antibody in TBS-Tween was overnight at 4° C, followed by 2 TBS washes. Recombinant protein A/G conjugated with horseradish peroxidase (Pierce) was applied for detection at 1:5000 in TBS-Tween for 1 hour at RT. After four TBS washes, membranes were developed with NBT-BCIP (Pierce).
Bacterial expression of human LPL
The 2.3 kb EcoRI-HindIII fragment encoding LPL was excised from pCMV-SPORT6-LPL (Open Biosystems) and inserted into the pProEx-HTa His-tag vector. Most of the fusion protein could not be solubilized, but that which could be recovered demonstrated reactivity with the anti-His4 antibody (Invitrogen). For immunodot assays 10 ng protein from cleared lysates of E. coli DH5α transformed with empty or LPL plasmid were spotted onto PVDF membranes, blocked, and incubated with antibody as described above.
Affinity isolation of LPL
Human milk and conditioned cell culture media were fractionated over heparin sepharose (Sigma) using a procedure modified from Hata and coworkers (17).
LPL activity
We employed the radiochemical assay of Nilsson-Ehle and coworkers (18) or a colorimetric assay based on determination of glycerol production (BioVision, Mountain View, CA). We used a protocol based on that of Cruz and coworkers (19) for determination of heparin-releasable LPL. Briefly, 5 × 106 cells from 75 cm2 flasks containing 5 × 106 cells were cultured for 72 h, and scraped pellets were washed x 3 in PBS with or without 100 U/ml heparin. Media and lysed cell pellets were assayed in triplicate for residual LPL activity.
Immunohistochemistry
This was performed as previously described (20). Anti-LPL monoclonal antibody clone 43 was used at a dilution of 1:10, with Citra Plus antigen retrieval (Biogenix, San Ramon, CA). CD36 was assessed using an affinity-purified rabbit polyclonal antibody (Thermo Scientific) according to the supplier’s protocol. The Internal Review Board (IRB)-approved the use of breast cancer tissue and the tissue microarray (TMA) containing 147 primary breast cancers from post-menopausal women, diagnosed between 2000 and 2007 at Dartmouth-Hitchcock Medical Center, Lebanon, NH. Each case was represented by one tissue core 1.0 mm in diameter. The liposarcoma TMA, also IRB approved, contained 26 liposarcomas diagnosed between 1995 and 2008 at Dartmouth-Hitchcock Medical Center. Each case was represented by 2–4 1.0 mm tissue cores. Prostate cancer specimens were acquired at the Katholieke Universiteit Leuven, Belgium, with IRB approval.
Results
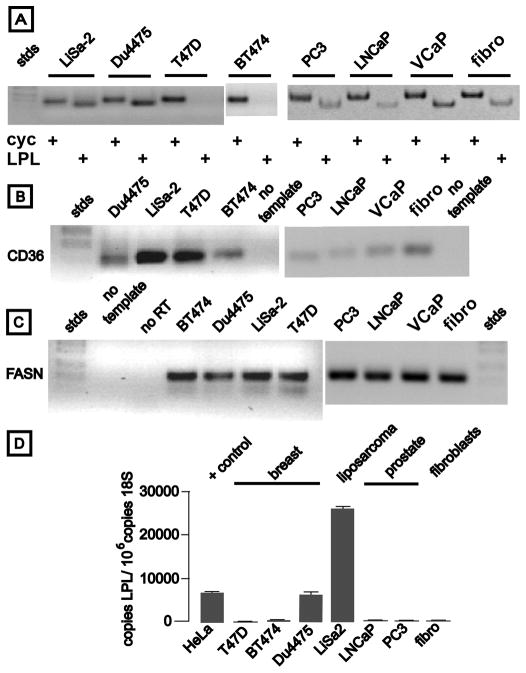
We used a cDNA microarray to screen 45 breast cancer-derived cell lines from the dataset of Neve and coworkers (11) for LPL gene expression, and for FASN mRNA as a marker for de novo fatty acid synthesis. We analyzed cell lines because breast tumor samples may contain adipocytes, which express high levels of LPL and FASN. We also sorted the breast cancer lines by their global gene expression signatures (21). These signatures include the luminal type (estrogen receptor (ER) +), the basal, or “triple negative” type that lacks receptors for estrogen, progestin, and trastuzumab (22), and the type with Her2/neu amplification. Only 6 breast cancer cell lines (HCC-2157, -1008, -1599, Du4475, SUM149, and SUM190) expressed high levels of LPL mRNA, and each of these exhibited the aggressive basal gene expression signature (Supplementary Fig. S1). Expression of LPL mRNA by selected cell lines was verified by RT-PCR (Fig. 1A), as was expression of CD36 mRNA (Fig. 1B). LiSa-2 liposarcoma cells, which we previously demonstrated to exhibit the lipogenic phenotype (23), also expressed LPL and CD36, as expected for a tumor cell derived from an adipocytic lineage. All of the cell types expressed substantial FASN mRNA (Fig. 1C), and in the breast cancer cell lines this did not vary among the gene expression signatures (Supplementary Fig. S1). Quantitative real time RT-PCR of representative lines confirmed that LiSa-2 liposarcoma and triple negative Du4475 breast cancer cells expressed the highest levels of LPL mRNA (Fig. 1D). In contrast, prostate cancer cells, which are highly lipogenic (24), expressed relatively low levels of LPL mRNA, and ER+ T47D and BT474 breast cancer cells expressed essentially none.
Figure 1.
