Abstract
MicroRNA (miRNA)-mediated gene regulation has become a major focus in many biological processes. GW182 and its long isoform TNGW1 are marker proteins of GW/P bodies and bind to Argonaute proteins of the RNA induced silencing complex. The goal of this study is to further define and distinguish the repression domain(s) in human GW182/TNGW1. Two non-overlapping regions, Δ12 (amino acids 896–1219) containing the Ago hook and Δ5 (amino acids 1670–1962) containing the RRM, both induced comparable silencing in a tethering assay. Mapping data showed that the RRM and its flanking sequences in Δ5, but not the Ago hook in Δ12, were important for silencing. Repression mediated by Δ5 or Δ12 was not differentially affected when known endogenous repressors RCK/p54, GW182/TNGW1, TNRC6B were depleted. Transfected Δ5, but not Δ12, enhanced Ago2-mediated repression in a tethering assay. Transfected Δ12, but not Δ5, released endogenous miRNA reporter silencing without affecting siRNA function. Alanine substitution showed that GW/WG motifs in Δ12 (Δ12a, amino acids 896–1045) were important for silencing activity. Although Δ12 appeared to bind PABPC1 more efficiently than Δ5, neither Δ5 nor Δ12 significantly enhanced reporter mRNA degradation. These different functional characteristics of Δ5 and Δ12 suggest that their roles are distinct, and possibly dynamic, in human GW182-mediated silencing.
INTRODUCTION
MicroRNAs (miRNA) are endogenous 20–25 nt RNAs largely transcribed from independent miRNA genes or gene clusters and play many key roles in a variety of normal and pathological cellular processes (1). MiRNAs are incorporated into the RNA-induced silencing complex (RISC) to effect translational repression or RNA degradation of their target mRNAs (2–6). The Argonaute protein family, a highly conserved key component of the RISC complex, is represented by four proteins (Ago1–Ago4) in mammals that are involved in miRNA-mediated translational silencing (7). Only Ago2 harbors RNase H-type activity in its C-terminal P-element induced wimpy testis (PIWI) domain and is known to function in small interfering RNA (siRNA)-mediated slicing of mRNA targets by endonucleolytic cleavage (8–10).
GW182 (Gene name TNRC6A) was first identified and characterized by our laboratories in 2002 as a novel protein recognized by an autoimmune serum from a patient with motor and sensory neuropathy (11). It is an 182-kDa protein characterized by multiple glycine (G) and tryptophan (W) motifs and is an essential component of GW bodies (also known as mammalian processing bodies, or P bodies) (6,12). Two isoforms of GW182, named TNGW1 (long isoform) and GW182 (short isoform) respectively, have been subsequently reported with TNGW1 being identical in sequences with GW182 but has additional N-terminal 253 amino acids containing trinucleotide glutamine-repeat (TNR Q-repeat) domain (13). In the GW182 family, there are three paralogs of TNRC6 (GW182-related) proteins comprising GW182/TNGW1, TNRC6B (containing three isoforms) and TNRC6C in mammal, a single Drosophila ortholog (dGW182, also known as Gawky) and two Caenorhabditis elegans orthologs AIN-1 and AIN-2. (1,5,14–16). They are known to play a critical role in the silencing and degradation of miRNA-targeted mRNAs across different species (13,16–35). Significant progress has been made in characterizing the 3′-UTR sequence element required for efficient targeting and regulation of miRNA (36,37) but the detailed molecular basis of the miRNA-mediated translational silencing and mRNA degradation, especially with respect to their role of human GW182/TNGW1, is not completely understood (1,5,14–16). The Argonaute proteins, including Ago1–Ago4, are the most highly characterized factors in the miRNA-induced silencing complex (miRISC), where they bind miRNA to mediate recognition of target mRNAs (38,39). Argonaute proteins artificially tethered to the mRNA 3′-UTR induce translational silencing (25,40,41). However, the Ago–miRNA/mRNA complex requires recruitment of additional protein factors to effect subsequent translational repression (13,21,42). Multiple candidates have been proposed to play an important role in the miRNA-mediated translational silencing. Among these, GW182 is a conserved factor that retains a key role in miRNA-mediated translational repression and mRNA degradation across different species, as evidenced by studying of GW182 proteins in humans (17,23–26,28–30,33), Drosophila (18–22,27,31,42) and C. elegans (35,43). An important feature of the GW182 family in this process is its conserved ability to bind with Ago proteins (17,20,21,24–26,28,31–34,43). In addition, the GW182 family is shown to induce translational silencing effect despite the absence of Ago2 (13,20,25,31). Knockdown of individual GW182 related proteins by specific siRNAs only partially rescue the repression indicating the functional redundancy among those paralogs (28). However, they appear not to have identical roles in repression as TNRC6B and TNRC6C form distinct protein complexes with the four human Argonaute proteins (17).
Significant efforts have been made to map the repression domains of human (17,24,28) and Drosophila GW182-related proteins (18–20,22). The C-terminal domain including the domain of unknown function (DUF), M-GW, RRM and C-GW is commonly identified as the ‘silencing domain’ in a variety of species. However, it is controversial and remains to be confirmed if the ‘N-terminal Ago-binding domain’ spanning the N-GW region possesses full silencing effects (18,19), is partially active (28) or completely inactive (17,20,24), albeit these studies use slightly different deletion construct boundaries and/or different species.
In the current study, mapping of the repression domain(s) of human GW182 was performed by generating a series of deletion constructs covering the full-length GW182 protein. Two non-overlapping domains, a middle GW182 fragment Δ12 (amino acids 896–1219) and a C-terminal GW182 fragment Δ5 (amino acids 1670–1962), were shown to trigger translational silencing when tethered to the 3′-UTR. We showed that these domains representing the minimum length of middle- and C-terminal deletion constructs caused comparable silencing effects on the reporter compared to two full-length GW182 isoforms, TNGW1 and GW182. The present study defined a novel silencing domain on human GW182 and the role of GW/WG motifs within this domain.
MATERIALS AND METHODS
Plasmids
The cDNAs of TNGW1, GW182, TNR, Δ1, Δ10, Δ12, Δ7, Δ8, Δ5, Ago2 and PIWI were constructed as described in our previous studies (6,13,25). The N-terminal construct ‘1–565’ was generated by restriction digestion using enzymes HpaI and SmaI to excise the 3′ fragment from the full-length construct pENTR-TNGW1; the construct was completed by T4 DNA ligase reaction to re-circularize the truncated linearized plasmid. Δ5a, Δ5b, Δ8 and Δ11 were generated by polymerase chain reaction (PCR) using GW182 cDNA as the template. The PCR cloning primers were: Δ5a, forward 5′-AAAAAGCAGGCTCCTCATCCTTGAACACCACG-3′, reverse 5′-AGAAAGCTGGGTTCAGCTGGAACCTGGGGTATATC T-3′; Δ5b, forward 5′-AAAAAGCAGGCTCCTCATCCTTGAACACCACG-3′, reverse 5′-AGAAAGCTGGGTTCAGTCTCCACAGTTAGTTCCCGACA-3′; Δ8, forward 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATTAGACAGAATGGCAATCC-3′, reverse 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTAGGCAACATCAAGGCATAG-3′; Δ11, forward 5′-AAAAAGCAGGCTTCACTTGGGGAAACAACATA-3′, reverse 5′-AGAAAGCTGGGTTCACTCTGGTGAGTCTCTCGAAAA-3′. Fragments Δ12a and Δ12b were directly synthesized by GenScript Corp (Piscataway, NJ, USA). The cDNA of Δ12a mutant (Δ12am, all GW or WG changed to AA) was also synthesized by GenScript. The PABPC1 expression vector was purchased from OriGene (Rockville, MD, USA). All of the variants used in current study were subcloned into the Gateway destination vector for GST, green fluorescence protein (GFP), or hemagglutinin tagged with a 22-amino-acid-long N peptide specifically recognizing the BoxB hairpin (NHA) (13) expression using the Gateway LR recombination reaction (Invitrogen, Carlsbad, CA). The tethering assay plasmids, including pCIneo-NHA vector, NHA-Ago2, Renilla luciferase (RL) and firefly luciferase (FL), with or without the 5BoxB structure in the 3′-UTR, were obtained from Dr Witold Filipowicz, Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland (40). The miR20 luciferase reporter RL-20 bulge and RL-20 perfect was obtained from Dr Phillip Sharp, Massachusetts Institute of Technology (44). All DNA constructs used in this study were confirmed by direct DNA sequencing.