LPL, CD36, and FASN gene expression in cancer cells. Ethidium-stained gel electrophoresis of RT-PCR products is shown in Panels A–C. Cell lines analyzed are listed above each lane. Stds indicates electrophoretic size standards; LiSa-2 is a liposarcoma line; Du4475 are breast cancer cells lacking receptors for sex steroids and trastuzumab; T47D are breast cancer cells with receptors for estrogen and progesterone, but not trastuzumab; BT474 are breast cancer cells with receptors for sex steroids and trastuzumab. PC3, LNCaP, and VCaP are prostate cancer lines; Fibro denotes human fibroblasts. Panel A: Primers corresponded to cyclophilin (cyclo) or LPL mRNAs. Panel B: Primers corresponded to the fatty acid translocase CD36. Panel C: Primers detected FASN mRNA. Panel D depicts real time RT-PCR quantitation of LPL mRNA (normalized to 18S rRNA, mean ± SEM, n = 3 wells/cell line). HeLa adenocarcinoma cells are included as a positive control, as we previously reported expression of LPL mRNA by this cell line (25).
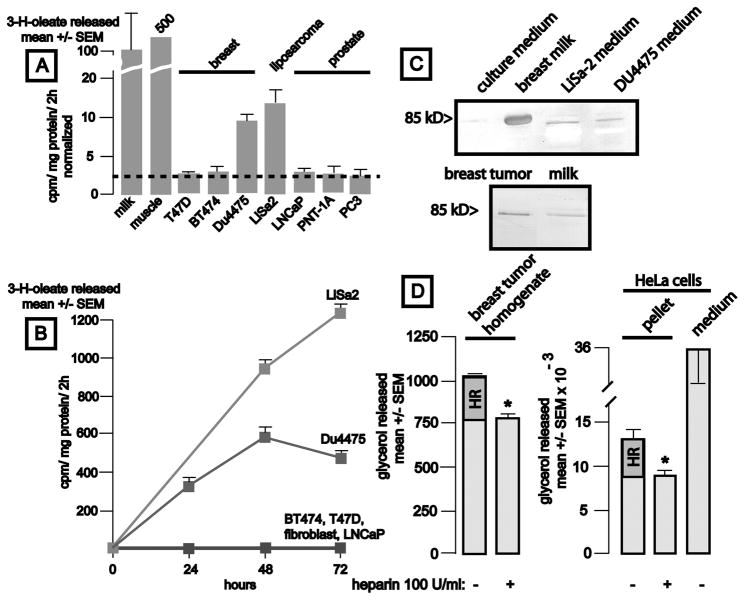
We examined conditioned tissue culture media for LPL enzyme activity, and it paralleled the levels of LPL mRNA (Fig. 2A). LPL activity accumulated over time in culture media of LiSa-2 liposarcoma and Du4475 breast cancer cells (Fig. 2B). In contrast, ER+ T47D, ER+ Her2/neu+ BT474 breast cancer cells, and fibroblasts did not secrete detectable lipase activity. Prostate cancer cells produced low levels of the enzyme. LPL activities in breast milk and murine striated muscle were substantially greater than those observed in any of the conditioned (72 h) media.
Figure 2.
Production of lipoprotein lipase activity by breast cancer, liposarcoma, and prostate cancer cells and in a breast cancer tissue sample. Panel A: Lipase activity is shown (mean ± SEM, 4 samples/group, corrected for cellular protein content and normalized to the value observed in milk (9 × 103 cpm/2h). Human breast milk (50 μl), mouse gastrocnemius muscle (50 mcg protein, 45 × 103 cpm/2h), or tissue culture media conditioned by the indicated cell lines x 3 d were assessed for lipase activity (mean ± SEM, 4 samples/group, corrected for cellular protein content and activity observed in unconditioned media). The dotted line denotes the LPL activity found in unconditioned culture medium. Panel B: Time course of accumulation of lipase activity in conditioned culture media. Media (50 μl) were removed from cultures at the indicated intervals (mean cpm/mg protein ± SEM, n = 4 wells/timepoint). Panel C, upper: Identification of LPL in conditioned cell culture media. LPL was heparin-sepharose affinity purified from 10 ml fresh culture medium, 1.0 ml human breast milk, or 10 ml culture media conditioned (72 h) by LiSa-2 liposarcoma or DU4475 “triple negative” breast cancer cells, eluted with 0.6–0.8 M NaCl, and analyzed by western blot using anti-human LPL clone 43 (1:200). The band from milk was verified to contain LPL by mass spectrometry. Panel C, lower: Western analysis of a breast tumor homogenate (50 mcg protein without affinity purification) and breast milk (10 μl) for LPL. A band of the appropriate size is apparent in the tumor sample. Panel D: Estimation of the heparin-releasable LPL pool in breast cancer tissue and HeLa cells. Panel D left: tumor associated LPL activity is significantly reduced by heparin treatment (p = 0.0001). Panel D: right: Heparin reduced LPL activity residing in HeLa cell pellets by 29% (p < 0.04).
We found that available antibodies were not sufficiently specific to analyze LPL protein by immunohistochemistry. We therefore raised a mouse monoclonal antibody using a peptide representing residues 20–36 of the human enzyme as antigen. This antibody is highly specific (Supplemental Fig. S2), and permitted detection of heparin sepharose-purified LPL from tissue culture media conditioned by Du4475 breast cancer and LiSa-2 liposarcoma cells (Fig. 2C upper). The band recognized by this antibody in western analysis of milk was verified to represent LPL by mass spectrometry. We could not detect LPL protein in media from ER+ breast or prostate cancer cells. Western analysis of a clinical breast tumor homogenate (50 mcg protein) without affinity purification revealed a single band exhibiting the same migration as that observed in milk (Fig. 2C lower).
It appeared possible that expression of heparanase could inactivate LPL, and thus could vitiate the metabolic relevance of LPL expression by tumors. We assessed expression of the heparanase gene (HPSE) using cDNA microarray data from 45 human breast cancer cell lines. This showed that the cells generally express very low levels of heparanase mRNA, as was the general case for LPL mRNA. We were intrigued to note that the subgroup of “triple negative” cell lines exhibiting substantial LPL expression also expressed the lowest levels of heparanase mRNA. Indeed, linear regression of the relationship between LPL and heparanase mRNAs in lines with the basal A signature revealed a statistically significant inverse correlation (p = 1.27 × 10−5,, R2 = 0.38). Thus, the coupling of high LPL with low heparanase expression appears to provide an advantage to the subset of cells that produce substantial LPL. Our examination of total and heparin-releasable LPL activity in a freshly-prepared breast tumor homogenate also reflects on this question, as heparin-releasable activity was readily detectable, arguing against depletion of a cell surface-bound LPL pool in breast tumors (see below).
We performed two experiments to determine whether cancer-associated LPL is bound to tumor cells by noncovalent interactions with cell surface heparan sulfate proteoglycans, using a protocol based on that of Cruz and coworkers (19). First, we homogenized freshly-resected invasive breast cancer tissue shown to contain LPL immunoreactivity (Fig. 2C lower), and extracted equal aliquots with buffer containing heparin or not. LPL activity in the control sample was 1032 ± 8 without heparin, 768 ± 4 with heparin treatment (mean ± SE, nanomoles glycerol produced/g tumor/h, measured in triplicate; p < 0.0001). This represented a heparin-releasable fraction of 26% of the total tumor-associated LPL activity (represented by the portion of the bar labeled HR, Fig. 2D left).
Second, we determined the heparin-releasable fraction of LPL in HeLa cells, and calculated turnover rates for cellular LPL pools (Fig. 2D right). Residual LPL activity in cell pellets was 13,260 ± 1,080 without, and 9,360 ± 820 with heparin exposure (units are nanomoles glycerol produced/flask/h, mean ± SEM, n= triplicate measurements/group; p < 0.04). We thus estimate that 29% of the HeLa cell-associated pool of LPL is heparin-releasable (indicated by HR on the graph), a fraction similar to that observed in the breast tumor sample. Measurement of LPL activity in culture media indicated that 36,000 ± 4,000 units of LPL activity were secreted/24 h. We therefore estimate that the total cellular LPL pool turns over 2.7 times/d, while the heparin-labile pool (3900 units/well) turns over, presumably by secretion, 9.2 times/d.
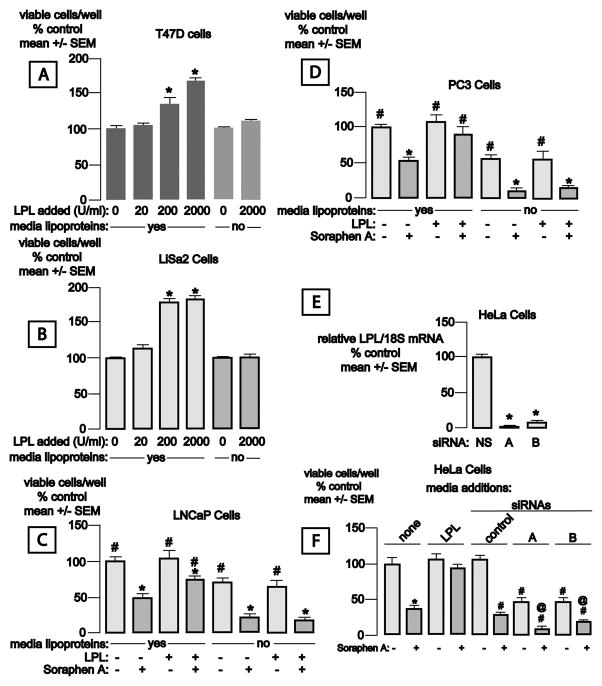
Our fetal calf serum (FCS) contained 660 mcg triglyceride/ml. LPL secreted by cells is removed when culture media are replaced, so the enzyme content in tissue culture never approaches that observed in tissues. We therefore assessed the functional significance of LPL by adding the enzyme to media containing 10% FCS and measuring cell accumulation. LPL activity under these culture conditions approximated that observed in mouse muscle. LPL enhanced the growth of T47D breast cancer cells, which do not express LPL, and of LiSa-2 liposarcoma cells, which do (Fig. 3A and B). This effect of LPL was greatly reduced in media containing FCS that was nearly depleted of trigyceride (20 mcg/ml).
Figure 3.