Antibodies
Rabbit polyclonal anti-GST and RCK/p54 were purchased from MBL International (Woburn, MA, USA). Mouse monoclonal anti-HA was purchased from Covance (Emeryville, CA, USA). Mouse monoclonal anti-tubulin was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Cell culture and transfection
HeLa, A549 and HEK293 cells were cultured in DMEM containing 10% fetal bovine serum in a 37°C incubator with 5% CO2. The plasmid transfection was performed using Lipofectamine 2000 (Invitrogen) as per the manufacturer’s instruction. The tethering assays were performed in a 24-well plate format. Six hundred nanograms of NHA-GW182 construct were co-transfected with either 10 ng RL-5BoxB/100 ng FL or 100 ng FL-5BoxB/10 ng RL in HEK293 cells. Cells were harvested 48 h after transfection for luciferase assays. For the GST pull-down assays, 2 µg of GST-tagged GW182 construct was co-transfected with 2 µg NHA-tagged construct into HeLa cells. HeLa cells were harvested 24 h after transfection, lysed by NET/NP40 buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris, pH 7.4, 0.3% NP40) with Complete Protease Cocktail Inhibitor (Roche Diagnostics, Indianapolis, IN, USA), and then applied to GST pull-down assays.
GST pull-down assays and western blot analysis
Cell lysates were sonicated at 20% amplitude for 10 s three times on ice and then centrifuged at 13 200 rpm for 5 min. A fraction of soluble lysate was mixed with Laemmli sample buffer as input for western blot analysis. Two hundred micro liters of the soluble fractions were incubated with Glutathione Sepharose 4B (GE Healthcare, Piscataway, NJ, USA) and mixed at 4°C for 2 h for GST pull-downs. After the incubation, the beads were washed with NET/0.3% NP40 buffer four times and the samples were eluted in Laemmli sample buffer. The input and GST pull-down samples were separated on 10% polyacrylamide gels and transferred to nitrocellulose and western blotting performed as described previously (45). The dilutions of primary antibodies were: 1:1000 for anti-GST, and 1:1000 for anti-GFP.
Micrococcal nuclease assay
Three parallel transfections in HeLa cells were set up with indicated combinations of GST-PIWI and NHA-tagged constructs. Whole cell lysates were harvested in EDTA-free lysis buffer containing 150 mM NaCl and EDTA-free Protease Cocktail Inhibitor (Roche Diagnostics, Indianapolis, IN, USA) 24 h after transfection. Samples (200 μl) were diluted in equal volume of the same buffer without NaCl to adjust to a final concentration of 75 mM NaCl. Untreated group was immediately transferred on ice until ready for the pull-down assay. Mock and micrococcal nuclease (MNase) treated groups were added 10 μl 0.1 M CaCl2. A total of 0.2 U MNase (Sigma, St. Louis, MO) was added only to the MNase treated group. Both Mock and MNase groups were incubated in 37°C for 10 min. Twenty microliters of 0.5 M EGTA was added to inactivate MNase. An aliquot of cell lysate from each group were separated for RNA extraction using mirVana total RNA isolation kit (Applied Biosystems, Foster City, CA, USA). All three groups were subjected to GST pull-down assay protocol described above.
Tethering assay and miRNA interference assay using a dual luciferase assay
HEK293 cells were harvested 48 h after transfection with tethering constructs and dual luciferase reporters. The FL and RL activities were measured using the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) following the manufacturer’s protocol. Relative luciferase activities were calculated as the ratio of targeted luciferase activities over control luciferase activities (40). The repression levels of experimental groups were calculated by the percentage reduction of relative luciferase activities compared with that in the NHA control group (13). The relative repression effects of all constructs were normalized to that of NHA, which was standardized as 1. Data for each construct were collected in 6–18 replicates. HeLa and A549 adenocarcinoma human alveolar basal epithelial cells were also used to address any cell specific effects. For miRNA interference assays, RL-20 bulge and RL-20 perfect, containing seven copies of miR-20 target sites at the 3′-UTR which form imperfect or perfect matches with endogenous miR-20 were co-transfected into HEK293, HeLa and A549 cells with FL internal control and tested constructs. Luciferase activities were measured as described above. Cell lysates from representative luciferase assays were mixed directly with Laemmli sample buffer and separated in 4–20% HCl–Tris Ready Gels (BioRad, Hercules, CA, USA) to quantify the expression levels of different NHA-tagged GW182 constructs. Samples were then transferred to nitrocellulose membranes and analyzed by western blot using anti-HA tag antibody. To avoid the relatively narrow dynamic range of traditional film systems, bands visualized by an enhanced chemiluminescence assay were captured with a Geliance 600 (PerkinElmer, Waltham, MA, USA) to obtain optimal images. The results were then analyzed by GeneTools software (PerkinElmer) or Image J (http://rsbweb.nih.gov/ij/) to quantify the amount of protein expressed in individual assays.
siRNA and qRT–PCR
HeLa cells were seeded at a concentration of 5 × 104 cells/well into 24-well plates in 0.5 ml culture medium. siGENOME SMARTpool siRNA for GFP, GW182 (NM_014494) and TNRC6B (NM_015088) or RCK/p54 (NM_004397) were purchased from Dharmacon, Inc (Lafayette, CO). The final concentration used for transfection was 100 nM. Four different duplexes of GW182 siGENOME siRNA were purchased separately from Dharmacon (Cat No. D-014107-01–D-014107-04). Since duplex 1 targets sequences common in Δ5, only duplexes 2–4 were used to knockdown endogenous GW182 in experiments when the co-expression of Δ12 or Δ5 was required. To monitor the efficiency of the siRNA knockdown, parallel experimental groups were set up. Total RNA samples were harvested using mirVana total RNA isolation kit (Applied Biosystems) 48 h after siRNA transfection following the manufacturer’s instructions. Reverse transcription was performed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The relative mRNA levels of target genes were measured in duplicate using TaqMan Fast Universal Master Mix (Applied Biosystems) with the corresponding TaqMan Gene Expression Assay (Applied Biosystems). Quantification of mRNA degradation using SYBR-Green quantitative real time polymerase chain reaction (qRT–PCR) was described previously (13).