LPL stimulates tumor cell growth in the presence of lipoproteins. Panel A: T47D breast cancer cells were grown x 72 h in media containing complete or lipoprotein-depleted fetal calf serum (triglyceride content 660 and 20 μg/ml, respectively) plus the indicated concentrations of LPL. Media were replaced at 24 h intervals. Data in this and other panels are mean ± SEM, normalized to the control group (seeded in 24 well plates at 20k cells/well, n = 6 wells/group). *p< 0.05 compared to control. Panel B: LiSa-2 liposarcoma cells were grown x 72 h in media containing complete or lipoprotein-depleted fetal calf serum plus the indicated concentrations of LPL were replaced at 24 h intervals. *p< 0.05 compared to control. Panel C: LnCaP prostate cancer cells were treated with the indicated concentrations of LPL +/− 100 nM Soraphen A to inhibit lipid synthesis. Comparisons are within the lipoprotein plus and minus groups. *p< 0.05 compared to no LPL or Soraphen A, #p< 0.05 compared to no LPL, + Soraphen A. Panel D: PC3 prostate cancer cells were treated as in Panel C. *p< 0.05 compared to no LPL or Soraphen A, #p< 0.05 compared to no LPL, + Soraphen A. Panel E: Two LPL siRNAs (A, B), but not a nonspecific siRNA (NS) cause a substantial decline in LPL mRNA. Data are mean LPL mRNA signal normalized to 18S RNA± SEM, 4 wells/group. RNA was harvested 48 h after transfection. *p<0.05 compared to the nonspecific siRNA. Panel F: LPL siRNA impairs the growth of HeLa cells, and augments the antiproliferative effect of Soraphen A. Data are viable cells/well, mean ± SEM (n = 4 wells/group). Cell growth was assessed 96 h after siRNA transfection. *p< 0.05 compared to the no siRNA, no Soraphen A, no LPL group, #p< 0.05 compared to the control siRNA, no Soraphen group, @p< 0.05 compared to the respective siRNA groups (A, B) without Soraphen A.
LNCaP prostate cancer cell growth was not accelerated by LPL addition. The ability of these cells to use exogenous triglyceride-derived fatty acids to maintain growth was revealed, however, in the presence of Soraphen A, a potent inhibitor of the lipogenic enzyme acetyl CoA-carboxylase (7). The cells were rescued from Soraphen A-induced cytotoxicity by provision of LPL in the presence, but not in the absence, of lipoproteins (Fig. 3C). Experiments using PC3 prostate cancer cells yielded similar results (Fig. 3D).
In complementary studies we assessed the impact of siRNA-mediated knockdown of LPL mRNA on the growth of HeLa cells, which we previously reported to express the LPL gene (25), and its interaction with inhibition of lipogenesis by Soraphen A. Two different siRNAs each caused > 90% disappearance of LPL mRNA, whereas a nonspecific siRNA was without effect (Fig. 3E). Soraphen A caused a major inhibition of HeLa cell accumulation, and this effect was prevented by provision of LPL to the cultures (Fig. 3F). Transfection of LPL siRNA A or B, but not of the nonspecific siRNA, significantly inhibited HeLa cell growth, and the anticancer effects of the two LPL siRNAs were further enhanced by exposure to Soraphen A.
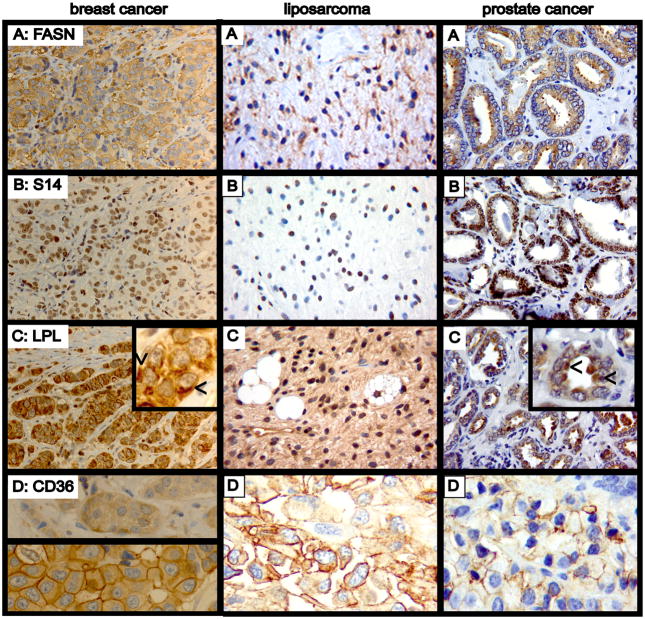
We employed immunohistochemistry to assess the relevance of our findings in cultured cells to human tumors. We assessed the expression of markers of de novo lipogenesis (FASN, THRSP (Spot 14, S14)), lipolysis (LPL), and exogenous FA uptake (CD36), in a panel of 147 breast, 24 liposarcoma, and 10 prostate tumor tissues (examples in Fig. 4). FASN was cytosolic, in agreement with previous studies. S14, which promotes expression of the FASN gene (26) (27), was primarily nuclear, as reported (20).
Figure 4.
Immunohistochemical analysis of markers of fatty acid metabolism in breast, liposarcoma, and prostate tumors. Slides from a representative invasive ductal carcinoma of the breast (left column), liposarcoma (middle column), and prostatic adenocarcinoma (right column) were immunostained for A: fatty acid synthase (FASN), B: THRSP (Spot 14, S14), C: lipoprotein lipase (LPL), or D: CD36. Original magnification was 40x. Detection was with peroxidase (brown pigment), and slides were counterstained with hematoxylin (blue pigment). FASN staining is cytosolic, and S14 is nuclear. LPL demonstrated an asymmetric, perinuclear distribution (arrows in insets) compatible with localization to the Golgi apparatus. Note that the well-visualized prostate tumor stroma does not express detectable LPL. CD36 exhibited two distinct patterns of subcellular localization in breast tumors. Only a cytosolic signal was seen in 29% of cases (row D, left, upper panel), whereas prominent cell surface staining was seen in 69% (lower panel), and only 2% were devoid of CD36 immunoreactivity. Staining was also seen in most liposarcoma and prostate cancers. This was primarily a cell surface pattern, and was not uniformly present across those tumors.