RESULTS
Non-overlapping GW182 fragments Δ12 and Δ5 harbored comparable repression effects to full-length GW182/TNGW1
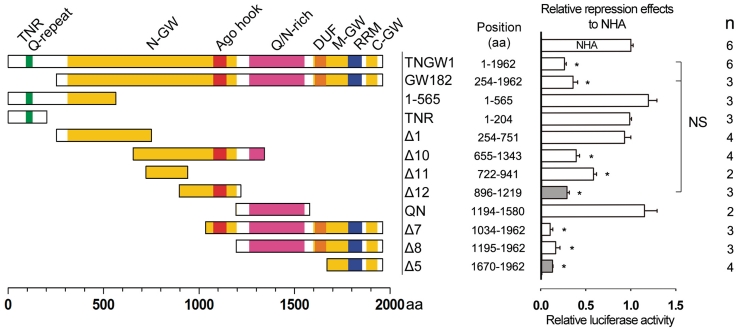
As shown in Figure 1A, human GW182 and its longer isoform TNGW1 (13) contain three glycine/tryptophan (GW)-rich regions in their N-terminal, middle and C-terminal domains (N-GW, M-GW and C-GW, respectively), as well as an RNA recognition motif (RRM, amino acids 1780–1853, cd00590). Another interesting domain is a short sequence element termed the ‘Argonaute hook’ (Ago hook, amino acids 1076–1144, pfam10427) that binds to PIWI domains of Ago proteins (34). There is also a short stretch of glutamine repeats in the N-terminal domain of TNGW1 [trinucleotide repeat (TNR) Q-repeat, amino acids 93–127] (13) and a glutamine/asparagine-rich region (Q/N-rich, amino acids 1264–1553) between the N-GW and M-GW regions (46). A conserved ubiquitin-associated domain (UBA) (31) and another DUF (amino acids 1604–1641) (23,28) are also shown in recent reviews (14,15). Right after reporting our findings that human GW182 induced silencing independent of Ago2 (13), the current study was initiated with multiple truncated constructs of GW182 spanning the full-length protein (Figure 1) to narrow down the region responsible for the repression effect in the tethering assay. These constructs were adapted to the tethering assay (13,40) and examined for their repression effects accordingly. As shown in the right panel, the relative repression effects observed for different GW182-truncated constructs was sorted into three categories: (i) no repression effect including 1–565, TNR, Δ1 and QN; (ii) high repression effect comparable to full length protein including Δ10, Δ12, Δ8, Δ7 and Δ5; (iii) Δ11, which had a low to moderate repression effect. Interestingly, the results revealed that there were more than one non-overlapping region able to induce a repression effect when tethered to the 3′-UTR of the reporter mRNA. GW182 fragment Δ12 and Δ5 were the smallest representative, non-overlapping constructs that retained the most repression effect of the full-length protein without being expressed significantly higher than other truncated constructs (Supplementary Figure S1). Both FL-5BoxB and RL-5BoxB reporters were used in these tethering assays and similar effects were observed from using either reporter. In subsequent studies, primarily the RL-5BoxB reporter was utilized. Since Δ12 and Δ5 were reported to bind to Ago proteins (25), an initial interpretation was that their repression activities were related to their binding of Ago proteins. Contradicting this hypothesis, Δ1, an N-terminal truncated construct of GW182 that strongly binds to all four human Ago proteins (25) did not show repression in the tethering assay. This finding is consistent with previous reports that Ago proteins were not the direct effectors of repression (20,24). In summary, a novel domain Δ12 was identified in human GW182/TNGW1 with a comparable silencing activity to the established silencing domain Δ5 and full-length GW182/TNGW1.
Figure 1.
Δ12 and Δ5 are two non-overlapping GW182 domains harboring repression in tethering function assay. The left panel shows GW182/TNGW1 and their series of truncation constructs. Amino acid residues of GW182 constructs are referenced to TNGW1, the longer isoform of GW182 (GenBank Accession NM_014494.2). TNR Q-repeat (green), glutamine repeat at the N-terminal domain of TNGW1; N-GW, M-GW and C-GW (yellow), three glycine/tryptophan-rich regions; Ago hook (red), a region reported to bind Ago protein; Q/N-rich (purple), glutamine/asparagine-rich region; DUF, DUF (orange); RRM (blue), RNA recognition motif. The right panel shows relative repression effects on reporter (either FL-5BoxB or RL-5BoxB; data combined as no difference was observed between reporters) by tethering the corresponding construct to the 3′-UTR of lucifearse mRNA. Their repression effects were normalized to NHA control, which was assigned as 1. Bar graphs show averages with standard errors (error bars); n, numbers of repeated experiments; Asterisk represents significant difference from NHA in t-test, P < 0.01; NS, no statistical significance.
Contribution of defined domains within Δ12 and Δ5 in reporter silencing and Ago2 binding
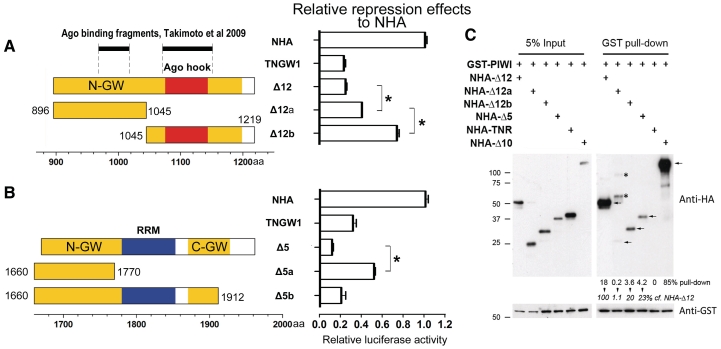
An intriguing finding was that the two identified GW182 domains with repression activities had different defined domains: the Ago hook domain in Δ12 (Figure 2A) and the RRM domain in Δ5 (Figure 2B). The Ago hook has been shown to bind to Ago2 independently in vitro but it is not conserved in all GW182-related proteins (14,20,34). The RRM is linked to RNA-binding activity (11) and is highly conserved in the GW182 family. Initial experiments were designed to examine whether the Ago hook or RRM was important for the repression effect of Δ12 or Δ5 in human GW182 respectively. To better evaluate the silencing roles of these two domains, truncated constructs were generated (Figure 2A and B). When Δ12 was expressed as Δ12a and Δ12b, which contain about equal distribution of GW/WG motifs, both truncated constructs had a significant reduction in repression activity compared to Δ12. Compared to the N-terminal half Δ12a, which lacked the Ago hook but retained most of the repression activity of Δ12, the C-terminal half Δ12b containing the Ago hook significantly lost the repression activity (Figure 2A). Thus, these results showed that the Ago hook was not critical for Δ12-induced repression in the tethering assay.
Figure 2.
Roles of Ago hook and RRM domains for repression in tethering function assay and binding to Ago2. (A) The Ago hook in Δ12 was not critical for its repression effect. Compared to Δ12a which still retained 60% repression compared to NHA, Δ12b had only 27% repression. Results are expressed as mean ± standard error from three independent experiments. Asterisk represents significant difference between Δ12a to Δ12 and Δ12a to Δ12b using t-test, P < 0.01. (B) RRM domain and its flanking sequences in Δ5 were required for maximal repression effect. The repression activity of Δ5a was significantly reduced to only 55% of Δ5 (Asterisk represents significantly different compared to Δ5 in t-test, P < 0.01). The repression effect of Δ5b was not significantly different from that of Δ5 (P = 0.12). Results are expressed as mean ± standard error from three independent experiments. (C) Semi-quantitative western blot analysis showed differential binding of NHA-GW182 domains to GST-PIWI in GST pull-down assay. Bands (arrows) in blotting data were quantified and normalized to the total input (left panel) to obtain percent pull-down as shown for each lane at the bottom of the anti-HA panel. NHA-Δ12a, NHA-Δ12b and NHA-Δ5 showed only 0.2, 3.6 and 4.2% pull-down, respectively, whereas NHA-Δ12 showed 18%. Thus NHA-Δ12a, NHA-Δ12b and NHA-Δ5 showed only 1.1, 20 and 23% binding, respectively, to GST-PIWI compared to NHA-Δ12 as 100%. NHA-TNR and NHA-Δ10 served as negative and positive controls respectively. Asterisk, non-specific bands.
To examine the importance of the RRM domain, two C-terminal deletion constructs of Δ5 were generated (Figure 2B). Fragment Δ5b, which contained the RRM domain, M-GW region and part of the C-GW region, retained repression activity when compared to Δ5 or GW182/TNGW1. The repression capacity of Δ5a, which contained M-GW but not the RRM domain, was significantly reduced compared to full-length Δ5 but still retained ∼50% of repression compared to NHA control (Figure 2B). Our data with human GW182 were consistent with recent reports on human TNRC6C (28) and dGW182 (14,15,20,22) showing that the RRM and its flanking sequences enriched in GW/WG motifs (M-GW and C-GW) were pivotal for the repression, whereas the Ago hook contributed little to the repression induced by Δ12.