In contrast to our findings in breast cancer cell lines, LPL immunoreactivity was observed in all of the breast tumors examined, and, also in contrast to the cell lines, expression was not limited to triple negative tumors. Similarly, all liposarcoma and prostate tumors examined expressed readily detectable LPL by immunohistochemistry. Intracellular LPL demonstrated an asymmetric, perinuclear distribution suggestive of localization to the Golgi apparatus, as predicted for a glycosylated and secreted protein (Fig. 4C insets). As expected, extracellular LPL was found on the luminal surfaces of capillaries (Supplemental Fig. S2C left). We stained tonsil tissue as a negative control, based on previous work showing undetectable LPL mRNA in lymphoid cells (25). The lymphoid stroma indeed showed no staining except for scattered isolated monocytes, whereas the highly proliferative basal (stem cell) layer of the mucosal epithelium overlying the tonsil unexpectedly showed a strong signal (Supplemental Fig. S2C right).
The majority of tumors also stained for CD36 (Fig. 4D). Interestingly, two distinct staining patterns were observed in breast cancer tissue. Of 144 evaluable cores, 42 exhibited diffuse cytoplasmic staining without accentuation at the plasma membrane (Fig. 4D, left upper), whereas 100 also demonstrated a strong cell surface signal (lower panel). Only 2 breast cancer cases were devoid of staining. Chi square analysis demonstrated a statistically significant difference in the prevalence of the membranous staining pattern between the “triple negative” and ER+ breast cancers (42 vs. 76%, p < 0.02).
Of 25 liposarcoma cases, 21 stained for CD36, almost all in a mixed cytoplasmic plus plasma membrane pattern (Fig. 4D, middle), including all 9 cases of well-differentiated liposarcoma. Of 9 evaluable prostate cancers, 4 showed focally positive staining in a mixed cytoplasmic and plasma membrane pattern (Fig. 4D, right), while 5 cases scored negative for CD36.
Expression of LPL by breast cancer cells suggested the possibility that the cells could use the enzyme not only to hydrolyze extracellular triglyceride, but also for receptor-mediated endocytosis of triglyceride-rich lipoproteins. This process employs LPL as a bridge between the cell surface receptor syndecan-1 and the lipoprotein (28). RT-PCR revealed readily detectable syndecan-1 mRNA from DU4475 breast cancer cells, while LiSa-2 and T47D exhibited a faint signal (Supplementary Fig. S3A). We incubated fibroblasts and DU4475 cells with fluorescently-labeled VLDL particles, and assessed for cellular uptake using confocal microscopy. Abundant uptake was observed in fibroblasts (Fig. S3B), but not in DU4475 cells (Fig. S3C). Occasional fluorescence was detected on the cell surface (Fig. S3D), but never within the breast cancer cells.
Discussion
Our data demonstrate that cancer cells may use two different mechanisms to acquire fatty acids to fuel proliferation. Breast and liposarcoma tumors are equipped for both lipid synthesis and for LPL-mediated extracellular lipolysis followed by fatty acid uptake via CD36. Prostate cancer cells, which have a very high capacity for de novo lipogenesis (24), express very little LPL. The low LPL expression could be explained in part by the reported loss of heterozygosity at the LPL locus in 47% of prostate tumors, owing to the presence of a nearby tumor suppressor gene (29). These cells, however, can acquire sufficient exogenous fatty acids to maintain growth in the face of fatty acid synthesis inhibition when they are supplied with LPL and triglyceride-rich lipoprotein particles.
LPL expression has been shown to be a marker of poor prognosis in chronic B cell lymphocytic leukemia (B-CLL) (30, 31). The single reported examination of the functional significance of LPL in B-CLL was difficult to interpret, however, because Orlistat, a compound that inhibits both LPL and FASN (4), was used to inhibit LPL in those studies (32). To our knowledge, these are the first experiments to demonstrate widespread expression of LPL by solid tumors. We find that, in contrast to cultured breast cancer cell lines, where substantial LPL is found only in a subset with a “triple negative” gene expression signature, the enzyme is a universal component of breast tumors, irrespective of biomarker status. Moreover, we also find that all liposarcoma and prostate tumors examined also express LPL.
Several plausible explanations exist for the discrepancy between cell lines and tumors with respect to LPL expression. First, the cell lines have been passaged over time in culture systems lacking vascular endothelium, which is the physiological site for LPL action, or reliably fixed concentrations of triglyceride-rich lipoprotein substrate, whereas cell culture media generally contain high concentrations of glucose. Thus de novo synthesis, rather than lipolysis or receptor-mediated endocytosis, may have been selected as the preferred mechanism for FA acquisition in cell culture. Second, it is possible that interactions with stroma elicit LPL expression. Third, each of the breast cancer cell lines that we find to express substantial levels of LPL are not only “triple negative”, but are also non-adherent to tissue culture plasticware. In view of reports that cellular detachment provokes major metabolic adaptations in Her2/neu-expressing breast cancer cells (33), we examined the hypothesis that cellular detachment (72 h) would provoke enhanced LPL mRNA expression. This proved, however, not to be the case (data not shown). Irrespective of the cause of the discrepancy, it is important to recognize that tissue culture experiments may not faithfully recreate in vivo physiology.
Efficient utilization by cancer cells of fatty acids released by extracellular lipolysis would require the expression of both LPL and CD36. It was therefore not surprising to find CD36 expression in the majority of tumor tissues examined. CD36 is known to traffic from cytoplasm to the plasma membrane in response to insulin stimulation of adipocytes (34). We observed cell surface localization in ~70% of breast cancers, whereas ~30% exhibited only a cytoplasmic signal. Based on our observation that cell surface staining was significantly less frequent in triple negative tumors, we speculate that CD36 trafficking may be driven by cell-surface acting growth factors and/or sex steroids in breast cancers.
Although further experiments are required to delineate the precise roles of lipogenesis and lipolysis in transformation, proliferation, and metastasis, recent studies have advanced this area. Previous work established a tight linkage of enhanced fatty acid synthesis to transformation (35), and recent studies have defined the role of an intracellular lipase, monoacyl glycerol lipase (MAGL), in promoting tumorigenesis. MAGL provides, by de-esterification, a stream of intracellular free fatty acids to fuel proliferation, growth, and migration (36). The current report demonstrates a complementary role for LPL, an extracellular lipase, in providing a stream of fatty acids to fuel cancer cell proliferation.
Various hypotheses have been proposed to explain the dependence of tumors on lipogenesis, but it is clear that the primary metabolic fate of fatty acids in proliferating tumor cells is incorporation into phospholipids destined for membrane biosynthesis (37, 38). As mitochondrial production and export of citrate are the key steps required to maintain de novo lipogenesis in the cytosol, this begs the question of how such mitochondrial metabolism may be maintained under the hypoxic (but not anoxic) conditions that prevail in tumors. Indeed, hypoxia induced factor-1 (HIF-1), a key mediator of the cellular response to hypoxia, reduces the fractional entry of glucose-derived carbon into mitochondria by down-regulating pyruvate dehydrogenase, thus driving the increased lactate production that is the most well-recognized aspect of intermediary metabolism in tumors (39). However, net flux of carbon through the glycolytic pathway is substantially elevated in glucose-avid tumor cells, because of increased uptake and trapping. The reduced amount of carbon directed to mitochondria is thus sufficient to provide an estimated 60–85% of the ATP generated (40). Brisk citrate export from mitochondria appears to be favored by incomplete combustion, as a consequence of the “truncated Krebs cycle” in tumor mitochondria (41), which also may serve to reduce oxygen use by reducing carbon flux through steps downstream from citrate in the cycle. Thus, the competing oxygen-sparing and anabolic demands on tumors are met by a balanced set of metabolic alterations, the former favored by HIF-1, and the latter driven by oncogenes (reviewed in (42)). Overall, it appears that the uptake of exogenous fatty acids, for which the current work demonstrates most tumors to be equipped, would be an advantageous response to the metabolic dilemma of hypoxic, proliferating cancer cells.
Our findings have several implications. First, therapeutic efforts aimed solely at inhibition of long-chain fatty acid synthesis may not be effective for tumors that are provided with LPL and express CD36. Such tumors may be sensitive to agents that inhibit the enzymes for both lipogenesis and lipolysis, such as Orlistat (4) or the dietary supplement conjugated linoleic acid, which can suppress the genes required for both pathways (8, 43). Efforts to target LPL will need to take into account the possibility that prolonged systemic suppression of LPL activity could result in hypertriglyceridemia and consequent pancreatitis, particularly if dietary fat intake is not curtailed. Second, the ability of nearby nonmalignant cells to provide LPL to the tumor microenvironment may favor the ability of tumor cells, particularly those with low lipogenic potential, to establish metastases in LPL-rich tissues such as lung or fatty bone marrow. In order to benefit from LPL provided by tumor stroma, the expression of CD36 by the tumor would be required. Third, the presence of LPL in the tumor vasculature may mediate the reported effects of dietary fat intake on outcome (9). In addition to the well-characterized lipogenic tumor phenotype, our studies indicate the expression of a previously unappreciated “lipolytic” pathway active in cancer cells as well.
Supplementary Material
Acknowledgments
MCK-LPL transgenic mice were kindly supplied by Ira Goldberg, Columbia School of Medicine, New York, NY. We thank Martin Wabitsch, University of Ulm, Germany, for the LiSa2 cells. Soraphen A was kindly provided by Klaus Gerth and Rolf Jansen, Helmholtz-Zentrum für Infektionsforschung, Braunschweig, Germany. Triglyceride measurements were kindly provided by Hong K. Lee, Department of Pathology, Dartmouth-Hitchcock Medical Center, Lebanon, NH. Special thanks to Rebecca O'Meara MT (ASCP), Pathology Translational Research Laboratory at Dartmouth Medical School for performing the immunohistochemistry.
Grant support: This work was supported by N.I.H. grant RO1CA126618 (WBK). N.I.H. Training Grant DK07508 (NBK), a Howard Hughes Medical Foundation Fellowship 52005870 (AJF), Norris Cotton Cancer Center Prouty grants (BLE, WBK), grant G.0590.08 (JVS) and a fellowship (ER) from the Research Foundation-Flanders (FWO), the N.C.I. Bay Area Breast Cancer SPORE P50 CA58207 (LAT), and the Program in Experimental and Molecular Medicine at Dartmouth Medical School (CJF).
Abbreviations employed
- ATP
adenosine triphosphate
- B-CLL
B-cell chronic lymphocytic leukemia
- ER
estrogen receptor
- FA
fatty acid
- FASN
fatty acid synthase
- HIF-1
hypoxia-inducible factor 1
- IRB
internal review board
- LPL
lipoprotein lipase
- MAGL
monoacylglycerol lipase
- RNA
ribonucleic acid
- RT-PCR
reverse transcriptase polymerase chain reaction
- VLDL
very low density lipoprotein
Footnotes
None of the authors reports any conflict of interest.