To determine whether the observed repression activity was correlated with the Ago-binding function, Δ12 or its deletion constructs were analyzed for Ago2-binding activity using a GST pull-down assay (Figure 2C). The results showed that NHA-Δ12, NHA-Δ12a, NHA-Δ12b and NHA-Δ5 were all pulled-down by GST-PIWI albeit at different efficiency. For a semi-quantitative analysis, when normalized to the pulled-down GST-PIWI levels and compared to NHA-Δ12, only 1.1 and 20% of the expressed NHA-Δ12a and NHA-Δ12b, respectively, were pulled down (Figure 2C). This estimation was consistent with the results showing that the Ago hook alone was insufficient to achieve maximum binding to Ago2 (24,26). Intriguingly, the integrity of Δ12 was important as it bound to the Ago2 PIWI domain substantially stronger than the Δ12a or Δ12b fragments alone, implying the full-length Δ12 formed a higher order structure to perform both Ago2-binding and silencing of the bound mRNA. The positive control NHA-Δ10 containing all three defined Ago-binding sites reported by Takimoto et al. (26) efficiently bound to GST-PIWI as evidenced by up to 85% pull-down of the input protein (Figure 2C). NHA-TNR, previously shown not to bind to GST-PIWI, served as a negative control in this experiment (25). When NHA-Δ5 was compared to NHA-Δ12, only 4.2 versus 18% bound to GST-PIWI representing 23% relative efficiency. The very weak binding of this C-terminal GW182 fragment Δ5 to Ago2 might explain why this interaction was not reported in previous studies (14). In order to further characterize whether the interaction between GST-PIWI and NHA-Δ12 or NHA-Δ5 was RNA dependent, whole cell lysates were harvested 24 h post-transfection and treated with or without MNase prior to the pull-down assay. RNA degradation was monitored by 18S and lamin A/C RNA levels using qRT–PCR (Supplementary Figure S2A). The interactions of NHA-Δ12 and -Δ5 with GST-PIWI were shown to be RNA independent (Supplementary Figure S2B). Taken together, these data showed that the Ago hook domain was neither critical for Δ12-induced repression in the tethering assay nor important for optimal binding to Ago2. Its full function may rely on the tertiary structure and its relationship to adjacent Ago-binding sites. The RRM in Δ5 contributed to the full repression activity and Δ5 interacted with Ago2 relatively weakly when compared with Δ12.
Reducing endogenous repressor levels did not affect reporter repression induced by Δ12 and Δ5
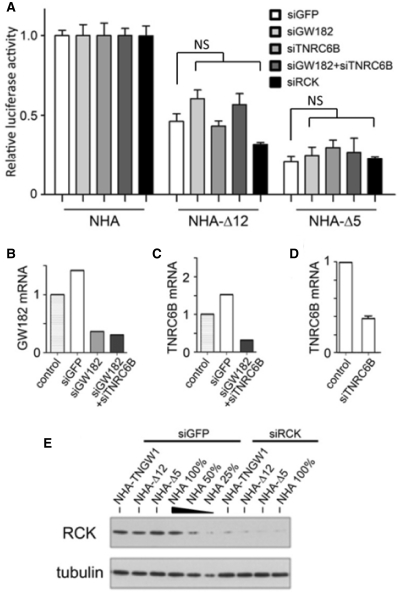
Although it was demonstrated earlier that Ago2 protein was not required for tethered GW182-mediated repression in the tethering function assay (13,25,31), it was still possible the repression mediated by Δ12 or Δ5 relied on recruitment of other important factors of the miRNA pathway machinery, including endogenous GW182 paralogs. This possibility was tested in a series of tethering assays in HeLa cells where siRNA knockdown was used as an approach to evaluating the roles of GW182 and/or TNRC6B in the repression mediated by Δ12 or Δ5. To knockdown endogenous GW182, a pool of three siRNAs targeted to GW182 mRNA, but not Δ12 and Δ5, was used as described in ‘Materials and Methods’ section. The other two factors examined were RCK/p54 and TNRC6B, both of which are reported to be important for miRNA function. For example, knockdown of RCK/p54 impaired miRNA function (47) and tethered RCK/p54 (48,49) or TNRC6B (17,24,28) induced translational repression effects. Since HeLa cells expressed both GW182 and TNRC6B but only very low levels of TNRC6C compared to A549 or HEK293 cells (Supplementary Figure S3), knockdown of both GW182 and TNRC6B together was also performed to create a ‘GW182-free’ background in HeLa cells. The luciferase activity in each knockdown experiment for NHA-Δ12 or NHA-Δ5 tethering was normalized to the same siRNA transfected control NHA group. With ∼70% reduction of endogenous GW182 or TNRC6B mRNA levels compared to mock transfected or siGFP transfected controls (Figure 3B, C and D), the repression effects of tethered Δ12 and Δ5 were not altered significantly (Figure 3A; t-test P-value > 0.05 between siGFP and other siRNA knockdown in both NHA-Δ12 and NHA-Δ5 groups). Efficient knockdown of GW182, TNRC6B and RCK/p54 was monitored by either qRT–PCR or western blot (Figure 3B–E). An addition control experiment was performed to monitor the effects of siGW182 and siGW182/siTNRC6B knockdown in the tethering assay using NHA-TNGW1, NHA-Δ12, or NHA-Δ5. Supplementary Figure S4 shows the siRNA transfection significantly affected the reporter silencing induced by NHA-TNGW1, but not NHA-Δ12, or NHA-Δ5, as would be expected since by design the siRNAs targeted endogenous GW182/TNGW1/TNRC6B and the NHA-TNGW1 construct but not NHA-Δ12 or NHA-Δ5. The apparently reduced repression effects in both Δ12 and Δ5 tethering assay compared to data shown in Figures 1 and 2 might be resulted from the introduction of siRNA transfection. In summary, knockdown of endogenous repressors GW182, TNRC6B and RCK/p54 did not significantly affect the repression activities of Δ12 and Δ5 in tethering assay. Our findings are in agreement with those previously described in Drosophila (18) and further emphasizes the independence of GW182 in inducing translation silencing in human cells.
Figure 3.
Knockdown of endogenous repressors did not affect repression activity of the two defined domains Δ12 and Δ5 in the tethering assay. (A) Repression by Δ12 and Δ5 were not significantly altered when GW182, TNRC6B, GW182/TNRC6B, or RCK/p54 was knocked down by respective siRNAs. Results are expressed as mean ± standard error from three independent experiments. There is no statistical significance difference comparing each knockdown to siGFP within NHA-Δ12 or -Δ5 group (NS, t-test). Efficiency of siRNA knockdown was monitored in each individual experiment using qRT–PCR for GW182 (B), TNRC6B (C–D) compared to the untreated HeLa cell control, or western blot analysis for RCK/p54 compared to siGFP transfected controls (E). The extract from cells transfected with NHA and siGFP was loaded at three concentrations (100, 50 and 25%) to demonstrate the semi-quantitative detection of RCK/p54.
Δ5, not Δ12, enhanced Ago2-mediated repression in tethering function assay
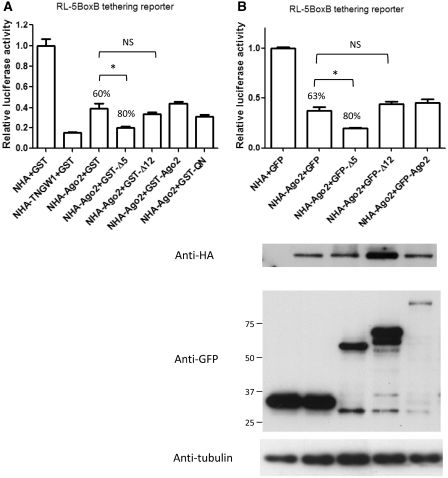
Since both Δ5 and Δ12 have comparable silencing activities when tethered to the luciferase reporter 3′-UTR (Figures 1 and 2) and none of the known endogenous factors tested was required for their repression function, it remained unclear whether their mechanisms of inducing repression were similar. Therefore, experiments were designed to determine whether the expression of Δ5 and Δ12 as a GST-tag fusion proteins could interfere with NHA-Ago2 tethered assays, as shown in Figure 4. As expected, the positive control NHA-TNGW1 typically showed 85–90% repression expressed as a reduction of luciferase activity compared to the NHA control when tethered to the RL-5BoxB reporter (13). There was also the typical ∼60% repression compared to the NHA control when NHA-Ago2 was tethered to the RL-5BoxB reporter (13,40). Interestingly, there was significantly (33%) enhanced repression with the co-expression of GST-Δ5 (Figure 4A, *, t-test, P < 0.01, n = 3). In contrast, significant differences were not observed for co-expression of GST-Δ12, -Ago2 or -QN (Figure 4A). Furthermore, neither GST-Δ5 nor –Δ12 co-expression affected repression in the tethering assay utilizing NHA-GW182 or NHA-TNGW1 (data not shown). Since both NHA-GW182 and NHA-TNGW1 consistently showed the highest levels of repression, it was not likely that any further enhancement could be observed for GST-Δ5 or GST-Δ12. Similar significantly enhanced repression for NHA-Ago2 was observed by co-expression of GFP-Δ5 but not GFP-Δ12 (Figure 4B) indicating the effect is not related to the GST tag. The Δ5-mediated enhancement in repression by NHA-Ago2 implied that this could be due to binding to other translational machinery or RNA decay factors that remain to be determined. Although Δ12 strongly bind to Ago2, it did not affect repression by tethered Ago2.