Supplementary Information accompanies the paper online at XX.
References
- 1.Kuhajda F. Fatty acid synthase and cancer: new application of an old pathway. Cancer Res. 2006;66:5977–5980. doi: 10.1158/0008-5472.CAN-05-4673. [DOI] [PubMed] [Google Scholar]
- 2.Menendez J, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 3.Alli P, Pinn M, Jaffee E, McFaden J, Kuhajda F. Fatty acid synthesis inhibitors are chemopreventive for mammary cancer in neu-N transgenic mice. Oncogene. 2005;24:39–46. doi: 10.1038/sj.onc.1208174. [DOI] [PubMed] [Google Scholar]
- 4.Kridel S, Axelrod F, Rozenkrantz N, Smith J. Orlistat is a novel inhibitor of fatty acid synthase with antitumor activity. Cancer Res. 2004;64:2070–2075. doi: 10.1158/0008-5472.can-03-3645. [DOI] [PubMed] [Google Scholar]
- 5.Weiss L, Hoffman G, Schreiber R, Andres H, Fuchs E, Korber E, et al. Fatty-Acid Biosynthesis in Man, a Pathway of Minor Importance. J Biol Chem. 1986;367:905–912. doi: 10.1515/bchm3.1986.367.2.905. [DOI] [PubMed] [Google Scholar]
- 6.Kuhajda F, Jenner K, Wood F, Hennigar R, Jacobs L, Dick J, et al. Fatty acid synthesis: a potential selective target for antineoplastic therapy. PNAS. 1994;91:6279–6383. doi: 10.1073/pnas.91.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beckers A, Organe S, Timmermans L, Scheys K, Peeters A, Brusselmans K, et al. Chemical inhibition of acteyl-CoA carboxylase induces growth arrest and cytotoxicity selectively in cancer cells. Cancer Res. 2007;67:8180–8187. doi: 10.1158/0008-5472.CAN-07-0389. [DOI] [PubMed] [Google Scholar]
- 8.Donnelly C, Olsen A, Lewis L, Eisenberg B, Eastman A, Kinlaw W. Conjugated Linoleic Acid (CLA) inhibits expression of the Spot 14 (THRSP) and fatty acid synthase genes and impairs the growth of human breast cancer and liposarcoma cells. Nutrition and Cancer. 2009;61:114–122. doi: 10.1080/01635580802348666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chlebowski RT, Blackburn GL, Thomson CA, Nixon DW, Shapiro A, Hoy MK, et al. Dietary fat reduction and breast cancer outcome: interim efficacy results from the Women's Intervention Nutrition Study. JNCI. 2006;98:1767–1776. doi: 10.1093/jnci/djj494. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg I, Eckel R, Abumrad N. Regulation of fatty acid uptake into tissues: lipoprotein lipase- and CD36-mediated pathways. J Lipid Research. 2009;50:S86–S90. doi: 10.1194/jlr.R800085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neve R, Chin K, Fridlyand J, Yeh J, Baehner F, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irizarry R, Hobbs B, Collin F, Beazer-Barclay Y, Antonellis K, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 1998;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 13.Eisen M, Spellman P, Brown P, Botstein D. Cluster analysis and display of genome-wide expression patterns. PNAS. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martel P, Bingham C, McGraw C, Baker C, Morganelli P, Meng M, et al. S14 protein in breast cancer cells: direct evidence for regulation by SREBP-1c, superinduction with progestin, and implication in cell growth. Experimental Cell Research. 2005;312:278–288. doi: 10.1016/j.yexcr.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 15.Mosmann T. Rapid colorometric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Meth. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein J, Basu S, Brown M. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods in Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 17.Hata A, Ridinger D, Sutherland S, Emi M, Shuhua Z, Myers R, Ren, et al. Binding of lipoprotein lipase to heparin. J Biol Chem. 1993;268:8447–8457. [PubMed] [Google Scholar]
- 18.Nilsson-Ehle P, Schotz M. A stable, radioactive substrate emulsion assay of lipoprotein lipase. J Lipid Res. 1976;17:536–541. [PubMed] [Google Scholar]
- 19.Cruz W, Kwon G, Marshall C, McDaniel M, Semenkovich C. Glucose and insulin stimulate heparin-releasable lipoprotien lipase activity in mouse islets and INS-1 cells. J Biol Chem. 2001;276:12162–12168. doi: 10.1074/jbc.M010707200. [DOI] [PubMed] [Google Scholar]
- 20.Wells W, Schwartz G, Morganelli P, Cole B, Chambers J, Kinlaw WB. Expression of “Spot 14” (THRSP) predicts disease free survival in invasive breast cancer: immunohistochemical analysis of a new molecular marker. Breast Cancer Research and Treatment. 2006;98:231–240. doi: 10.1007/s10549-005-9154-z. [DOI] [PubMed] [Google Scholar]
- 21.Sorlie T, Perou C, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. PNAS. 2001;98:10896–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cleator S, Heller W, Coombes R. Triple-negative breast cancer: therapeutic options. Lancet Oncol. 2007;8:235–244. doi: 10.1016/S1470-2045(07)70074-8. [DOI] [PubMed] [Google Scholar]