Figure 4.
Δ5, but not Δ12, enhanced Ago2-mediated repression in the tethering function assay. (A) Co-expressing GST-Δ5 with NHA-Ago2 that was tethered to the RL-5BoxB reporter enhanced the NHA-Ago2-mediating repression from 60 to 80% (33% enhanced repression). In contrast, co-expression of GST-Δ12, -QN, or -Ago2 did not show significant change. Results are expressed as mean ± standard error from three independent experiments. Asterisk represents t-test compared NHA-Ago2+GST-Δ5 with NHA-Ago2 alone, P < 0.01. (B) Identical experiments as in (A) except GFP-tagged proteins were used in place of GST fusion proteins. Results are expressed as mean ± standard error from three independent experiments. Representative cell lysates were analyzed by western blot to demonstrate expression of NHA-Ago2 and GFP fusion proteins with tubulin expression shown as loading controls. Asterisk represents significant difference in t-test compared NHA-Ago2+GFP-Δ5 with NHA-Ago2 alone, P < 0.01.
Δ12, not Δ5, interfered with endogenous miRNA repression
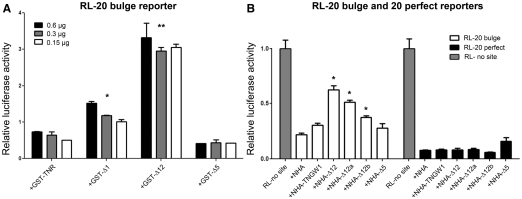
In our previous study, it was shown that multiple GW182 regions (Δ1, Δ5 and Δ12) were able to bind Ago proteins (25). Thus, the next experiment was to examine whether these three Ago-binding regions would interfere with endogenous miRNA function in a dominant-negative manner. The RL reporter for miR-20 (RL-20 bulge), which contains seven miR-20 target sites and forms bulge structures with miR-20 (44), was used to monitor the cellular miR-20 functional status. In normal conditions, the expression of RL-20 bulge is inhibited by the endogenous basal miR-20 (44, data not shown). To our surprise, when GST-TNR, –Δ1, –Δ12, or –Δ5 were co-transfected with the RL-20 bulge reporter using the dual luciferase system, only GST-Δ12 increased the expression of RL-20 bulge in all three conditions when increasing amounts of GST plasmids were used (Figure 5A). The results showed that although Δ1, Δ12 and Δ5 were able to bind Ago protein, only Δ12 impaired miR20-induced repression with >3-fold increase in luciferase activity (P < 0.0001) and Δ1 showed mild release of RL-20 bulge reporter silencing (P < 0.01).
Figure 5.
Δ12 significantly interfered with endogenous miRNA repression. (A) GST-Δ12, but not GST-Δ5, interfered with miR-20-mediated repression. Three different amounts (0.6, 0.3 and 0.15 µg) of GST-TNR, -Δ1, -Δ12 and -Δ5 plasmids were co-transfected with the RL-20 bulge reporter into HEK293 cells. The RL-20 bulge expression was significantly increased when cells co-expressed Δ12. Overexpression of Δ1 also mildly interfered with miRNA function. Asterisk represents significant difference in t-test compared with +GST-TNR, P < 0.01, n = 3; Double asterisks represent highly significant difference in t-test compared with +GST-TNR, P < 0.0001, n = 3. (B) Δ12 and its deletion constructs only interfered with miRNA but not siRNA mediated repression. NHA tag, NHA-TNGW1, -Δ12, -Δ12a, -Δ12b and -Δ5 were co-transfected with reporters RL-20 bulge/FL or RL-20 perfect/FL in HEK293 cells. Compared to the NHA control, the relative activity of RL-20 bulge was significantly increased in cells expressing NHA-Δ12 and -Δ12a/b but not in cells expressing -TNGW1 or -Δ5. NHA-Δ12 and its deletion constructs did not interfere with RL-20 perfect reporter that repressed by siRNA pathway. Results are expressed as mean ± standard error from three independent experiments. Asterisk represents significant difference in t-test compared with +NHA, P < 0.01.
As already shown in Figure 2, the Ago hook in Δ12 was neither critical for Δ12-induced repression in a tethering assay nor responsible for maximum binding with Ago2. Therefore, the next question was whether the Ago hook domain could be responsible for the miRNA interference effect or de-repression observed for Δ12. To also avoid any potential tag-dependent effects when only GST-tag was used, NHA-tagged constructs including Δ12a and Δ12b were employed to repeat the experiment. Meanwhile, another RL reporter with miR-20 target sites forming perfect match with endogenous miR-20, thus acting as reporter for the siRNA pathway, was utilized to determine whether this interference also applies to the siRNA pathway. The results clearly showed that Δ12a, which lacked the Ago hook, retained almost the same capability to interfere with RL-20 bulge reporter miRNA function (Figure 5B, left panel, P < 0.001). However, Δ12b, which contained the Ago hook domain, mildly altered the miR20-induced repression (Figure 5B, left panel). This interference was only observed with the RL-20 bulge but not the RL-20 perfect reporter, thus demonstrating overexpression of Δ12 and its deletion constructs impaired reporter silencing in a miRNA-specific manner. Compared to the RL-20 bulge reporter, the RL-20 perfect reporter showed enhanced repression effects from efficient mRNA degradation induced by the siRNA machinery and it therefore served as a useful functional control. Note that Δ12b bound to Ago2 more efficiently than Δ12a (Figure 2C) and yet its miRNA interference effect was weaker, indicating there were additional factors involving in interfering this miRNA-mediated silencing. This led us to further examine the role of GW/WG motifs residing in this fragment by generating mutations of those motifs.
Substitution of GW/WG motifs with alanines in GW1Δ12a hindered its tethering assay activity, as well as its interference with endogenous miRNA repression
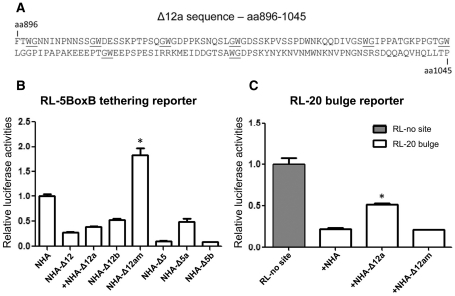
Since the results showed that the Ago hook was not critical for function but Δ12a retained almost the same capability to interfere with RL-20 bulge reporter, it was postulated that the GW/WG motifs in Δ12a might be important for functions besides binding to Ago2 (Figure 6A). A Δ12a mutant (Δ12am) was then generated to replace all GW/WG residues in Δ12a with alanine–alanine (AA) residues. In a tethering assay using RL-5BoxB, NHA-Δ12am was devoid of translation silencing when compared to NHA-Δ12a and other relevant controls (Figure 6B). When Δ12am was analyzed in the interference of RL-20 bulge reporter repression, the ability of Δ12a to release miR-20 activity was abolished in Δ12am (Figure 6C). These data suggested that the GW/WG motifs in Δ12a were important for the silencing in the tethering assay and interference in RL-20 bulge reporter function. Since it was shown that Δ12a did not bind efficiently to Ago2 (Figure 2C), the GW/WG motifs in Δ12a might be responsible for both mediating strong repression and impairing miRNA-mediated silencing. It appeared that the GW/WG motifs in different regions of GW182 have different functional roles. For example, Δ1 also possessed multiple GW/WG motifs and a defined Ago-binding site but it was not efficient in both tethering and RL-20 bulge interference assays compared to Δ12. Furthermore, mutation of some of the GW motifs on Δ1 did not abolish the Ago binding (25). However, it is acknowledged that these mutations may influence the global folding of the fragment and more detailed mutagenesis is needed in future studies to further define the roles of these GW/WG motifs. Collectively, GW/WG motifs in Δ12a region showed significant effects in silencing tethered mRNA and impaired miRNA-induced repression. GW/WG motifs located in different regions of GW182 might have different functional preferences or formed a particular 3D structure that requires further investigation.
Figure 6.
Substitution of glycine (G) and tryptophan (W) residues with alanine (A) in GW1Δ12a interfered with its repression on reporter and interference in miRNA repression activity. (A) Amino acid sequence of Δ12a shown with GW and WG residues are underlined; these residues are substituted with AA to generate a mutant Δ12am. (B) Δ12am had no repression activity in RL-5BoxB tethering assay. NHA-Δ5 and its deletion constructs served as positive controls. Asterisk represents significant different in t-test compared with NHA-Δ12, P < 0.01, n = 3. (C) GW/GW mutated Δ12a (Δ12am) no longer affected miRNA mediated repression. Experiment was performed as described in Figure 5B. Asterisk represents significant difference in t-test compared NHA-Δ12a with NHA, P < 0.01.
Δ12 and Δ5 bound to PABPC1 but only mildly affecting mRNA degradation
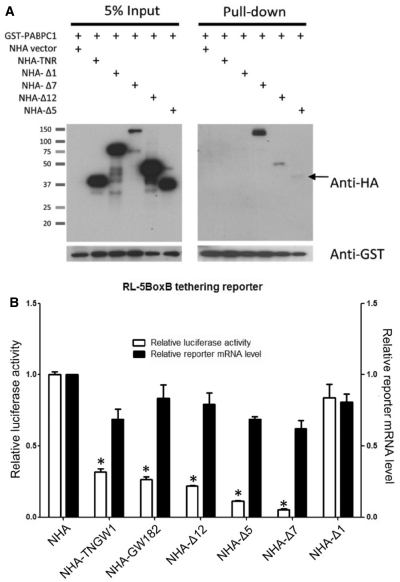
In the eukaryotic cap-dependent translation initiation step, mRNAs usually form circularized structures facilitated by the binding between the cap-binding complex eIF4E/4G and Poly-A-binding protein PABPC1 to favor association with the 40S ribosome (50). Thus, PABPC1 has been shown to play a critical role in mRNA degradation (23). To determine whether the two GW182 silencing domains Δ12 and Δ5 play differential roles in mRNA degradation, GST pull-down experiments were designed to investigate the interaction between GW182 fragments with PABPC1. The positive control Δ7 harboring the C-terminal half of TNWG1, including the PAM2 (DUF) domain, bound strongly to GST-PABPC1 (Figure 7A). This data is consistent with the results observed with TNRC6C that the PAM2 domain is the major binding site to PABPC1. Δ5 showed weak binding (arrow) for PABPC1 and this is consistent with a recent report (29) describing weak binding activity in the C-terminal region of GW182. It is noteworthy that Δ12 showed intermediate binding activity to PABPC1 compared to Δ7 and Δ5. In order to further characterize whether the interaction between GST-PABPC1 and NHA-Δ12 or NHA-Δ5 was RNA dependent, whole cell lysates were harvested 24 h post-transfection and treated with or without MNase prior to the pull-down assay as described in Supplementary Figure S2. The interactions of NHA-Δ12 and -Δ5 with GST-PABPC1 were shown to be RNA independent (Supplementary Figure S5). To further fine mapping the PABPC1-binding sites on two GW182 repression domains, additional experiments were performed to show that subclones NHA-Δ12b and NHA-Δ5b were positive in pull-down assays with GST-PABPC1 (Supplementary Figure S6). Note that the interaction of Δ12 and Δ5 with endogenous PABPC1 was not observed potentially reflecting that these are weak interactions (data not shown).
Figure 7.
Binding of Δ12 and Δ5 to PABPC1 did not significantly affect reporter mRNA degradation. (A) Differential binding of GW182 fragments Δ12, Δ7 and Δ5 to PABPC1. GST-PABPC1 was co-transfected with different NHA-tagged constructs into HeLa cells as shown above the panels for the designed GST pull-down assay. After 24 h, cell lysates were harvested and analyzed by GST pull-down followed by western blot analysis. GST-PABPC1 strongly pulled down NHA-Δ7 compared to NHA-Δ12 and -Δ5 (arrow). (B) Both tethered Δ12 and Δ5 induced primarily translational repression with only moderate reporter mRNA degradation. To determine the mRNA level of the reporter in tethering assay, a pair of each RL and FL primers was utilized in SYBR-Green qRT–PCR. The RL mRNA level was normalized to FL mRNA. All results are expressed as mean ± SD from three independent experiments. Asterisk represents significant difference in t-test compared with NHA, P < 0.01. No significant difference observed in mRNA degradation for any of the constructs compared with NHA.
A qRT–PCR was utilized to measure the level of degradation of the RL-B5Boxb reporter mRNA in tethering assay normalized to the FL mRNA level (13). As shown in Figure 7B, both NHA-Δ12 and -Δ5 induced comparable, mild mRNA degradation when tethered to the reporter comparable to the activities of NHA-GW182 and -TNGW1, respectively. The observation that NHA-Δ12 induced mild reporter mRNA degradation is consistent with the recent report that a Δ12-comparable dGW182 silencing domain (amino acids 205–490, cf. dGW182) also induced mRNA degradation in tethering assay (19). This difference in the extent in mRNA degradation may be explained by variations in methodology used to detect degradation [i.e. qRT–PCR in this study versus northern blot (19)], as well as differences in the activity of human GW182 versus the Drosophila orthologs.
DISCUSSION
Δ12 and Δ5 mapped as two non-overlapping domains mediating repression
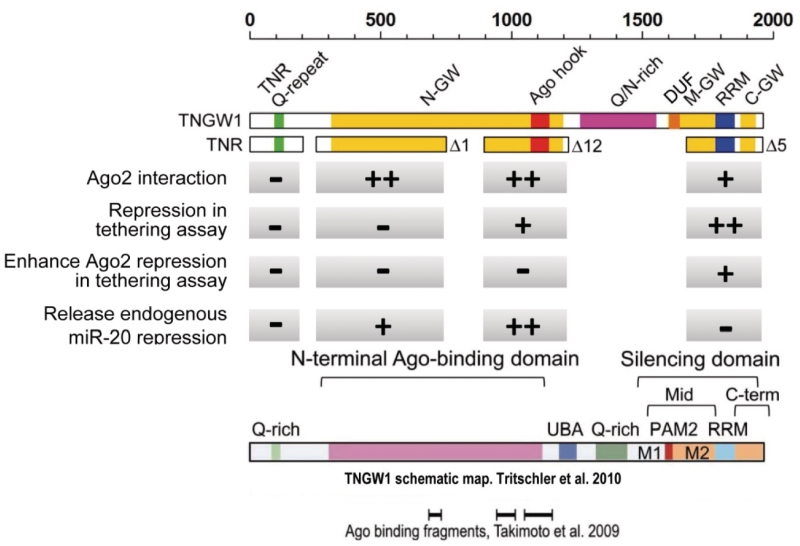
Since its discovery in 2002 (11), increasing and compelling evidence from different laboratories has shown that GW182 and its paralogs have important functions in miRNA-mediated translational repression, as well as mRNA degradation (13,17–34). A summary of the functional characteristics of different domains of GW182 in this study is shown in Figure 8, where their differential Ago2 binding, repression in tethering assays, enhancing Ago2-mediated repression and releasing endogenous miR-20 repression are compared and contrasted. Our previous data showed translational repression of tethered Ago2 to reporter 3′-UTR required GW182 and that tethered GW182 exerted a stronger repression than tethered Ago2 (13). As exemplified in the present study, these observations led to further mapping of the functional repression domain(s) of human GW182 by generating a series of deletion constructs using tethering assays. Recent reports have shown that the C-terminal domain of human GW182/TNGW1, TNRC6B and TNRC6C (17,24,28) and Drosophila GW182 (18–20,22) exert strong translational repression when tethered to the 3′-UTR of the reporter mRNA. Our current data from human GW182 truncated constructs also support the conclusion that GW182 C-terminal fragments Δ7, Δ8 and Δ5 indeed inhibit luciferase activity of the 5BoxB reporter when tethered to its 3′-UTR. Among those, Δ5 (amino acids 1670–1962) was the minimum domain that retained the full activity. It was well established by Izaurralde and colleagues that the M-GW and C-GW domains in the C-terminus of Drosophila GW182 act as a bipartite silencing domain and the RRM contributed to, but was not required for, silencing in tethering and complementation assays (14,15,20,22). In a study of TNRC6C by Filipowicz and co-workers (28), C-terminal mutation and deletion constructs were generated to elucidate the importance of the integrity of M-GW and C-GW in silencing domains; only RRM mutations mildly affected the repression tethered to 3′-UTR. Our data of human GW182 supported these observations by showing the repression activity of Δ5a with the RRM and C-GW deletion was impaired but still maintained ∼50% repression through the M-GW. Δ5b with the C-terminal Δ5 deletion but containing intact M-GW, RRM and C-GW retained full repression activity.