- 23.Hughes D, Martel P, Kinlaw W, Eisenberg B. The synthetic triterpenoid CDDO-Im inhibits fatty acid synthase expression and has antiproliferative and proapoptotic effects in human liposarcoma cells. Cancer Invest. 2007;26:118–127. doi: 10.1080/07357900701522612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swinnen J, Brusselmans K, Verhoeven G. Increased lipogenesis in cancer cells: new players, novel targets. Curr Opin Clin Nutr Metab Care. 2006;9:358–365. doi: 10.1097/01.mco.0000232894.28674.30. [DOI] [PubMed] [Google Scholar]
- 25.Kinlaw W, Quinn J, Wells W, Roser-Jones C, Moncur J. S14 in breast cancer: a marker of aggressive disease and a potential therapeutic target. Endocrinology. 2006;147:4048–4055. doi: 10.1210/en.2006-0463. [DOI] [PubMed] [Google Scholar]
- 26.Kinlaw W, Church J, Harmon J, Mariash C. Direct evidence for a role of the “spot 14” protein in the regulation of lipid synthesis. J Biol Chem. 1995;270:16615–16618. doi: 10.1074/jbc.270.28.16615. [DOI] [PubMed] [Google Scholar]
- 27.Moreau A, Teruel C, Beylot M, Albalea V, Tomasi V, Umbdenstock T, et al. A novel pregnane X receptor and S14-mediated lipogenic pathway in human hepatocyte. Hepatology. 2009;49:2068–2079. doi: 10.1002/hep.22907. [DOI] [PubMed] [Google Scholar]
- 28.Obunike J, Edwards I, Rumsey S, Curtiss L, Wagnerf W, Deckelbaum R, Goldberg I. Cellular differences in lipoprotein lipase-mediated uptake of low density lipoproteins. J Biol Chem. 1994;269:13129–13135. [PubMed] [Google Scholar]
- 29.Bova G, Carter B, Bussemakers M, Emi M, Fujiwara Y, Kyprianou N, et al. Homozygous deletion and frequent allelic loss of chromosome 8p22 loci in human prostate cancer. Cancer Research. 1993;53:3869–3873. [PubMed] [Google Scholar]
- 30.Heintel D, Kienle D, Shehata M, Krober A, Kroemer E, Schwarzinger I, et al. High expression of lipoprotein lipase in poor risk B-cell chronic lymphocytic leukemia. Leukemia and Lymphoma. 2005;19:1216–1223. doi: 10.1038/sj.leu.2403748. [DOI] [PubMed] [Google Scholar]
- 31.van't Veer M, Broojimans A, Langerak A, Verhaaf B, Goudswaard C, Graveland W, et al. The predictive value of lipoprotein liase for survival in chronic lymphocytic leukemia. Lymphoproliferative Disorders. 2006;91:56–63. [PubMed] [Google Scholar]
- 32.Pallasch C, Schwamb J, Konigs S, Schulz A, Debey S, Kofler D, et al. Targeting lipid metabolism by the lipoprotein lipase inhibitor orlistat results in apoptosis of B-cell chronic lymphocytic leukemia cells. Leukemia. 2008;22:585–592. doi: 10.1038/sj.leu.2405058. [DOI] [PubMed] [Google Scholar]
- 33.Schafer Z, Grassian A, LS, Jiang Z, Gerhart-Hines Z, Irie H, Gao S, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luiken J, Dyck D, Han X, Tandon N, Arumugum Y, Glatz J, et al. Insulin induces the translocation of the fatty acid transporter FAT/CD36 to the plama membrane. Am J Physiol Endocrinol Metab. 2002;282:E491–E495. doi: 10.1152/ajpendo.00419.2001. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Han W, Morin P, Chrest F, Pizer E. Activation of fatty acid synthase during neoplastic transformation: role of mitogen-activated protein kinase and phophatidylinositol 3-kinase. Exp Cell Res. 2002;279:80–90. doi: 10.1006/excr.2002.5600. [DOI] [PubMed] [Google Scholar]
- 36.Nomura D, Long J, Niessen S, Hoover H, Ng S, Cravatt B. Monoacylglycerol lipase regulates a fatty acid network the promotes cancer pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pizer E, Wood F, Pasternack G, Kuhajda F. Fatty acid synthase (FAS): a target for cytotoxic antimetabolites in HL60 promyelocytic leukemia cells. Cancer Res. 1996;56(4):745–751. [PubMed] [Google Scholar]
- 38.Swinnen J, Van Veldhoven P, Timmermans L, De Schrijver E, Brusselmans K, Vanderhoydonc F, et al. Fatty acid synthase drives the synthesis of phospholipids partitioning into detergent-resistant membrane microdomains. Bioche Biophys Res Commun. 2003;302:898–903. doi: 10.1016/s0006-291x(03)00265-1. [DOI] [PubMed] [Google Scholar]
- 39.Kim J, Tchernyshyov I, Semenza G, Dang C. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metabolism. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Pedersen P. Tumour mitochondria and the bioenergetics of cancer cells. Prog Exp Tumour Res. 1978;22:190–274. doi: 10.1159/000401202. [DOI] [PubMed] [Google Scholar]
- 41.Parlo R, Coleman P. Enhanced rate of citrate export from cholesterol-rich hepatoma mitochondria. J Biol Chem. 1984;259 [PubMed] [Google Scholar]
- 42.Gordan J, Thompson C, Simon M. HIF and c-Myc: Sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harvatine K, Bauman D. SREBP1 and thyroid hormone responsive spot 14 (S14) are involved in the regulation of bovine mammary lipid synthesis during diet-induced milk fat depression and treatment with CLA. J Nutrition. 2006;136:2468–2474. doi: 10.1093/jn/136.10.2468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.