Figure 8.
Summary of GW182 domain functional characteristics. DUF, sequence identified to be important for PABPC1 binding. A reference schematic map of TNGW1 in one recent review (15) is included for comparison. UBA, Ubiquitin-associated domain; PAM2, PABP-interacting motif 2. Regions M1 and M2 together with PAM2 formed the Mid region. Middle region and C-terminal region but not RRM defined the bipartite silencing domain.
In contrast to reports to date, the current study demonstrated that another repression region Δ12 (amino acids 896–1219), in the middle region of human GW182 appear to be crucial for this repression function. This is the first time that the functional repression domain Δ12 is identified within the described ‘N-terminal Ago-binding domain’ (Figure 8) of human GW182. However, it should be noted that some TNRC6C fragments such as amino acids 1–405 or 1–1304 have been shown to be partially active with ∼50% repression activity in tethering assays (28). Our data ruled out the Ago hook as a critical region for repression activity. Since the Ago hook domain was defined in binding the Ago PIWI domain both in vitro and in vivo (34), a number of recent studies showed that additional Ago hook-independent GW182 fragments could efficiently bind Ago protein in human GW182, TNRC6B and TNRC6C as well as dGW182 (17,20,24–26). For example, the report of Takimoto et al. (26) identified three defined Ago2-binding domains that corresponded to amino acids 697–739, amino acids 969–1031 and amino acids 1059–1163 (Figure 8). It has also been shown that deletion of the Ago hook in GW182 and TNRC6C did not totally abolish the binding to Ago proteins (24). Chekulaeva et al. (18) observed that three dGW182 regions were responsible for the repression in Drosophila. The first repression region was the C-terminal domain of dGW182; the second repression region was the N-terminal domain amino acids 1–605 (cf. dGW182); the third region (amino acids 605–830, cf. dGW182), including the QN rich region, also triggered repression. We note that comparable activity for the corresponding region in human GW182 was not observed in our present study, an inconsistency that could be due to paralog- or species- specific effects. During the preparation of this manuscript, Chekulaeva et al. (19) published a report showing the functional significances of GW/WG repeats on dGW182 repression domain (amino acids 205–490) that induced reporter silencing. These investigators showed alignment of their repression domain with other GW182 homologs and found that mutation of certain conserved amino acid residues abolished repression induced by these dGW182 fragments. Interestingly, the alignment showed the second dGW182 repression domain corresponded closely to Δ10 (amino acids 655–1343). In agreement with this finding, Δ10 induced repression in our tethering assay was clearly observed (Figure 1). In addition, the Δ12 repression domain defined in our study represents a new core repression region with somewhat higher repression than in Δ10 (Figure 1, Δ12 versus Δ10).
In order to investigate whether the repression effects caused by these fragments were direct or indirect, knockdown-tethering experiments utilizing endogenous GW182, TNRC6B, or RCK/p54 did not dramatically impair the 5BoxB reporter repression tethered by Δ12 or Δ5. Thus, our study shows that there are two functionally independent repression domains in human GW182 with differences discussed further in the next paragraph. It is possible that Δ5 has a more important and more direct repression function than Δ12 because of the apparent higher tethering activity (Figures 1 and 3A) and its ability to enhance Ago2-mediated repression in a tethering assay (Figure 4). However, since it is still unclear how these domains bind in vivo, it can be argued that Δ12 can play an equally important role.
GW182 has been shown to bind multiple Ago–miRNA complexes (20,24–26). Studies have showed that closely spaced miRNA target sites often act synergistically and result in stronger repression than those separated by greater distances (36). The demonstration that there is more than one repression domain in each molecule of GW182 as described in this study and described by others in Drosophila (18,19), suggests that re-examination is in order to elucidate how enhanced repression may be triggered when miRNA sites are approximated to each other in 3′-UTRs. The functional advantages of the dual-repression domain GW182 with Δ12 and Δ5 having their own functional bias requires elucidation. It is conceivable that GW182, and perhaps its paralogs, function best to regulate mRNA with multiple miRNA-binding sites. Future studies will need to address the molecular mechanism how mRNAs with multiple putative miRNA-binding sites will benefit the efficiency of regulation in concerted manners.
GW182 mediated translational repression distinguished from mRNA deadenylation and decay
MicroRNAs bind primarily to the 3′-UTR of their target mRNA, and mediate translational repression and/or mRNA decay, although the detailed molecular mechanism is still not completely understood. Several models have been proposed indicating that translation repression is achieved by inhibition of translational initiation, elongation, or mRNA deadenylation (1,5). A recent report utilizing in vitro translation extracts from mouse Krebs-2 ascites cells showed miRNA-mediated deadenylation occurred 1–2 h after initial translational inhibition (23). These investigators and others also showed that the C-terminal domain of GW182 bound to deadenylases through the DUF conserved domain located between the QN-rich domain and the RRM domain (23,29) [also known as PAM2 domains (15), see Figure 8] and recruited deadenylases to the target mRNA. This deadenylation process requires binding of Ago2 and GW182 paralogs and this binding appears to be RNA independent (23). Similar results have been observed in mouse 3T3 cells where it was shown that when mRNA is bound by miRNA, a two-step deadenylation takes place followed by decapping (30). In Drosophila S2 cells, the DUF domain of dGW182 binds to PABPC1 and thus competes for eIF4G binding and leading to the disruption of the circularized mRNA structure that would normally favor translation (27). It is proposed that PAM2, M2 (between PAM2 and RRM) and C-terminal region on dGW182 together define a binding region to PABPC1 (15,27) (Figure 8). In contrast, PAM2 on TNRC6C appears to be the major binding site for its interaction with PABPC1 (15,23). Another weak PABPC1-binding site was subsequently identified on the TNRC6C C-terminal domain downstream from the RRM (29). These recent data indicate that there are differences in GW182–PABPC1 complex formation when different GW182-related proteins are compared. Of relevance to the present study, both Δ5 and Δ12 domains lack the known PABPC1-binding domain DUF (Figure 8) but still caused remarkable repression in a tethering assay and exhibited enhanced Ago2-mediated repression when compared to Δ7 or Δ8 that contains the DUF domain. GST pull-down assays indicated Δ7 strongly bound to PABPC1 whereas both Δ12 and Δ5 bind weakly to PABPC1 (Figure 7B); nevertheless, these reactivities with PABPC1 appeared specific as several other controls including Δ1, which binds Ago2, were negative. Thus, it is interesting that the two defined repression domains, lacking DUF, somehow still associated with PABPC1. It can be speculated that when mRNA is bound by specific miRNA–Ago complex, translational inhibition occurs relatively rapidly and is mediated by one or the other repression domains of GW182 (or from its paralogs), but the deadenylation step can be delayed. This process may be reversible within a narrow time frame and the repressed mRNA still may be released for further translation. Whether Δ12 and Δ5 play a role in this control will need further study. Our study demonstrated that human GW182 repression domains can be clearly separated from the putative PABPC1-binding domain DUF.
Distinct characteristics of Δ12 and Δ5 implying differential functions for GW182?
It has been shown by us (25,32) and others (20,24,26) that the N-terminal and middle region of the GW182 family and dGW182 possess multiple Ago2-binding sites. Our co-Immunoprecipitation experiments showed that Δ12 bound strongly to Ago2, while Δ5 had a substantially lower affinity for Ago2. Co-expression of Δ12, but not other GW182 fragments such as Δ5, Δ1, Δ11, or TNR, significantly inhibited miRNA rather than siRNA activity, as determined by the difference between activity of RL-20 bulge and RL-20 perfect miRNA reporters, although Δ1, Δ5 and Δ11 still bind to Ago2. The impairment of miRNA function following overexpression of dGW182 or human GW182 paralog fragments that bind Ago has been reported. For example, overexpression of the N-terminal half of Drosophila GW182, which bind Ago1 protein, impaired miRNA function and the impairment was rescued by overexpression of Ago protein (21). Other data showed that synthetic peptides or recombinant proteins corresponding to the Ago hook domain impaired translational efficiency in an in vitro translation assay (34) or impaired miRNA-mediated deadenylation (23). In particular, let-7-mediated translational repression is inhibited by a GW182 fragment containing multiple Ago2-binding sites in vitro (26). As summarized in Figure 8, clearly there are distinct properties of Δ12 and Δ5.
Why did Δ5 enhance Ago2 repression effects in the tethering assay but have no apparent effect on miR-20 repression? Why did Δ12 abolish miR-20 repression, but not the siRNA-like repression by miR-20, or affect Ago2 repression in tethering assay? Clearly, more studies are needed to address these questions. Speculations on the potential implication of these questions will undoubtedly stimulate discussion, debate and future research directions. GW182 was identified as a marker for cytoplasmic foci GW/P bodies (11) and it is likely that there is a certain amount of ‘soluble’ cytoplasmic pool versus ‘GW/P body-bound insoluble’ pool of this protein. The distribution between the two putative pools has not been studied extensively as there are many factors that may influence this dynamic process. For example, transfection of siRNA into culture cells (45) or lipopolysaccharide stimulated monocytes (51) cause an increase in number and size of these foci and probably switch GW182 from the soluble to the insoluble pool. In contrast, induced cell quiescence (12) or blocking the biogenesis of miRNA (52) lead to disassembly of GW/P bodies and increases in the soluble pool of GW182. A technical consideration is that, the separation of soluble versus insoluble pool also likely depends on the composition of the lysis buffer including whether commonly used detergents are used. The significance of the two distinct repression domains in GW182 and their relationship to two or more cytoplasmic pools also deserves consideration. Whether certain GW182 can exert inhibition of repression as demonstrated by transfected Δ12 in miR-20 repression will also need to be explored.
In conclusion, this study identified two distinct repression domains in GW182 using tethering assays and showed their characteristics in different functional assays. Observations that GW182 is characterized by having multiple Ago-binding sites with different binding affinities, as well as two distinct repression domains, is highly suggestive of its role in stabilizing multiple ‘repressed’ Ago–miRNA–mRNA complexes or in aggregating Ago–miRNA–mRNA complexes to establish an efficient repressed state. Alternatively, our data also suggest that GW182 may regulate the fate of repressed mRNA and potentially direct the repressed complex to decay or reversal to a translational state.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (Grant AI47859); the Andrew J. Semesco Foundation, Ocala, FL; the Canadian Institutes for Health Research (Grant MOP-38034); National Institute of Dental and Craniofacial Research oral biology training (grant T32 DE007200 to S.L. and S.L.L.); Howard Hughes Medical Institute science education grant to the University of Florida (to G.X.A.); Arthritis Society Research Chair at the University of Calgary (to M.J.F.). Funding for open access charge: NIH grant.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Witold Filipowicz (Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland) for providing tethering assay plasmids and Dr Phillip Sharp (Massachusetts Institute of Technology, Cambridge, MA) for the miR20 luciferase reporters.
REFERENCES
- 1.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 5.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 6.Jakymiw A, Pauley KM, Li S, Ikeda K, Lian S, Eystathioy T, Satoh M, Fritzler MJ, Chan EK. The role of GW/P-bodies in RNA processing and silencing. J. Cell Sci. 2007;120:1317–1323. doi: 10.1242/jcs.03429. [DOI] [PubMed] [Google Scholar]
- 7.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 9.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Jaronczyk K, Carmichael JB, Hobman TC. Exploring the functions of RNA interference pathway proteins: some functions are more RISCy than others? Biochem. J. 2005;387:561–571. doi: 10.1042/BJ20041822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Jakymiw A, Wood MR, Eystathioy T, Rubin RL, Fritzler MJ, Chan EK. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J. Cell. Sci. 2004;117:5567–5578. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- 13.Li S, Lian SL, Moser JJ, Fritzler ML, Fritzler MJ, Satoh M, Chan EK. Identification of GW182 and its novel isoform TNGW1 as translational repressors in Ago2-mediated silencing. J. Cell. Sci. 2008;121:4134–4144. doi: 10.1242/jcs.036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eulalio A, Tritschler F, Izaurralde E. The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. RNA. 2009;15:1433–1442. doi: 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tritschler F, Huntzinger E, Izaurralde E. Role of GW182 proteins and PABPC1 in the miRNA pathway: a sense of deja vu. Nat. Rev. Mol. Cell Biol. 2010;11:379–384. doi: 10.1038/nrm2885. [DOI] [PubMed] [Google Scholar]
- 16.Ding L, Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007;17:411–416. doi: 10.1016/j.tcb.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Baillat D, Shiekhattar R. Functional dissection of the human TNRC6 (GW182-related) family of proteins. Mol. Cell Biol. 2009;29:4144–4155. doi: 10.1128/MCB.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chekulaeva M, Filipowicz W, Parker R. Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA. 2009;15:794–803. doi: 10.1261/rna.1364909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chekulaeva M, Parker R, Filipowicz W. The GW/WG repeats of Drosophila GW182 function as effector motifs for miRNA-mediated repression. Nucleic Acids Res. 2010;38:6673–6683. doi: 10.1093/nar/gkq501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA. 2009;15:1067–1077. doi: 10.1261/rna.1605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 22.Eulalio A, Tritschler F, Buttner R, Weichenrieder O, Izaurralde E, Truffault V. The RRM domain in GW182 proteins contributes to miRNA-mediated gene silencing. Nucleic Acids Res. 2009;37:2974–2983. doi: 10.1093/nar/gkp173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazzaretti D, Tournier I, Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA. 2009;15:1059–1066. doi: 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lian SL, Li S, Abadal GX, Pauley BA, Fritzler MJ, Chan EK. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA. 2009;15:804–813. doi: 10.1261/rna.1229409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takimoto K, Wakiyama M, Yokoyama S. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA. 2009;15:1078–1089. doi: 10.1261/rna.1363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15:781–793. doi: 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat. Struct. Mol. Biol. 2010;17:238–240. doi: 10.1038/nsmb.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen CY, Zheng D, Xia Z, Shyu AB. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell. Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 35.Ding XC, Grosshans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 2009;28:213–222. doi: 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 38.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 40.Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu L, Fan J, Belasco JG. Importance of translation and nonnucleolytic ago proteins for on-target RNA interference. Curr. Biol. 2008;18:1327–1332. doi: 10.1016/j.cub.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding L, Spencer A, Morita K, Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol. Cell. 2005;19:437–447. doi: 10.1016/j.molcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lian S, Fritzler MJ, Katz J, Hamazaki T, Terada N, Satoh M, Chan EK. Small interfering RNA-mediated silencing induces target-dependent assembly of GW/P bodies. Mol. Biol. Cell. 2007;18:3375–3387. doi: 10.1091/mbc.E07-01-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minshall N, Kress M, Weil D, Standart N. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol. Biol. Cell. 2009;20:2464–2472. doi: 10.1091/mbc.E09-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tritschler F, Braun JE, Eulalio A, Truffault V, Izaurralde E, Weichenrieder O. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol. Cell. 2009;33:661–668. doi: 10.1016/j.molcel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 50.Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pauley KM, Satoh M, Pauley BA, Dominguez-Gutierrez PR, Wallet SM, Holliday LS, Cha S, Reeves WH, Chan EK. Formation of GW/P bodies as marker for microRNA-mediated regulation of innate immune signaling in THP-1 cells. Immunol. Cell Biol. 2010;88:205–212. doi: 10.1038/icb.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pauley KM, Eystathioy T, Jakymiw A, Hamel JC, Fritzler MJ, Chan EK. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–910. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.